Molecular Evolution of Multiple Arylalkylamine N-Acetyltransferase (AANAT) in Fish
Abstract
:1. Introduction
2. Results and Discussion
2.1. Kinetic Characterization of Mutated AANAT1a and AANAT2
2.1.1. β5 Mutations
2.1.2. α2 Mutation
2.1.3. β4 Mutation
2.1.4. αa Mutation
2.2. Inhibition Experiments
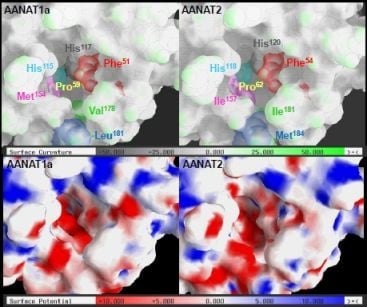
2.3. AANAT1a and AANAT2 Have Similar Predicted Binding Capacities
2.4. In Silico Assessment of Binding Stability
3. Experimental Section
3.1. Mutant Design
3.2. Preparation of Wild Type and Mutant Proteins
3.3. Assay of AANAT Activity
3.4. Structural Analysis
3.5. Statistical Analysis
4. Conclusions
Supplementary Data
marinedrugs-09-00906-s001.pdfAcknowledgments
- Samples Availability: Available from the authors.
References
- Neuwald, AF; Landsman, D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci 1997, 22, 154–155. [Google Scholar]
- Dyda, F; Klein, DC; Hickman, AB. GCN5-related N-acetyltransferases: A structural overview. Annu Rev Biophys Biomol Struct 2000, 29, 81–103. [Google Scholar]
- Ferry, G; Ubeaud, C; Dauly, C; Mozo, J; Guillard, S; Berger, S; Jimenez, S; Scoul, C; Leclerc, G; Yous, S; Delagrange, P; Boutin, JA. Purification of the recombinant human serotonin N-acetyltransferase (EC 2.3.1.87): Further characterization of and comparison with AANAT from other species. Protein Expr Purif 2004, 38, 84–98. [Google Scholar]
- Khalil, EM; de Angelis, J; Cole, PA. Indoleamine analogs as probes of the substrate selectivity and catalytic mechanism of serotonin N-acetyltransferase. J Biol Chem 1998, 273, 30321–30327. [Google Scholar]
- Coon, SL; Roseboom, PH; Baler, R; Weller, JL; Namboodiri, MA; Koonin, EV; Klein, DC. Pineal serotonin N-acetyltransferase: Expression cloning and molecular analysis. Science 1995, 270, 1681–1683. [Google Scholar]
- Voisin, P; Namboodiri, MA; Klein, DC. Arylamine N-acetyltransferase and arylalkylamine N-acetyltransferase in the mammalian pineal gland. J Biol Chem 1984, 259, 10913–10918. [Google Scholar]
- Ferry, G; Loynel, A; Kucharczyk, N; Bertin, S; Rodriguez, M; Delagrange, P; Galizzi, JP; Jacoby, E; Volland, JP; Lesieur, D; Renard, P; Canet, E; Fauchere, JL; Boutin, JA. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J Biol Chem 2000, 275, 8794–8805. [Google Scholar]
- Coon, SL; Klein, DC. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol Cell Endocrinol 2006, 252, 2–10. [Google Scholar]
- Iyer, LM; Aravind, L; Coon, SL; Klein, DC; Koonin, EV. Evolution of cell-cell signaling in animals: Did late horizontal gene transfer from bacteria have a role? Trends Genet 2004, 20, 292–299. [Google Scholar]
- Klein, DC; Coon, SL; Roseboom, PH; Weller, JL; Bernard, M; Gastel, JA; Zatz, M; Iuvone, PM; Rodriguez, IR; Begay, V; Falcon, J; Cahill, GM; Cassone, VM; Baler, R. The melatonin rhythm-generating enzyme: Molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res 1997, 52, 307–357, discussion 357–308. [Google Scholar]
- Ganguly, S; Coon, SL; Klein, DC. Control of melatonin synthesis in the mammalian pineal gland: The critical role of serotonin acetylation. Cell Tissue Res 2002, 309, 127–137. [Google Scholar]
- De Angelis, J; Gastel, J; Klein, DC; Cole, PA. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87). J Biol Chem 1998, 273, 3045–3050. [Google Scholar]
