Abstract
The diversity of symbiotic fungi associated with the gorgonian coral Echinogorgia rebekka from the Weizhou coral reef in the South China Sea was investigated. Combined with morphologic traits, ITS-rDNA sequences revealed 18 fungal strains from this gorgonian. All of the 18 fungi belonged to the phylum Ascomycota and were distributed among seven genera in five orders: Eurotiales (Aspergillus and Penicillium), Pleosporales (Alternaria), Capnodiales (Cladosporium), Trichosphaeriales (Nigrospora) and Hypocreales (Hypocrea and Nectria). Antibacterial activities of these fungal strains were investigated with five pathogenic bacteria. All of the 18 fungal strains displayed different levels of antibacterial activities, most of which exhibited moderate to high antibacterial activities to the Gram-positive pathogens Staphylococcus aureus and Micrococcus tetragenus, and showed relatively low bioactivities to other three pathogenic bacteria. Several fungal strains in the genera Penicillium and Cladosporium with strong antibacterial activities provide potential for further research on isolation of bioactive secondary metabolites.
1. Introduction
Coral reefs are widely recognized as one of the most diverse ecosystems on Earth, but little attention has been paid to the microbial communities associated with coral reefs. Complex microbial communities are known to be extremely important members of many ecosystems, and also have significant influence over coral reef ecosystems [1–3]. In fact, the coral reef provides a structurally and environmentally complex array of habitats supporting a broad microbial diversity that influences both host physiology and ultimately ecosystem processes [4]. Marine microbes have been shown to produce prolific novel bioactive natural products that are not found in terrestrial strains [5–7], and symbiotic microbes are believed to be the actual producers or take part in the biosynthesis of some bioactive marine secondary metabolites isolated from the marine organism hosts [8,9]. The large number of symbiotic microbes in corals is considered to be a promising source of novel drugs which have been largely unexplored [10,11]. Limited information is known about the microbial diversity associated with marine coral reefs, despite the vital role that microorganisms may play in coral reef ecosystems. As part of our ongoing investigation on antibacterial symbiotic microorganisms from marine invertebrates in the South China Sea, the fungi isolated from a gorgonian Echinogorgia rebekka attracted our attention because the fungal strains exhibited relatively high diversity and the ethyl acetate (EtOAc) extracts of the fungal cultures showed rather high antibacterial activities. Herein we report the fungal isolation and identification as well as the antibacterial activities of fungal extracts from this gorgonian. The diversity of symbiotic fungi was examined based on morphologic observations and phylogenetic relationships of ITS-rDNA sequences of the 18 isolated fungal strains. Antibacterial activities of the fungal extracts were investigated with five pathogenic bacteria including Escherichia coli ATCC25922, S. aureus ATCC6538P, Bacillus subtilis 20019, M. tetragenus ATCC13623 and Vibrio anguillarum ATCC19019.
2. Results and Discussions
2.1. Diversity of Culturable Fungi Derived from the Gorgonian Echinogorgia rebekka
Cultivation of fungi from the tissue of the gorgonian coral Echinogorgia rebekka yielded a total of 53 isolates. Redundant isolates were excluded under the guidance of observation on morphological characteristics and 18 distinct isolates were identified (WZL001–WZL018; Figure 1, Table 1). There was no dominant morphotypes covering most of the strains, but strains from Pennicillium sp., Aspergillus sp. and Cladosporium sp. account for a large proportion of the 53 isolates. The 18 isolates were cultured for genomic DNA extraction and sequencing analysis. Of the 18 strains, most were distinct from each other and could be identified in morphology and/or in DNA sequences, with the exception of the WZL007 and WZL009 (Figure 1, Table 1). These two strains were highly similar in the sequences of ITS region, differing by only one nucleotide, and they were also similar in clonal and microscopic morphology (Figure 1). We cultivated these two isolates under the same culture conditions for a long period of time, and it was consistent that the WZL007 could produce conidia spores while WZL009 could not breed spores. This distinguishable characteristic in production indicates that these two isolates likely belong to different species.
