Molecular Tools for the Selective Detection of Nine Diatom Species Biomarkers of Various Water Quality Levels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Environmental Sampling

2.2. Diatoms Isolation and Culture Conditions
2.3. Morphological Identification
2.4. Isolation of DNA and DNA Amplification
2.5. Probe Design and Application of Microarray Experiments to Pure Diatom Cultures
3. Results
3.1. Isolation and Identification of Diatom Cells from Environmental Samples
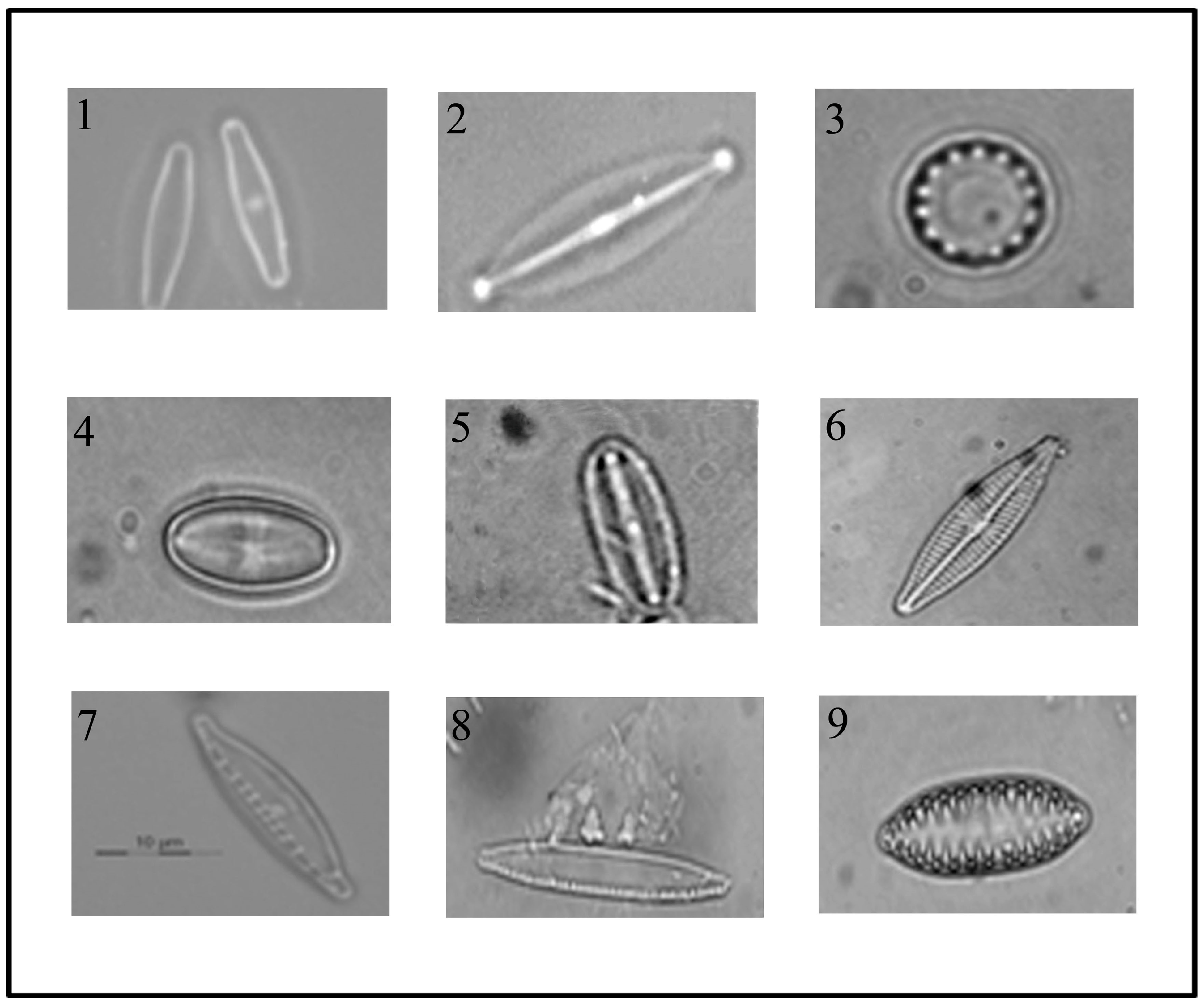
| Diatom Species | Water Quality Levels According to EPI-D Index | Water Quality Levels According to ICMI Index | |
|---|---|---|---|
| Index of Sensitivity to Pollutants (IPS_S) | Trophic Index (TI_TW) | ||
| 1. Achnanthidium minutissimum (Kützing) Czarnecki | I–II Excellent–Good | 5 Good–High | 1,2 Good–High |
| 2. Craticula halophila (Grunow ex Van Heurck) Mann | IV–V Bad–Very Bad | 2 Bad–Poor | 3,4 Bad–Poor |
| 3. Cyclotella meneghiniana (Kützing) | II–III Good - Mediocre | 2 Bad–Poor | 2,8 Poor–Sufficient |
| 4. Eolimna minima (Grunow) Lange-Bertalot | IV Bad | 2,2 Poor–Sufficient | 2,9 Poor–Sufficient |
| 5. Mayamaea atomus var. permitis (Hustedt) Bruder & Medlin | IV–V Bad–Very Bad | 2,3 Poor–Sufficient | 3,1 Bad–Poor |
| 6. Navicula veneta (Kützing) | IV–V Bad–Very Bad | 1 Bad–Poor | 3,5 Bad–Poor |
| 7. Nitzschia dissipata (Kützing) Grunow | II–III Good–Mediocre | 4,5 Good–High | 2,4 Sufficient–Good |
| 8. Nitzschia palea (Kützing) W Smith | IV Bad | 1 Bad–Poor | 3,3 Bad–Poor |
| 9. Surirella angusta Kützing | III Mediocre | 4 Sufficient–Good | 3,7 Bad–Poor |
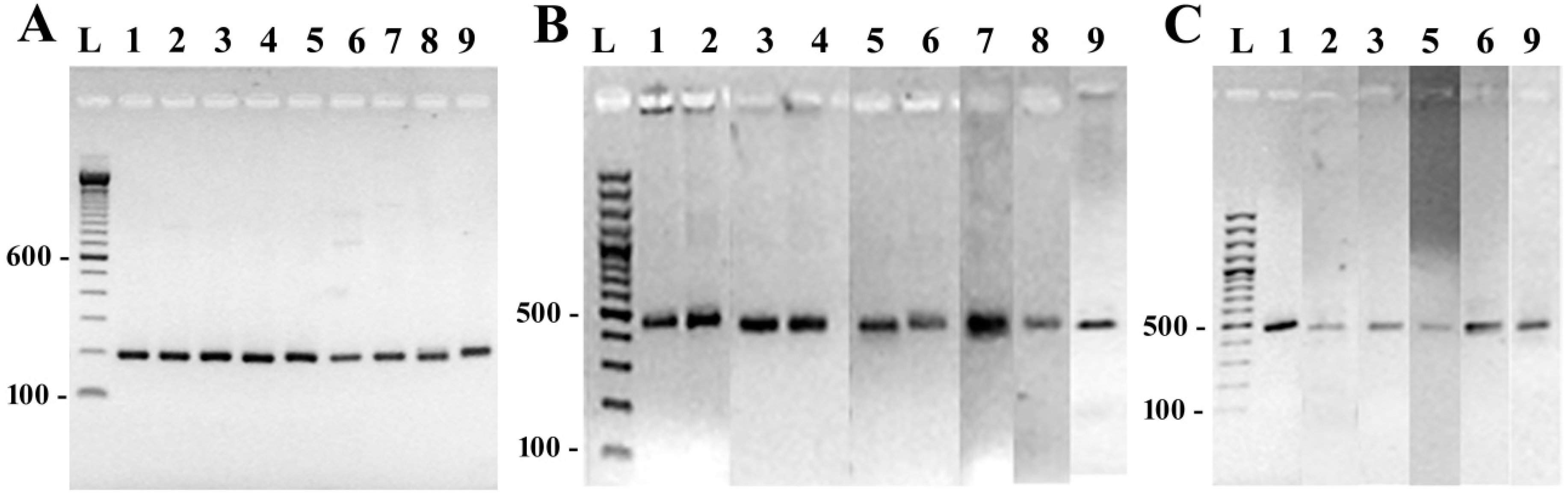
3.2. DNA Amplification and Sequencing of Conserved Diatom Genes
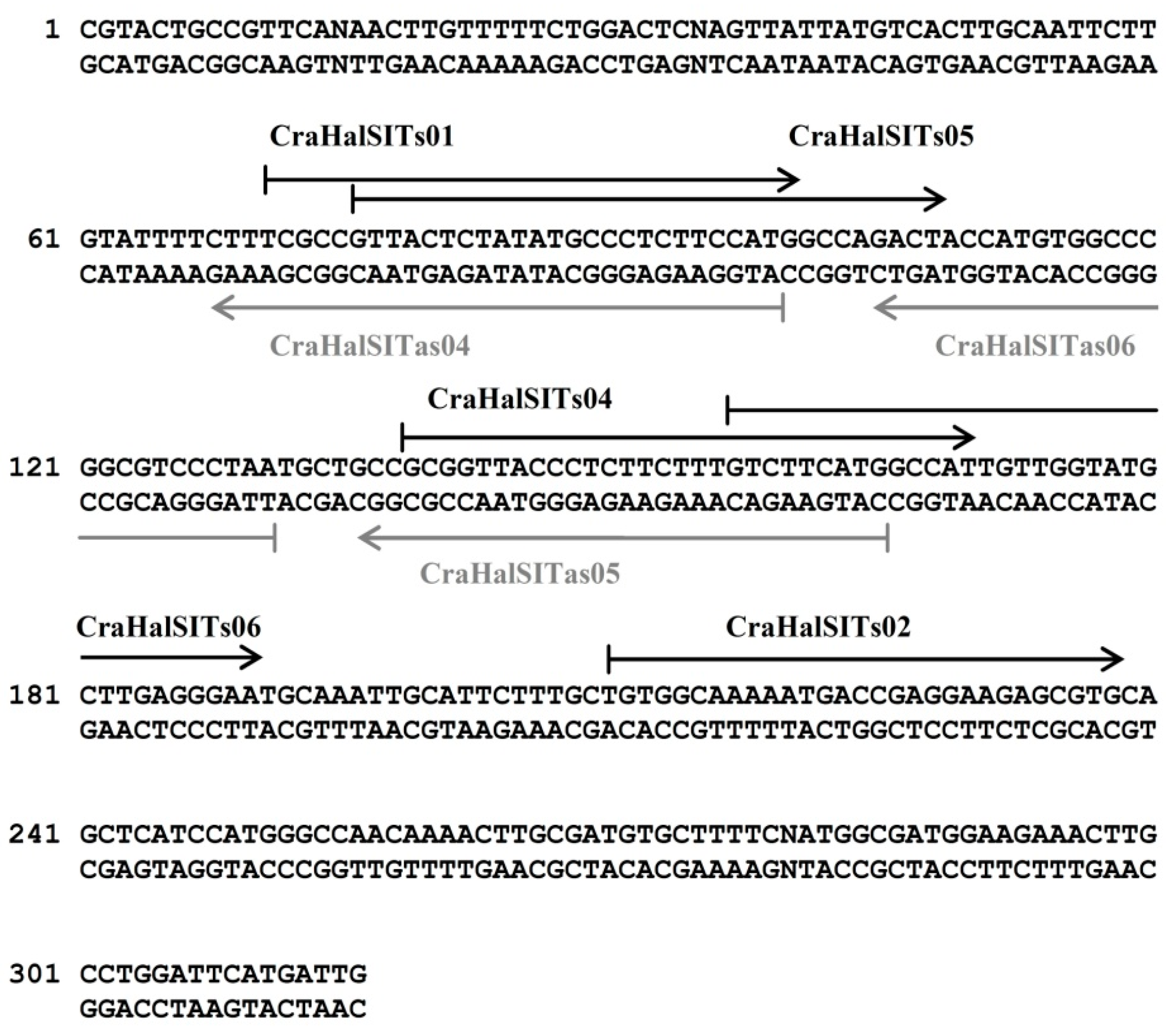