- Scheibner, KA; de Angelis, J; Burley, SK; Cole, PA. Investigation of the roles of catalytic residues in serotonin N-acetyltransferase. J Biol Chem 2002, 277, 18118–18126. [Google Scholar]
- Hickman, AB; Klein, DC; Dyda, F. Melatonin biosynthesis: The structure of serotonin N-acetyltransferase at 2.5 A resolution suggests a catalytic mechanism. Mol Cell 1999, 3, 23–32. [Google Scholar]
- Hickman, AB; Namboodiri, MA; Klein, DC; Dyda, F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: Enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell 1999, 97, 361–369. [Google Scholar]
- Obsil, T; Ghirlando, R; Klein, DC; Ganguly, S; Dyda, F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell 2001, 105, 257–267. [Google Scholar]
- Wolf, E; De Angelis, J; Khalil, EM; Cole, PA; Burley, SK. X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition. J Mol Biol 2002, 317, 215–224. [Google Scholar]
- Coon, SL; Begay, V; Deurloo, D; Falcon, J; Klein, DC. Two arylalkylamine N-acetyltransferase genes mediate melatonin synthesis in fish. J Biol Chem 1999, 274, 9076–9082. [Google Scholar]
- Gothilf, Y; Coon, SL; Toyama, R; Chitnis, A; Namboodiri, MA; Klein, DC. Zebrafish serotonin N-acetyltransferase-2: Marker for development of pineal photoreceptors and circadian clock function. Endocrinology 1999, 140, 4895–4903. [Google Scholar]
- Mizusawa, K; Iigo, M; Masuda, T; Aida, K. Photic regulation of arylalkylamine N-acetyltransferase 1 mRNA in trout retina. Neuroreport 2000, 11, 3473–3477. [Google Scholar]
- Falcon, J; Gothilf, Y; Coon, SL; Boeuf, G; Klein, DC. Genetic, temporal and developmental differences between melatonin rhythm generating systems in the teleost fish pineal organ and retina. J Neuroendocrinol 2003, 15, 378–382. [Google Scholar]
- Zilberman-Peled, B; Benhar, I; Coon, SL; Ron, B; Gothilf, Y. Duality of serotonin-N-acetyltransferase in the gilthead seabream (Sparus aurata): Molecular cloning and characterization of recombinant enzymes. Gen Comp Endocrinol 2004, 138, 139–147. [Google Scholar]
- Falcon, J; Besseau, L; Fuentes, M; Sauzet, S; Magnanou, E; Boeuf, G. Structural and functional evolution of the pineal melatonin system in vertebrates. Ann NY Acad Sci 2009, 1163, 101–111. [Google Scholar]
- Aparicio, S. Vertebrate evolution: Recent perspectives from fish. Trends Genet 2000, 16, 54–56. [Google Scholar]
- Volff, JN. Genome evolution and biodiversity in teleost fish. Heredity 2005, 94, 280–294. [Google Scholar]
- True, JR; Carroll, SB. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol 2002, 18, 53–80. [Google Scholar]
- Falcon, J; Migaud, H; Munoz-Cueto, JA; Carrillo, M. Current knowledge on the melatonin system in teleost fish. Gen Comp Endocrinol 2010, 165, 469–482. [Google Scholar] [Green Version]
- Benyassi, A; Schwartz, C; Coon, SL; Klein, DC; Falcon, J. Melatonin synthesis: Arylalkylamine N-acetyltransferases in trout retina and pineal organ are different. Neuroreport 2000, 11, 255–258. [Google Scholar]
- Zilberman-Peled, B; Ron, B; Gross, A; Finberg, JP; Gothilf, Y. A possible new role for fish retinal serotonin-N-acetyltransferase-1 (AANAT1): Dopamine metabolism. Brain Res 2006, 1073–1074, 220–228. [Google Scholar]
- Klein, DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland—A tale of conflict and resolution. J Biol Rhythms 2004, 19, 264–279. [Google Scholar]