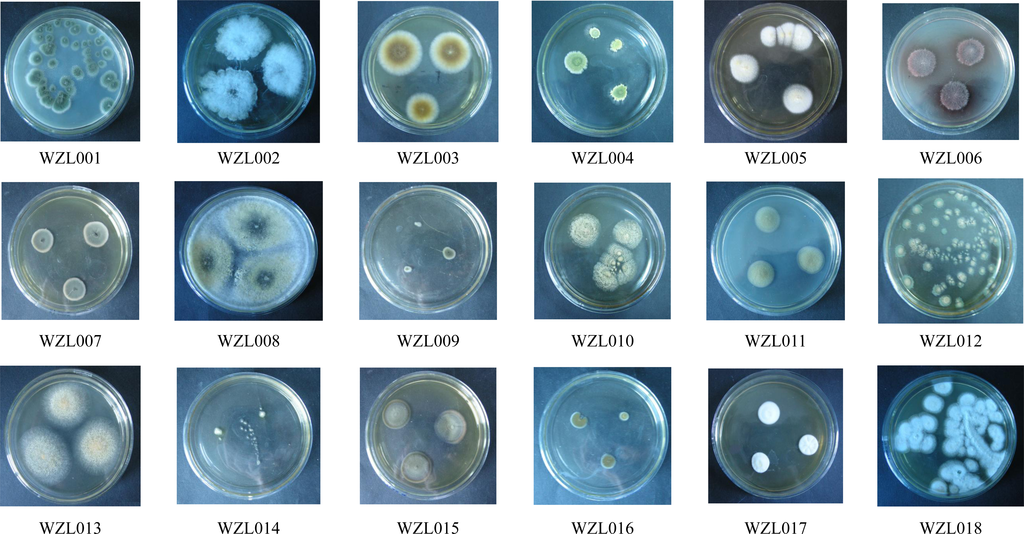
Figure 1.
Morphological photos of 18 gorgonian-derived fungal strains (WZL001–WZL018).

Table 1.
Identification of fungal strains isolated from the gorgonian Echinogorgia rebekka sample based on morphological characteristics as well as DNA analysis of the internal transcribed spacer (ITS) region. Closest relatives to fungal strains according to BLAST search are presented.
The rooted NJ tree of partial ITS-rDNA gene sequences of fungal isolates is shown in Figure 2. In total, seven genera of ascomycetes were recognized from 18 fungal strains including Aspergillus (Eurotiales, Eurotiomycetes), Peniillium (Eurotiales, Eurotiomycetes), Alternaria (Pleosporales, Dothideomycetes), Cladosporium (Capnodiales, Dothideomycetes), Nigrospora (Trichosphaeriales, Sordariomycetes), Hypocrea (Hypocreales, Sordariomycetes) and Nectria (Hypocreales, Sordariomycetes) (Figure 2). The most abundant fungi were observed in the genera Aspergillus and Cladosporium (5 strains for each). Relatively high diversity, i.e., a significant number of different strains found in a genus, was also detected in the genera Penicillum and Cladosporium (4 strains for each). For the other four genera, only one strain was found (Table 2, Figure 2).
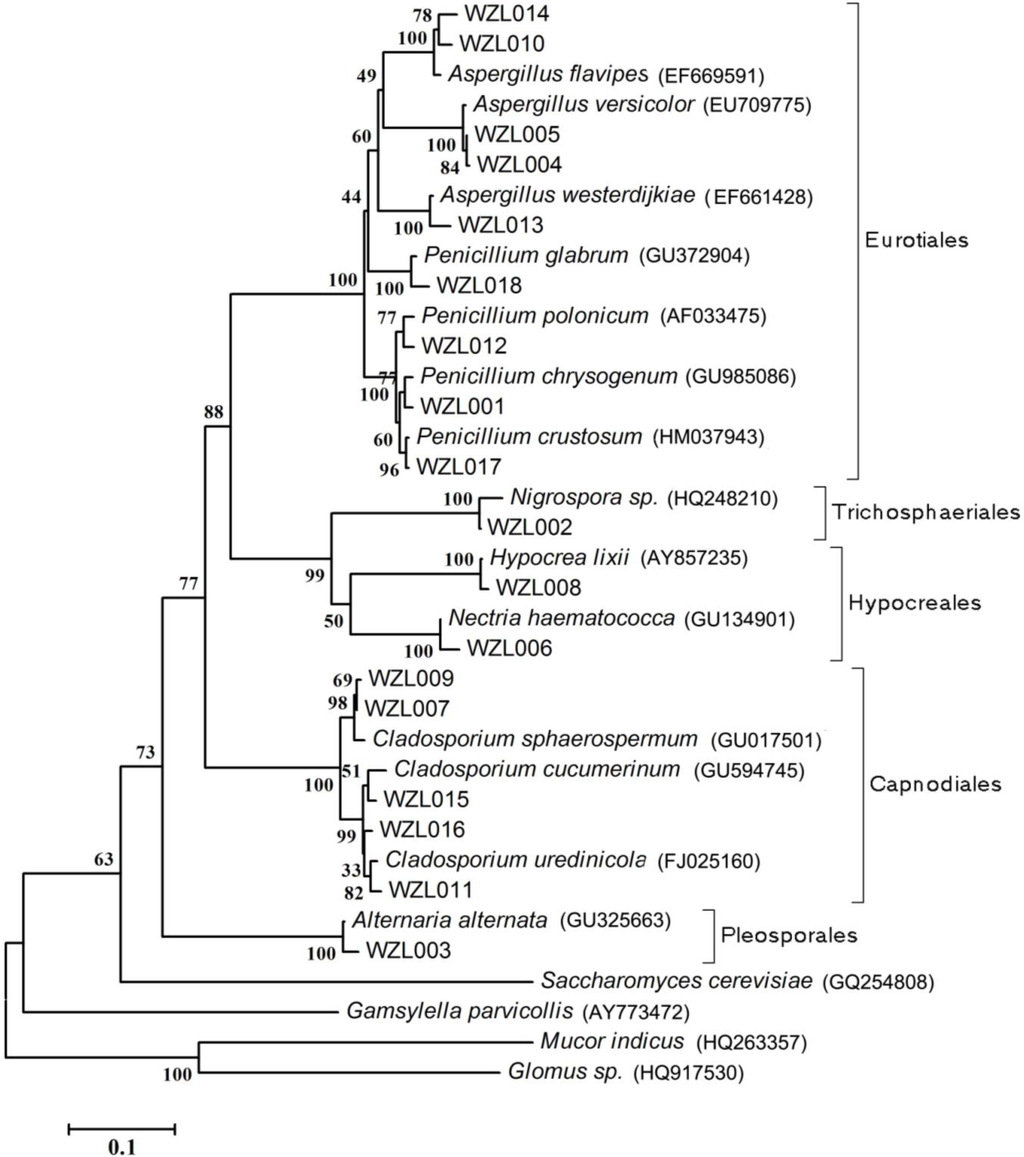
Figure 2.