3.3. Species-Specific Oligonucleotide Probe Design
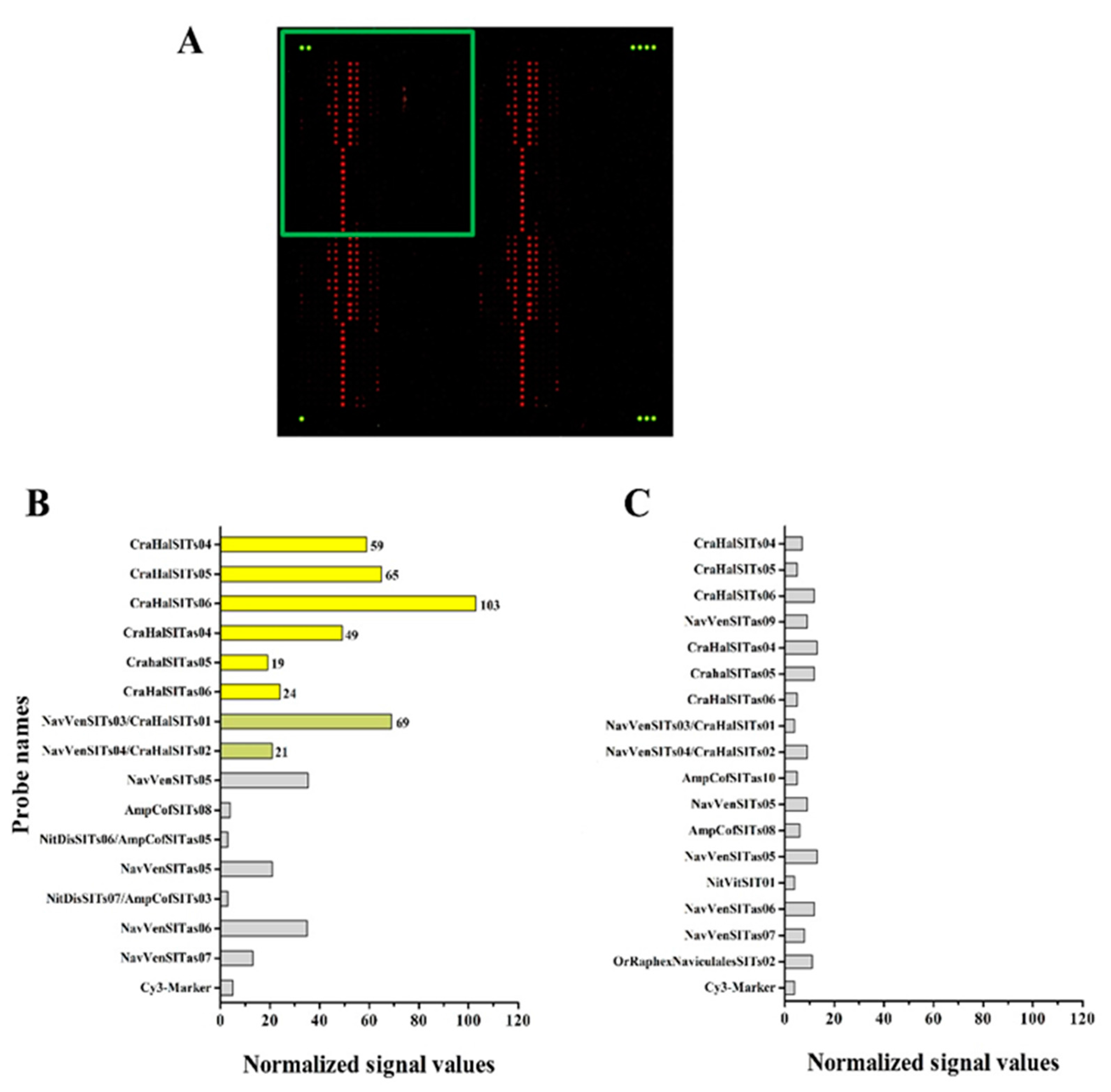
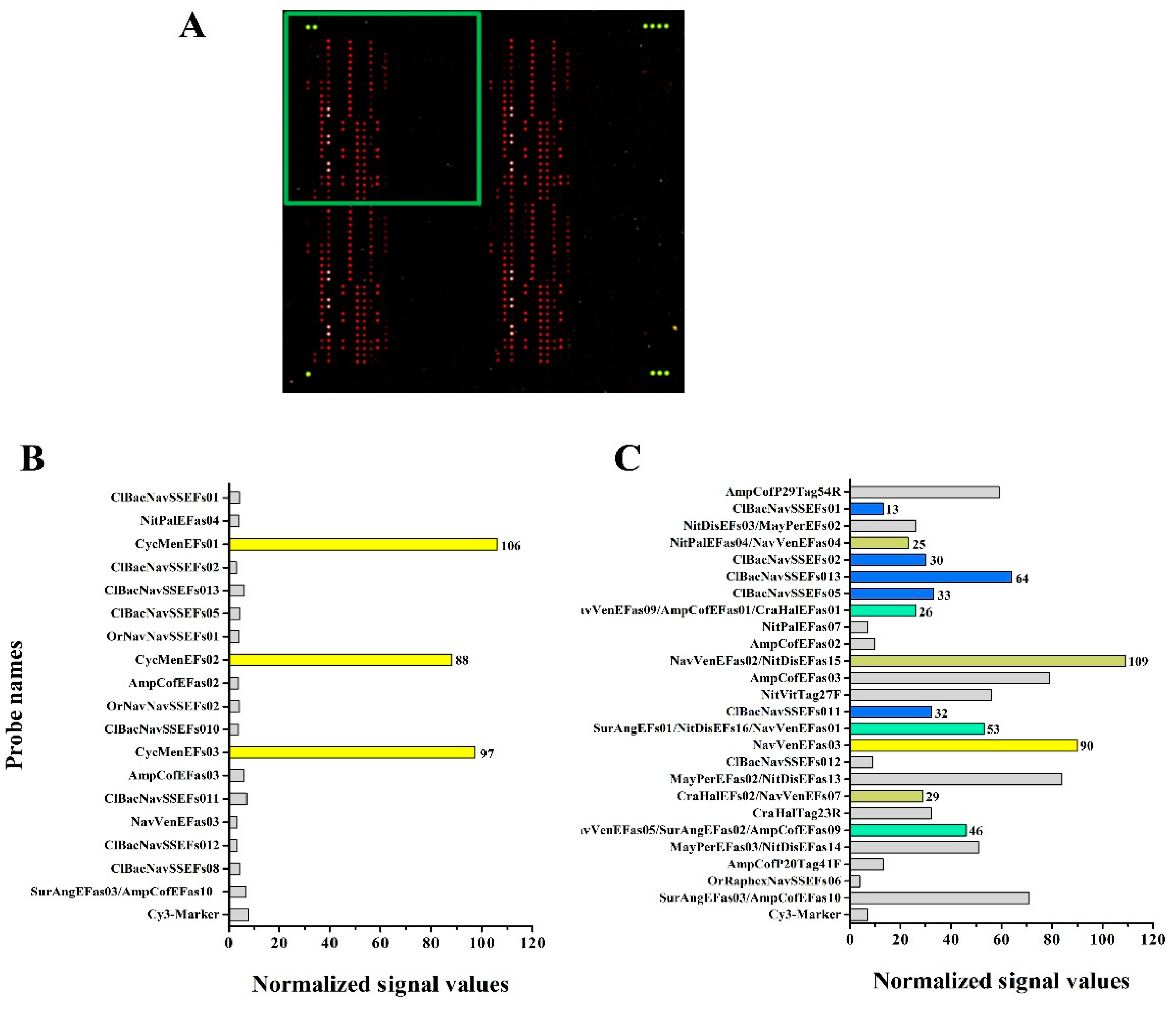
3.4. Hybridization Experiments on Microarrays
| Diatom Species | Target (eEF1-a) | Target (SIT) | Total Probes |
|---|---|---|---|
| Achnanthidium minutissimum | 3 * | 3 | |
| Craticula halophila | 2 * | 6 + 2 * | 10 |
| Cyclotella meneghiniana | 3 | 3 | |
| Eolimna minima | 3 | 3 | |
| Mayamaea permitis | 1 * | 1 | |
| Navicula veneta | 1 + 1 * | 2 | |
| Nitzschia dissipata | 6 * | 6 | |
| Nitzschia palea | 2 * | 2 | |
| Surirella angusta | 4 + 2 * | 6 |
4. Discussion and Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nelson, D.M.; Treguer, P.; Brzezinski, M.A.; Leynaert, A.; Queguiner, B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycle 1995, 9, 359–372. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Barber, R.T.; Smetacek, V. Biogeochemical controls and feedbacks on ocean primary production. Science 1998, 281, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.G. The species concept in diatoms. Phycologia 1999, 38, 437–495. [Google Scholar] [CrossRef]
- Falciatore, A.; Bowler, C. Revealing the molecular secrets of marine diatoms. Ann. Rev. Plant Physiol. Plant Mol. Biol. 2002, 53, 109–130. [Google Scholar] [CrossRef]
- Allen, A.E.; Vardi, A.; Bowler, C. An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Plant Biol. 2006, 9, 264–273. [Google Scholar]
- Gevrey, M.; Rimet, F.; Park, Y.S.; Giraudel, J.L.; Ector, L.; Lek, S. Water quality assessment using diatom assemblages and advanced modelling techniques. Freshw. Biol. 2004, 49, 208–220. [Google Scholar] [CrossRef]