- Klein, DC. Evolution of the vertebrate pineal gland: The AANAT hypothesis. Chronobiol Int 2006, 23, 5–20. [Google Scholar]
- Klein, DC. Arylalkylamine N-acetyltransferase: “The Timezyme”. J Biol Chem 2007, 282, 4233–4237. [Google Scholar]
- Appelbaum, L; Vallone, D; Anzulovich, A; Ziv, L; Tom, M; Foulkes, NS; Gothilf, Y. Zebrafish arylalkylamine-N-acetyltransferase genes—Targets for regulation of the circadian clock. J Mol Endocrinol 2006, 36, 337–347. [Google Scholar]
- Vuilleumier, R; Boeuf, G; Fuentes, M; Gehring, WJ; Falcon, J. Cloning and early expression pattern of two melatonin biosynthesis enzymes in the turbot (Scophthalmus maximus). Eur J Neurosci 2007, 25, 3047–3057. [Google Scholar]
- Pavlicek, J; Coon, SL; Ganguly, S; Weller, JL; Hassan, SA; Sackett, DL; Klein, DC. Evidence that proline focuses movement of the floppy loop of arylalkylamine N-acetyltransferase (EC 2.3.1.87). J Biol Chem 2008, 283, 14552–14558. [Google Scholar]
- Vetting, MW; de Carvalho, LPS; Yu, M; Hegde, SS; Magnet, S; Roderick, SL; Blanchard, JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys 2005, 433, 212–226. [Google Scholar]
- Isorna, E; El M’rabet, A; Confente, F; Falcon, J; Munoz-Cueto, JA. Cloning and expression of arylalkylamine N-acetyltranferase-2 during early development and metamorphosis in the sole Solea senegalensis. Gen Comp Endocrinol 2009, 161, 97–102. [Google Scholar]
- Shi, Q; Ando, H; Coon, SL; Sato, S; Ban, M; Urano, A. Embryonic and post-embryonic expression of arylalkylamine N-acetyltransferase and melatonin receptor genes in the eye and brain of chum salmon (Oncorhynchus keta). Gen Comp Endocrinol 2004, 136, 311–321. [Google Scholar]
- Yu, EZ; Hallenbeck, JM; Cai, D; McCarron, RM. Elevated arylalkylamine-N-acetyltransferase (AA-NAT) gene expression in medial habenular and suprachiasmatic nuclei of hibernating ground squirrels. Brain Res Mol Brain Res 2002, 102, 9–17. [Google Scholar]
- Velarde, E; Cerda-Reverter, JM; Alonso-Gomez, AL; Sanchez, E; Isorna, E; Delgado, MJ. Melatonin-synthesizing enzymes in pineal, retina, liver, and gut of the goldfish (Carassius): mRNA expression pattern and regulation of daily rhythms by lighting conditions. Chronobiol Int 2010, 27, 1178–1201. [Google Scholar]
- Riddles, PW; Blakeley, RL; Zerner, B. Reassessment of Ellman’s reagent. Meth Enzymol 1983, 91, 49–60. [Google Scholar]
- Czaplicki, J; Cornelissen, G; Halberg, F. GOSA, a simulated annealing-based program for global optimization of nonlinear problems, also reveals transyears. J Appl Biomed 2006, 4, 87–94. [Google Scholar]
- Sugden, D; Namboodiri, MA; Weller, JL; klein, DC. Melatonin synthesizing enzymes: Serotonin N-acetyltransferase and hydroxyindol-O-methyltransferase. In Methods in Biogenic Amine Research; Parvez, H, Parvez, S, Nagatsu, T, Eds.; Elsevier/North Holland Biomedical Press: Amsterdam, The Netherlands, 1983; pp. 567–572. [Google Scholar]
- Needleman, SB; Wunsch, CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 1970, 48, 443–453. [Google Scholar]
- Karuskal, JB. An overview of sequence comparison. In Time Warps, String Edits and Macromolecules: The Theory and Practice of Sequence Comparison; Sankoff, D, Kruskal, JB, Eds.; Addison Wesley: Boston, MA, USA, 1983; pp. 1–44. [Google Scholar]