Phylogenetic tree of partial ITS-rDNA sequences of gorgonian-derived fungal strains. Reference sequences were downloaded from NCBI with the accession numbers indicated in parentheses.

Table 2.
Antibacterial activities of ethyl acetate (EtOAc) extracts from fermentation broth and mycelia of the gorgonian-derived fungal strains.
The 18 fungal strains derived from the gorgonian Echinogorgia rebekka were shown to be distributed in seven genera of the phylum Ascomycota. In contrast with the the Nectria and Hypocrea, the other five fungal genera were known to be marine-derived (Nigrospora [12]; Alternaria [13]; Aspergillus, Penicillium, and Cladosporium [14]). The ascomycetes represent those fungi that are readily cultivable and could be easily recovered when culture-dependent techniques are applied [15]. Representative strains of these fungi have previously been isolated from marine algae, seagrasses, sponges and cnidarians [16]. The ascomycetes have been reported as invertebrate symbiotic fungi and as a prolific source of biologically active secondary metabolites, such as polyketides with antitumoral and/or antimicrobial activities [15–18]. Our investigation revealed that the diversity of symbiotic fungi (Aspergillus, 5 strains; Penicillium, 4 strains; Cladosporium, 5 strains) isolated from the gorgonian Echinogorgia rebekka was high.