- Hering, D.; Johnson, R.K.; Kramm, S.; Schmutz, S.; Szoszkiewicz, K.; Verdonschot, P.F.M. Assessment of European streams with diatoms, macrophytes, macroinvertebrates and fish: A comparative metric-based analysis of organism response to stress. Freshw. Biol. 2006, 51, 1757–1785. [Google Scholar] [CrossRef]
- Descy, J.P. A new approach to water quality estimation using diatoms. Nova Hedwig. 1979, 64, 305–323. [Google Scholar]
- Coring, E. Situation and developments of algal (diatom)-based techniques for monitoring rivers in Germany. In Use of Algae Monitoring Rivers III; Prygel, J., Whitton, B.A., Bukowska, J., Eds.; Agence de l’Eau Artois-Picardie: Douai, France, 1999; pp. 122–127. [Google Scholar]
- Rimet, F.; Cauchie, H.M.; Hoffmann, L.; Ector, L. Response of diatom indices to simulated water quality improvements in a river. J. Appl. Phycol. 2005, 17, 119–128. [Google Scholar] [CrossRef]
- Kelly, M.; Juggins, S.; Guthrie, R.; Pritchard, S.; Jamieson, J.; Rippey, B.; Hirst, H.; Yallop, M. Assessment of ecological status in UK rivers using diatoms. Freshw. Biol. 2008, 53, 403–422. [Google Scholar]
- Bennion, H.; Kelly, M.G.; Juggins, S.; Yallop, M.L.; Burgess, A.; Jamieson, J.; Krokowski, J. Assessment of ecological status in UK lakes using benthic diatoms. Freshw. Sci. 2014, 33, 639–654. [Google Scholar] [CrossRef]
- European Commission. Directive 2000/60/EC of the European Parliament and of the Council of 23rd October 2000 establishing a framework for Community action in the field of water policy. Off. J. Eur. Commun. 2000, 327, 1–72. [Google Scholar]
- Foster, D.; Wood, A.; Griffiths, M. The EC Water Framework Directive and its implications for the Environment Agency. Freshw. Forum 2001, 16, 4–28. [Google Scholar]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.M.; Berzano, M.; Gualerzi, C.O.; Spurio, R. Development of molecular tools for the detection of freshwater diatoms. J. Microbiol. Meth. 2011, 84, 33–44. [Google Scholar] [CrossRef]