- Petrey, D; Xiang, Z; Tang, CL; Xie, L; Gimpelev, M; Mitros, T; Soto, CS; Goldsmith-Fischman, S; Kernytsky, A; Schlessinger, A; Koh, IY; Alexov, E; Honig, B. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 2003, 53(Suppl 6), 430–435. [Google Scholar]
- Laskowski, RA; MacArthur, MW; Moss, DS; Thornton, JM. PROTHECK: A program to check the steriochemical quality of protein structures. J Appl Cryst 1993, 26, 283–291. [Google Scholar]
- Pieper, U; Eswar, N; Braberg, H; Madhusudhan, MS; Davis, FP; Stuart, AC; Mirkovic, N; Rossi, A; Marti-Renom, MA; Fiser, A; Webb, B; Greenblatt, D; Huang, CC; Ferrin, TE; Sali, A. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucl Acids Res 2004, 32, D217–D222. [Google Scholar]
- Gordon, JC; Myers, JB; Folta, T; Shoja, V; Heath, LS; Onufriev, A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 2005, 33, W368–371. [Google Scholar]
- Petrey, D; Honig, B. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Meth Enzymol 2003, 374, 492–509. [Google Scholar]
- Duhovny, D; Nussinov, R; Wolfson, HJ. Efficient unbound docking of rigid molecules. In Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI); Rome, Italy, 17–21 September 2002; Guigo, R, Gusfield, D, Eds.; Springer: Berlin, Germany, 2002; Volume 2452. [Google Scholar]
- Schneidman-Duhovny, D; Inbar, Y; Polak, V; Shatsky, M; Halperin, I; Benyamini, H; Barzilai, A; Dror, O; Haspel, N; Nussinov, R; Wolfson, HJ. Taking geometry to its edge: Fast unbound rigid (and hinge-bent) docking. Proteins 2003, 52, 107–112. [Google Scholar]




| Mutation sites | |||
|---|---|---|---|
| Mutation region/name | AANAT1a | AANAT2 | Sheep AANAT |
| β5 | M154I | I157M | M159 |
| α2 | - | L69R + N70H + G73T + Q74L | Q71 + H72 + T75 + L76 |
| β4 | - | S123A | A125 |
| αa | - | M107L | L109 |
| AANAT1a | AANAT2 | ||||||
|---|---|---|---|---|---|---|---|
| Wild type | β5 | Wild type | β5 | α2 | β4 | αa | |
| Serotonin | 61.36 ± 4.39 | 64.15 ± 3.49 | 6.60 ± 0.54 | 11.34 ± 0.93 | 17.65 ± 1.06 | 18.58 ± 1.41 | 7.29 ± 0.54 |
| Tryptamine | 23.37 ± 1.59 | 14.84 ± 0.60 | 3.19 ± 0.25 | 8.50 ± 0.69 | 6.62 ± 0.33 | 9.06 ± 0.78 | 1.46 ± 0.003 |
| Dopamine | 7.30 ± 0.54 | 1.86 ± 0.49 | 1 | 2.40 ± 0.10 | 3.80 ± 0.07 | 2.48 ± 0.18 | 0.65 ± 0.03 |
| Phenylethylamine | 32.86 ± 2.74 | 5.05 ± 0.61 | 1 | 2.52 ± 0.26 | 4.33 ± 0.56 | 3.03 ± 0.09 | 0.61 ± 0.03 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zilberman-Peled, B.; Bransburg-Zabary, S.; Klein, D.C.; Gothilf, Y. Molecular Evolution of Multiple Arylalkylamine N-Acetyltransferase (AANAT) in Fish. Mar. Drugs 2011, 9, 906-921. https://doi.org/10.3390/md9050906
Zilberman-Peled B, Bransburg-Zabary S, Klein DC, Gothilf Y. Molecular Evolution of Multiple Arylalkylamine N-Acetyltransferase (AANAT) in Fish. Marine Drugs. 2011; 9(5):906-921. https://doi.org/10.3390/md9050906
Chicago/Turabian StyleZilberman-Peled, Bina, Sharron Bransburg-Zabary, David C. Klein, and Yoav Gothilf. 2011. "Molecular Evolution of Multiple Arylalkylamine N-Acetyltransferase (AANAT) in Fish" Marine Drugs 9, no. 5: 906-921. https://doi.org/10.3390/md9050906
APA StyleZilberman-Peled, B., Bransburg-Zabary, S., Klein, D. C., & Gothilf, Y. (2011). Molecular Evolution of Multiple Arylalkylamine N-Acetyltransferase (AANAT) in Fish. Marine Drugs, 9(5), 906-921. https://doi.org/10.3390/md9050906