2.2. Antibacterial Activities of the Extracts from Fungal Broth and Mycelia
Antibacterial activities of the extracts from the fermentation supernatant and mycelia of 18 isolated fungi were assayed by using four terrestrial pathogenic bacteria (Escherichia coli ATCC25922, S. aureus ATCC6538P, Bacillus subtilis 20019 and M. tetragenus ATCC13623) and one marine pathogenic bacterium (Vibrio anguillarum ATCC19019). E. coli, S. aureus, B. subtilis, and M. tetragenus are common pathogens and model bacteria for this type of work. V. anguillarum is a harmful bacterium in aquaculture. The inhibition activities to these 5 bacterial can provide useful data to find promising natural products candidates. All of the 18 fungi showed different levels of antibacterial activities to at least one pathogen with their fermentation broth and/or mycelia (Table 2). Out of the 18 fungal strains, 12 were found to show moderate to high level of antibacterial activities to S. aureus, while 9 had moderate to very high activities to M. tetragenus (Table 2). It is worth pointing out that the mycelia EtOAc extract of the Penicillium strain WZL018 displayed exceptionally high antibacterial activities to M. tetragenus (growth inhibition diameters: larger than 15 mm). In addition, the mycelia EtOAc extract of the Penicillium WZL017 and the Cladosporium strain WZL015 exhibited high activities to the pathogen B. subtilis with growth inhibition diameters between 10 and 15 mm, while the broth EtOAc extract of Cladosporium strain WZL009 displayed similar activities to the pathogen S. aureus (Table 2). Microbes can secrete a wide variety of metabolites including intracellular and extracellular products [19]. From the data in the present study, we found that some broth and/or mycelia EtOAc extracts of fungi have the antibacterial activities. This suggested that different fungi could produce intracellular bioactive metabolites or secrete extracellular bioactive compounds. In addition, the results indicated that the fungal extracts exhibited higher inhibition activities to Gram-positive bacteria, such as B. subtilis, S. aureus and M. tetragenus, than to Gram-negative bacteria, which might be a result of different cell wall compositions of Gram-positive and Gram-negative bacteria. Many diseases are known to be caused by gram-positive, such as the infection of respiratory tract and skin. The gram-positive bacteria can produce exotoxin which can infect human bodies. The fungi with better bioactivity toward gram-positive bacteria will be a possible source of medicine to treat these diseases. Our results indicate that the gorgonian-derived fungi isolated in this study, especially the Penicillium strains WZL018, WZL017, and the Cladosporium strains WZL015, WZL009, exhibited potential for the isolation of antibacterial natural products.
3. Experimental Section
3.1. Coral Materials
The sample of a gorgonian coral Echinogorgia rebekka was collected from the Weizhou coral reef in the South China Sea in September 2008. The coral sample was transferred directly to a sterile plastic bag without seawater. Seawater samples from the collection sites were collected to serve as controls. The samples were stored immediately at 4 °C and sent to the laboratory as soon as possible for the isolation of fungi. Latex gloves were worn during collection.
3.2. Fungi Isolation
The coral sample was rinsed three times with sterile artificial seawater to get rid of nonspecific fungal propagules that stick to the coral surface from seawater colony, and the surface of the coral was disinfected with 70% ethanol [20]. Then the coral was cut into small pieces and homogenized using a blender containing 20 mL sterile natural seawater under aseptic conditions. The resulting homogenate was diluted with sterile seawater at three dilutions (1:10, 1:100, and 1:1000). For fungi cultivation, 100 μL of each dilution was plated in quadruplicate onto the four plates of the following medium: potato dextrose agar (PDA) medium (200 g of washed but unpeeled potatoes slices were boiled in 1 L seawater for 30 min, 20 g dextrose and 20 g agar powder was then added into the potato soup), Rose Bengal Medium (RBM) (peptone 5 g, glucose 10 g, KH2PO4 1 g, MgSO4·7H2O 0.5 g, Rose Bengal 0.033 g, agar 20 g, seawater 1 L), Czapek-Dox medium (NaNO3 3 g, KCl 0.5 g, K2HPO4 0.1 g, MgSO4·7H2O 0.5 g, FeSO4 0.01 g, sucrose 30 g, agar 20 g, seawater 1 L, pH 6.7), and Luria Bertani (LB) agar medium. The antibiotics of penicillin and streptomycin (100 mg/mL for each) were added to each plate described above. The plates were incubated at 28 °C for 1–3 weeks until the morphology of fungi could be distinguished. The fungi isolates were identified morphologically according to Wei’s morphological criteria [21]. The morphological traits (e.g., morphology and color of spore and mycelia) of the pure fungal colonies were examined by microscopy to exclude redundant fungal isolates and were used to group the strains into different morphotypes. Photos were taken for distinct isolates. Each isolate was picked and transferred onto a new corresponding agar plate containing PDA, RBM, Czapek-Dox, or LB medium with penicillin and streptomycin. The resulting plates were incubated at 28 °C for pure culture.