- Gentry, T.J.; Wickham, G.S.; Schadt, C.W.; He, Z.; Zhou, J. Microarray Applications in Microbial Ecology Research. Microb. Ecol. 2006, 52, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Metfies, K.; Medlin, L.K. Ribosomal RNA probes and microarrays: Their potential use in assessing microbial biodiversity. Methods Enzymol. 2005, 395, 258–278. [Google Scholar] [PubMed]
- Metfies, K.; Medlin, L.K. Refining cryptophyte identification with DNA-microarrays. J. Plankton Res. 2007, 29, 1071–1075. [Google Scholar] [CrossRef]
- Galluzzi, L.; Cegna, A.; Casabianca, S.; Penna, A.; Saunders, N.; Magnani, M. Development of an oligonucleotide microarray for the detection and monitoring of marine dinoflagellates. J. Microbiol. Meth. 2011, 84, 234–242. [Google Scholar] [CrossRef]
- Gescher, G.; Metfies, K.; Medlin, L.K. The ALEX Chip—Development of a DNA chip for identification and monitoring of Alexandrium. Harmful Algae 2008, 7, 485–494. [Google Scholar] [CrossRef]
- Small, J.; Call, D.R.; Brockman, F.J.; Straub, T.M.; Chandler, D.P. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 2001, 67, 4708–4716. [Google Scholar] [CrossRef] [PubMed]
- Goberna, M.; Gadermaier, M.; Schoen, M.A.; Sperl, D.; Franke-Whittle, I.H.; Wett, B.; Insam, H. Fingerprinting the Microbial Communities in Organic Wastes Using Oligonucleotide Microarrays and Real-Time PCR. In Soil Enzymology in the Recycling of Organic Wastes and Environmental Restoration. Environmental Science and Engineering; Trasar-Cepeda, C., Hernández, T., García, C., Rad, C., González-Carcedo, S., Eds.; Springer: Berlin, Germany, 2012; pp. 285–298. [Google Scholar]
