Genotypes and Antibiotic Resistances of Campylobacter jejuni Isolates from Cattle and Pigeons in Dairy Farms
Abstract
:1. Introduction
2. Experimental Section
2.1. Samples
2.2. Genotyping and Determination of Antibiotic Resistance
2.3. Data Analysis
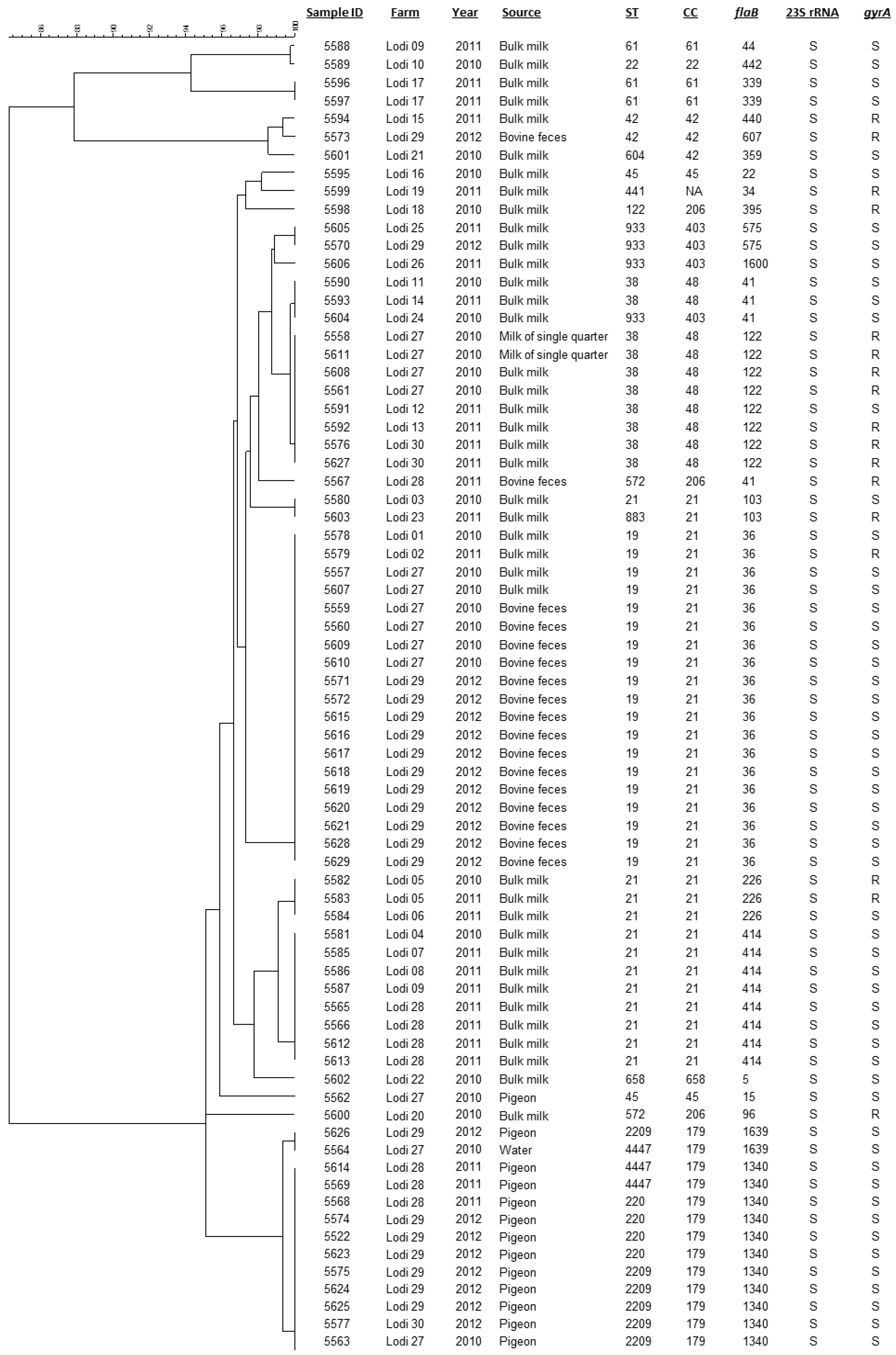
3. Results and Discussion
3.1. Typing
3.2. Genotypic Antibiotic Resistance
| Source | No. of Samples | No. of Resistant Isolates (%) | |
|---|---|---|---|
| Quinolones | Macrolide | ||
| Bulk tank milk | 40 | 13 (32.5) | 0 |
| Milk of single quarter | 2 | 2 (100) | 0 |
| Bovine feces | 17 | 2 (11.8) | 0 |
| Pigeon intestine | 13 | 0 | 0 |
| Water point | 1 | 0 | 0 |
| Total | 73 | 17 (23.3) | 0 |
3.3. Association between Genotype and Antibiotic Resistance
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Humphrey, T.; O’Brien, S.; Madsen, M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 2007, 117, 237–257. [Google Scholar]
- European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 2014, 12, 99–107. [Google Scholar]
- Blaser, M.J.; Engberg, J. Clinical Aspects of Campylobacter jejuni and Campylobacter coli Infections. In Campylobacter; Nachamkin, I., Szymanski, C.M., Blaser, M.J., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 99–121. [Google Scholar]
- Ge, B.; Wang, F.; Sjölund-Karlsson, M.; McDermott, P.F. Antimicrobial resistance in Campylobacter: Susceptibility testing methods and resistance trends. J. Microbiol. Methods 2013, 95, 57–67. [Google Scholar] [CrossRef]
- Iovine, N.M. Resistance mechanisms in Campylobacter jejuni. Virulence 2013, 4, 230–240. [Google Scholar]
- Wieczorek, K.; Osek, J. Antimicrobial resistance mechanisms among Campylobacter. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Bianchini, V.; Borella, L.; Benedetti, V.; Parisi, A.; Miccolupo, A.; Santoro, E.; Recordati, C.; Luini, M. Prevalence in bulk tank milk and epidemiology of Campylobacter jejuni in dairy herds in Northern Italy. Appl. Environ. Microbiol. 2014, 80, 1832–1837. [Google Scholar] [CrossRef]
- Taboada, E.N.; Clark, C.G.; Sproston, E.L.; Carrillo, C.D. Current methods for molecular typing of Campylobacter species. J. Microbiol. Methods 2013, 95, 24–31. [Google Scholar] [CrossRef]
- Korczak, B.M.; Zurfluh, M.; Emler, S.; Kuhn-Oertli, J.; Kuhnert, P. Multiplex strategy for multilocus sequence typing, fla typing, and genetic determination of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolates collected in Switzerland. J. Clin. Microbiol. 2009, 47, 1996–2007. [Google Scholar] [CrossRef]
- Campylobacter MLST. Available online: http://pubmlst.org/campylobacter/ (accessed on 9 July 2014).
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar]
- Griekspoor, P.; Colles, F.M.; McCarthy, N.D.; Hansbro, P.M.; Ashhurst-Smith, C.; Olsen, B.; Hasselquist, D.; Maiden, M.C.; Waldenström, J. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol. Ecol. 2013, 22, 1463–1472. [Google Scholar] [CrossRef]