3.3. Extraction of Genome DNA from Cultured Fungi
The distinct fungi isolates described in the above paragraph were cultured in GPY broth (glucose 20 g, yeast extract paste 10 g, peptone 20 g, dissolved in 1 L seawater, pH 6.5) at 28 °C for 2–5 days. The mycelia was harvested by using vacuum filtration and dried with two layers of paper towel. The resulting mycelial mat was ground into powder with liquid nitrogen.
The fungal DNA was extracted according to the procedure described by Fredricks with modifications [22]. In brief, the mycelial powder was transferred to a 1.5 mL Eppendorf tube containing 500 μL TE buffer (10 mM Tris, 0.1 mM EDTA, pH 7.3). Lysozyme was then added, and the mixture was incubated at 37 °C for 30 min. Later, 10% SDS (1:10, w:v) and proteinase K (1:100, w:v) were added and incubated at 56 °C for 3 h. An equal volume of buffer equilibrated phenol was added followed by brief mixing and the mixture was centrifugation at 12,000 rpm for 10 min at 4 °C. The upper aqueous phase was gently transferred to a new tube, and an equal volume of chloroform/phenol (1:1, v:v) was added. The mixture was separated by centrifugation at 12,000 rpm for 10 min and the aqueous phase was removed. Then, 3 mol/L sodium acetate (1:10, v:v) and RNaseA were added to remove RNA. DNA was precipitated by adding 2.5 volumes of absolute ethanol. The DNA pellet was washed with 75% ethanol twice and resuspended in 50 μL of sterile water to be dissolved.
3.4. Amplification of Fungal ITS-rDNA Fragments
The genomic DNA was used as the template to amplify fungal ITS-rDNA fragments using the primers ITS1 (5′-TCCGTAGGTGAACCTGCG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [23] which were synthesized by Shanghai Sangon Biological Technology and Service Co., Ltd. The reaction mixture for PCR amplification contained 5 μL of 10× reaction buffer with 15 mM MgCl2 (TaKaRa), 2 μL of 2.5 mM dNTPs, 0.5 μL of 10 μM each primer, 4 μL of fungal DNA, 0.3 μL of Taq DNA polymerase (5 U·μL−1, TaKaRa), and 39.7 μL of H2O. PCR conditions included an initial denaturation at 94 °C for 5 min followed by 32 cycles of denaturation at 94 °C for 50 s, annealing at 52.5 °C for 50 s, and elongation at 72 °C for 1 min, with a final elongation at 72 °C for 10 min. PCR products were purified using the Agarose Gel DNA Purification Kit (TaKaRa) and sequenced in Shanghai Sangon Biological Engineering Technology and Service Co., Ltd.
3.5. Sequence and Phylogenetic Analysis
For preliminary identification, sequences of fungal ITS-rDNA regions obtained from the gorgonian coral Echinogorgia rebekka were compared with related sequences in NCBI [24]. Fungal ITS-rDNA sequences acquired in this study were edited and aligned with the best n-BLAST hits from GenBank in the Clustal X (version 1.83) program [25], and further manually adjusted using BioEdit software [26]. The sequences obtained in this study were deposited in the GenBank/EMBL/DDBJ under the accession numbers HQ588304–HQ588321. The program MEGA 5 [27] was applied to calculate the base composition of the fungal sequences. To analyze the relationship between the fungal species, MEGA 5 was also used to reconstruct phylogenetic trees using the neighbor-joining (NJ) method. A maximum likelihood statistical method implemented in MEGA 5 was first used to determine the best-fit nucleotide substitution models for ITS sequences based on Bayesian Information Criterion (BIC); BIC values revealed that the best substitution model was K2 + G model [28]. Then this model was employed to reconstruct phylogenetic trees with 10,000 bootstrap replicates. ITS sequences of related fungi were downloaded from NCBI as references.