- Wang, Z.; Orlandi, P.A.; Stenger, D.A. Simultaneous detection of four human pathogenic microsporidian species from clinical samples by oligonucleotide microarray. J. Clin. Microbiol. 2005, 43, 4121–4128. [Google Scholar] [CrossRef] [PubMed]
- Cleven, B.E.; Palka-Santini, M.; Gielen, J.; Meembor, S.; Kronke, M.; Krut, O. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J. Clin. Microbiol. 2006, 44, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Theriot, E.C.; Cannone, J.J.; Gutell, R.R.; Alverson, A.J. The limits of nuclear encoded SSU rDNA for resolving the diatom Phylogeny. Eur. J. Phycol. 2009, 44, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Korczak, B.M.; Stieber, R.; Emler, S.; Burnens, A.P.; Frey, J.; Kuhnert, P. Genetic relatedness within the genus Campylobacter inferred from rpoB sequences. Int. J. Syst. Evolut. Microbiol. 2006, 56, 937–945. [Google Scholar] [CrossRef]
- Sander, P.; Alcaide, F.; Richter, I.; Frischkorn, K.; Tortoli, E.; Springer, B.; Telenti, A.; Böttger, E.C. Inteins in mycobacterial GyrA are a taxonomic character. Microbiology 1998, 144, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Abdelbaqi, K.; Menard, A.; Prouzet-Mauleon, V.; Bringaud, F.; Lehours, P.; Megraud, F. Nucleotide sequence of the GyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol. Med. Microbiol. 2007, 49, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.T.; Lee, F.L.; Tai, C.J.; Kasai, H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA-DNA hybridization in the Bacillus subtilis group. Int. J. Syst. Evol. Microbiol. 2007, 57, 1846–1850. [Google Scholar] [CrossRef] [PubMed]