- European Food Safety Authority. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. EFSA J. 2013, 11, 152–198. [Google Scholar]
- Pezzotti, G.; Serafin, A.; Luzzi, I.; Mioni, R.; Milan, M.; Perin, R. Occurrence and resistance to antibiotics of Campylobacter jejuni and Campylobacter coli in animals and meat in northeastern Italy. Int. J. Food Microbiol. 2003, 82, 281–287. [Google Scholar] [CrossRef]
- Parisi, A.; Lanzilotta, S.G.; Addante, N.; Normanno, G.; di Modugno, G.; Dambrosio, A.; Montagna, C.O. Prevalence, molecular characterization and antimicrobial resistance of thermophilic Campylobacter isolates from cattle, hens, broilers and broiler meat in South-Eastern Italy. Vet. Res. Commun. 2007, 31, 133–123. [Google Scholar] [CrossRef]
- Habib, I.; Miller, W.G.; Uyttendaele, M.; Houf, K.; De Zutter, L. Clonal population structure and antimicrobial resistance of Campylobacter jejuni in chicken meat from Belgium. Appl. Environ. Microbiol. 2009, 75, 4264–4272. [Google Scholar]
- Wirz, S.E.; Overesch, G.; Kuhnert, P.; Korczak, B.M. Genotype and antibiotic resistance analyses of Campylobacter isolates from ceca and carcasses of slaughtered broiler flocks. Appl. Environ. Microbiol. 2010, 76, 6377–6386. [Google Scholar] [CrossRef]
- Han, J.; Sahin, O.; Barton, Y.W.; Zhang, Q. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 2008, 4. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bianchini, V.; Luini, M.; Borella, L.; Parisi, A.; Jonas, R.; Kittl, S.; Kuhnert, P. Genotypes and Antibiotic Resistances of Campylobacter jejuni Isolates from Cattle and Pigeons in Dairy Farms. Int. J. Environ. Res. Public Health 2014, 11, 7154-7162. https://doi.org/10.3390/ijerph110707154
Bianchini V, Luini M, Borella L, Parisi A, Jonas R, Kittl S, Kuhnert P. Genotypes and Antibiotic Resistances of Campylobacter jejuni Isolates from Cattle and Pigeons in Dairy Farms. International Journal of Environmental Research and Public Health. 2014; 11(7):7154-7162. https://doi.org/10.3390/ijerph110707154
Chicago/Turabian StyleBianchini, Valentina, Mario Luini, Laura Borella, Antonio Parisi, Romie Jonas, Sonja Kittl, and Peter Kuhnert. 2014. "Genotypes and Antibiotic Resistances of Campylobacter jejuni Isolates from Cattle and Pigeons in Dairy Farms" International Journal of Environmental Research and Public Health 11, no. 7: 7154-7162. https://doi.org/10.3390/ijerph110707154
APA StyleBianchini, V., Luini, M., Borella, L., Parisi, A., Jonas, R., Kittl, S., & Kuhnert, P. (2014). Genotypes and Antibiotic Resistances of Campylobacter jejuni Isolates from Cattle and Pigeons in Dairy Farms. International Journal of Environmental Research and Public Health, 11(7), 7154-7162. https://doi.org/10.3390/ijerph110707154