3.6. Assay of Antibacterial Activity
The fungi was inoculated into 500 mL Erlenmeyer flasks containing 150 mL of liquid medium (glucose 10 g, yeast extract paste 1 g, peptone 3 g, dissolved in 1 L seawater, pH 6.5), followed by shaking incubation at 28 °C with 165 rpm for nine days. The fermented whole broth was filtered through cheese cloth to separate the supernatant and the mycelia. The supernatant layer was concentrated to about a quarter of the original volume under reduced pressure, and then extracted with ethyl acetate (EtOAc) three times. The EtOAc phase was evaporated to gain EtOAc extract. The mycelia were extracted firstly with acetone three times, and the solvent was evaporated in vacuo to give a residue, which was extracted with EtOAc three times to yield another EtOAc extract. The obtained EtOAc extracts were stored after freeze drying.
Five pathogenic microorganisms were used for the antibacterial assay, including four terrestrial bacteria (E. coli ATCC25922, S. aureus ATCC6538P, B. subtilis 20019, M. tetragenus ATCC13623) and one marine bacterium Vibrio anguillarum ATCC19019. After overnight culture, each tested bacterium was adjusted to 2 × 108–5 × 108 colony forming units per mL, and 0.1 mL of such a culture was spread on medium of Petri dishes (Φ 9 cm). V. anguillarum were grown in Tryptic Soy Brothor Agar (TSB, TSA) and others were grown in Nutrient Agar (NA). Holes (Φ 6 mm) were then drilled on the agar plates. The lyophilized EtOAc extract described in the above paragraph was dissolved in DMSO with a final concentration of 100 μg/mL and used as a test solution. And 5 μL of the test solution was added to each hole on the plates. The dishes were incubated at 37 °C for 16–24 h. The diameters of the inhibition zone surrounding the holes were measured. DMSO and streptomycin were used as negative control and positive control, respectively. The experiments were carried out in triplicates.
4. Conclusions
A total of 18 symbiotic Ascomycota fungi were isolated from the gorgonian coral Echinogorgia rebekka. They were distributed among seven genera in five orders. These ascomycetes displayed different levels of antibacterial activities to pathogenic bacteria, and some strains in the genera Penicillium and Cladosporium showed strong growth inhibition to B. subtilis, S. aureus and M. tetragenus. The combined data gathered from this investigation contribute to broadening the knowledge of microbial diversity and antibacterial bioactivities associated with corals. As corals are an important source of marine bioactive natural products and drugs, and the coral resources are decreasing and limited, the isolation of symbiotic microbes from the gorgonian corals and the screening for bioactivities will have important significance for further research to discover new bioactive natural products. Meanwhile, due to the emergence of antibiotic resistance of traditional natural products and drugs, there is a pressing need for the development of novel antimicrobial therapies. Recently, researchers have invested their interest in the development of antimicrobial peptides (AMPs) [29,30], and more efforts are needed to be done in this area.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 40976077; 30901879), the Research Fund for the Doctoral Program of Higher Education, Ministry of Education of China (No. 20090132110002), the Basic Research Program of Science and Technology, Ministry of Science and Technology of China (No. 2007FY210500), and Program for Changjiang Scholars and Innovative Research Team in University, Ministry of Education of China (No. IRT0944).
- Samples Availability: Available from the authors.