- Hannula, M.; Hanninen, M.L. Phylogenetic analysis of Helicobacter species based on partial gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Cantoni, C.; Comi, G. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 2002, 68, 6273–6282. [Google Scholar] [CrossRef] [PubMed]
- Volokhov, D.; Rasooly, A.; Chumakov, K.; Chizhikov, V. Identification of Listeria species by microarray-based assay. J. Clin. Microbiol. 2002, 40, 4720–4728. [Google Scholar] [CrossRef] [PubMed]
- Volokhov, D.; Chizhikov, V.; Chumakov, K.; Rasooly, A. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 2003, 41, 4071–4080. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.S.Y.; Chow, A.W. Identification of enteric pathogens by heat shock protein 60 kDa (HSP60) gene sequences. FEMS Microbiol. Lett. 2002, 206, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Soltis, P.S.; Milligan, B.G. Molecular Systematics of Plants; Soltis, P.S., Soltis, D.E., Doyle, J.J., Eds.; Chapman and Hall: New York, NY, USA, 1992; pp. 117–150. [Google Scholar]
- Van der Schoot, J.; Pospiskova, M.; Vosman, B. Development and characterization of microsatellite markers in black poplar (Populus nigra L.). Theor. Appl. Genet. 2000, 101, 317–322. [Google Scholar] [CrossRef]
- Dhar, B.D.; Cimarelli, L.; Singh, K.S.; Brandi, L.; Brandi, A.; Puccinelli, C.; Marcheggiani, S.; Spurio, R. Molecular detection of a potentially toxic diatom species. Int. J. Environ. Res. Public Health 2015, 12, 4921–4941. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.G.; Cazaubon, A.; Coring, E.; Dell’Uomo, A.; Ector, L.; Goldsmith, B.; Guasch, H.; Hürlimann, J.; Jarlman, A.; Kawecka, B.; et al. Recommendations for the routine sampling of diatoms for water quality assessments in Europe. J. Appl. Phycol. 1998, 10, 215–224. [Google Scholar] [CrossRef]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (Cleve). Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [PubMed]
- Cox, E.J. Identification of Freshwater Diatoms from Live Material; Chapman & Hall: London, UK, 1996; p. 158. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. Teil: Naviculaceae; Bacillariaceae, Epithemiaceae, Surirellaceae; Centrales, Fragilariaceae, Eunotiaceae; Achnathaceae. In Kritische Erganzungen zu Navicula und Gomphonema; Ettl, H., Gärtner, G., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Süβwasserflora von Mitteleuropa, 2/1–5; Gustav Fischer Verlag: Stuttgart, Germany, 1986, 1988, 1991a, 1991b, 2000.
- Lange-Bertalot, H. Diatoms of Europe: Diatoms of the European Inland Waters and Comparable Habitat; Gantner Verlag: Ruggell, Liechtenstein, 2001; Volume 2. [Google Scholar]
- Lange-Bertalot, H. Diatomeen im Süßwasser—Benthos von Mitteleuropa. In Bestimmungsflora Kieselalgen für die Ökologische Praxis; Koeltz Scientific Books: Koenigstein, Germany, 2012. [Google Scholar]
- Leiconte, C.; Coste, M.; Prygiel, J. “OMNIDIA” software for taxonomy, calculation of diatom indices and inventories management. Hydrobiologia 1993, 269, 509–513. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, 43–46. [Google Scholar] [CrossRef]
- Dittami, S.M.; Edvardsen, B. GPR-Analyzer: A simple tool for quantitative analysis of hierarchical multispecies microarrays. Environ. Sci. Pollut. Res. Int. 2013, 20, 6808–6815. [Google Scholar] [CrossRef] [PubMed]
- Dell’Uomo, A. Assessment of Water Quality of an Appennine River as a Pilot Study for Diatom-Based Monitoring of Italian Watercourses. In Proceedings of the International Symposium Use of Algae for Monitoring Rivers II, Institut fur Botanik, Universität Innsbruck, Innsbruck, Austria, 17–19 September 1995; pp. 65–72.