References
- Balser, TC; McMahon, KD; Bart, D; Bronson, D; Coyle, DR; Craig, N. Bridging the gap between micro- and macro-scale perspectives on the role of microbial communities in global change ecology. Plant Soil 2006, 289, 59–70. [Google Scholar]
- Gutknecht, JLM; Goodman, RM; Balser, TC. Linking soil process and microbial ecology in fresh water wetland ecosystems. Plant Soil 2006, 289, 17–34. [Google Scholar]
- Schimel, J; Balser, T; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar]
- Ainsworth, TD; Thurber, RV; Gates, RD. The future of coral reefs: A microbial perspective. Trends Ecol. Evol 2009, 25, 233–240. [Google Scholar]
- Blunt, JW; Copp, BR; Hu, WP; Munro, MHG; Northcote, PT; Prinsep, MR. Marine natural products. Nat. Prod. Rep 2008, 25, 35–94. [Google Scholar]
- König, GM; Kehraus, S; Seiber, SF; Abdel-Lateff, A; Muller, D. Natural products from marine organisms and their associated microbes. Chem. Biochem 2006, 7, 229–238. [Google Scholar]
- Jensen, PR; Fenical, W. Secondary Metabolites from Marine Fungi. In Fungi in Marine Environments; Hyde, KD, Ed.; Fungal Diversity Press: Hong Kong, China, 2002; pp. 293–315. [Google Scholar]
- Li, Z. Advances in marine microbial symbionts in the China Sea and related pharmaceutical metabolites. Mar. Drugs 2009, 7, 113–129. [Google Scholar]
- Proksch, P; Edrada-Ebel, R; Ebel, R. Drugs from the sea—opportunities obstacles. Mar. Drugs 2003, 1, 5–17. [Google Scholar]
- Bhatnagar, I; Kim, S-K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar]
- Radjasa, OK. Marine invertebrate-associated bacteria in coral reef ecosystems as a new source of bioactive compounds. J. Coast. Dev 2004, 7, 65–70. [Google Scholar]
- Trisuwan, K; Rukachaisirikul, V; Sukpondma, Y; Preedanon, S; Phongpaichit, S; Rungjindamai, N; Sakayaroj, J. Epoxydons and a pyrone from the marine-derived fungus Nigrospora sp. PSU-F5. J. Nat. Prod 2008, 71, 1323–1326. [Google Scholar]
- Arvanitidou, M; Kanellou, K; Constantinidis, T; Katsouyannopoulos, V. Higher prevalence of Alternaria spp. in marine and river waters than in potable samples. Microbiol. Res 2000, 155, 49–51. [Google Scholar]
- Koh, LL; Tan, TK; Chou, LM; Goh, NKC. Fungi Associated with Gorgonians in Singapore. Proceedings of the 9th International Coral Reef Symposium, Bali, Indonesia, 23–27 October 2000.
- Baker, PW; Kennedy, J; Dobson, ADW; Marchesi, JR. Phylogenetic diversity and antimicrobial activities of fungi associated with Haliclona simulans isolated from Irish coastal waters. Mar. Biotechnol 2009, 11, 540–547. [Google Scholar]
- Da Silva, M; Passarini, MRZ; Bonugli, RC; Sette, LD. Cnidarian-derived filamentous fungi from Brazil: Isolation, characterization and RBBR decolourization screening. Environ. Technol 2008, 29, 1331–1339. [Google Scholar]
- Holler, U; Wright, AD; Matthee, GF; Konig, GM; Draeger, D; Aust, HJ. Fungi from marine sponges: Diversity, biological activity and secondary metabolites. Mycol. Res 2000, 104, 1354–1365. [Google Scholar]
- Bugni, TS; Ireland, CM. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep 2004, 21, 143–163. [Google Scholar]
- Konings, WN; Poolman, B; Driessen, AM. Can the excretion of metabolites by bacteria be manipulated? FEMS Microbiol. Lett 1992, 2, 93–108. [Google Scholar]
- Li, Q; Wang, G. Diversity of fungal isolates from three Hawaiian marine sponges. Microbiol. Res 2009, 164, 233–241. [Google Scholar]
- Wei, JC. The Identification Manual of Fungi; Shanghai Science and Technology Press: Shanghai, China, 1979. [Google Scholar]
- Fredricks, DN; Smith, C; Meier, A. Comparison of six DNA extraction methods for recovery of fungal DNA as assessed by quantitative PCR. J. Clin. Microbiol 2005, 43, 5122–5128. [Google Scholar]
- White, TJ; Bruns, T; Lee, S; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Application; Innis, MA, Gelfand, DH, Sninsky, JJ, White, TJ, Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov (accessed on 10 June 2011).
- Thompson, JD; Gibson, TJ; Plewniak, F; Jeanmougin, F; Higgins, DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25, 4876–4882. [Google Scholar]
- Hall, TA. BioEdit: A user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser 1999, 41, 95–98. [Google Scholar]
- Tamura, K; Peterson, D; Peterson, N; Stecher, G; Nei, M; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol 1980, 16, 111–120. [Google Scholar]
- Gottler, LM; Ramamoorthy, A. Structure, membrane orientation, mechanism, and function of pexiganan—A highly potent antimicrobial peptide designed from magainin. Biochim. Biophys Acta 2009, 1788, 1680–1686. [Google Scholar]
- Marsh, EN; Buer, BC; Ramamoorthy, A. Fluorine—A new element in the design of membrane-active peptides. Mol. BioSyst 2009, 5, 1143–1147. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).