- Dell’Uomo, A. L’indice Diatomico di Eutrofizzazione/Polluzione (EPI-D) nel Monitoraggio delle Acque Correnti. 2004. Available online: http://www.cisba.eu/attachments/article/75/IndiceEPI_D.pdf (accessed on 22 May 2015).
- Prygiel, J.; Coste, M. Guide Methodologique Pour la Mise en Oeuvre de l’Indice Biologique Diatomées. 2000. Available online: http://www.eau-artois-picardie.fr/IMG/BaseDoc/aegis/2496/B_16478.pdf (accessed on 21 May 2015).
- Ector, L.; Rimet, F. Using bioindicators to assess rivers in Europe: An overview. In Modelling Community Structure in Freshwater Ecosystems; Lek, S., Scardi, M., Verdonschot, P.F.M., Descy, J.P., Park, Y.S., Eds.; Springer: Berlin, Germany, 2005; pp. 7–19. [Google Scholar]
- Mancini, L.; Sollazzo, C. Metodo per la valutazione dello stato ecologico delle acque correnti: Comunità diatomiche. Roma, Istituto Superiore di Sanità. Available online: http://www.iss.it/binary/publ/cont/0919web.pdf (accessed on 22 May 2015).
- Berzano, M.; Marcheggiani, S.; Rombini, S.; Spurio, R. The application of oligonucleotide probes and microarrays for the identification of freshwater diatoms. Hydrobiologia 2012, 695, 57–72. [Google Scholar] [CrossRef]
- Hillis, D.M. Inferring complex phylogenies. Nature 1996, 383, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.S.; Thual, D.; Spurio, R.; Cannata, N. SaDA: From sampling to data analysis—An extensible open source infrastructure for rapid, robust and automated management and analysis of modern ecological high-throughput data. Int. J. Environ. Res. Public Health 2015. in review. [Google Scholar]
- Rhee, S.K.; Liu, X.; Wu, L.; Chong, S.C.; Wan, X.; Zhou, J. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 2004, 70, 4303–4317. [Google Scholar] [CrossRef] [PubMed]
- Ballarini, A.; Segata, N.; Huttenhower, C.; Jousson, O. Simultaneous quantification of multiple bacteria by the BactoChip microarray designed to target species-specific marker genes. PLoS One 2013, 8, 1–11. [Google Scholar]
- Moniz, M.B.J.; Kaczmarska, I. Barcoding of diatoms: Nuclear encoded ITS revisited. Protist 2010, 161, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Cejudo-Figueiras, C.; Tudesque, L.; Becares, E.; Hoffmann, L.; Ector, L. Are diatom diversity indices reliable monitoring metrics? Hydrobiologia 2012, 695, 199–206. [Google Scholar] [CrossRef]
- Martin, G.; de los Reyes Fernandez, M. Diatoms as indicators of water quality and ecological status: Sampling, analysis and some ecological remarks. Ecol. Water Qual. 2012, 9, 183–204. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cimarelli, L.; Singh, K.S.; Mai, N.T.N.; Dhar, B.C.; Brandi, A.; Brandi, L.; Spurio, R. Molecular Tools for the Selective Detection of Nine Diatom Species Biomarkers of Various Water Quality Levels. Int. J. Environ. Res. Public Health 2015, 12, 5485-5504. https://doi.org/10.3390/ijerph120505485
Cimarelli L, Singh KS, Mai NTN, Dhar BC, Brandi A, Brandi L, Spurio R. Molecular Tools for the Selective Detection of Nine Diatom Species Biomarkers of Various Water Quality Levels. International Journal of Environmental Research and Public Health. 2015; 12(5):5485-5504. https://doi.org/10.3390/ijerph120505485
Chicago/Turabian StyleCimarelli, Lucia, Kumar Saurabh Singh, Nguyen Thi Nhu Mai, Bidhan Chandra Dhar, Anna Brandi, Letizia Brandi, and Roberto Spurio. 2015. "Molecular Tools for the Selective Detection of Nine Diatom Species Biomarkers of Various Water Quality Levels" International Journal of Environmental Research and Public Health 12, no. 5: 5485-5504. https://doi.org/10.3390/ijerph120505485
APA StyleCimarelli, L., Singh, K. S., Mai, N. T. N., Dhar, B. C., Brandi, A., Brandi, L., & Spurio, R. (2015). Molecular Tools for the Selective Detection of Nine Diatom Species Biomarkers of Various Water Quality Levels. International Journal of Environmental Research and Public Health, 12(5), 5485-5504. https://doi.org/10.3390/ijerph120505485





