Abstract
Since sponges, as typical filter-feeders, are exposed to a high load of attacking prokaryotic and eukaryotic organisms, they are armed with a wide arsenal of antimicrobial/cytostatic low-molecular-weight, non-proteinaceous bioactive compounds. Here we present the first sponge agent belonging to the group of ASABF-type antimicrobial peptides. The ASABF gene was identified and cloned from the demosponge Suberites domuncula. The mature peptide, with a length of 64 aa residues has a predicted pI of 9.24, and comprises the characteristic CSα β structural motif. Consequently, the S. domuncula ASABF shares high similarity with the nematode ASABFs; it is distantly related to the defensins. The recombinant peptide was found to display besides microbicidal activity, anti-fungal activity. In addition, the peptide lyses human erythrocytes. The expression of ASABF is upregulated after exposure to the apoptosis-inducing agent 2,2′-dipyridyl. During the process of apoptosis of surface tissue of S. domuncula, grazing gastropods (Bittium sp.) are attracted by quinolinic acid which is synthesized through the kynurenine pathway by the enzyme 3-hydroxyanthranilate 3,4-dioxygenase (HAD). Finally, the gastropods are repelled from the sponge tissue by the ASABF. It is shown that the effector peptide ASABF is sequentially expressed after the induction of the HAD gene and a caspase, as a central enzyme executing apoptosis.
1. Introduction
Sponges, as sessile filter feeders, are exposed to a huge number of microorganisms, including bacteria, fungi, and viruses, which are eliminated during the passage through the aquiferous sponge canal system [1]. It had been calculated that some sponge species filter up to 0.84 cm3 of water per second per cubic centimeter of sponge tissue through their canals to extract edible material [2]. In order to resist against those microorganisms that could be harmful for the sponge specimens, these animals have developed a wide array of bioactive compounds [3] that act highly specifically and efficiently. Among those are secondary metabolites interfering with distinct pathways of higher metazoans, such as tyrosine kinases (e.g., aeroplysinin), and replication of viruses, e.g., herpes simplex viruses (arabinofuranosyl adenine) or retroviruses (e.g., avarol) [4,5].
In addition to those directly acting inhibitory, secondary metabolites sponges have a very effective immune system [6]. Nevertheless it is well established that sponges are not provided with an antigen-specific adaptive immune system, even though they express proteins composed of two characteristic immunoglobulin (Ig)-like domains, e.g., in Geodia cydonium [7]. However, these animals compensate this lack with a highly diverse innate immune system against non-specific as well as specific targets [8]. Most of the biological and molecular biological studies on immune responses in sponges have been performed with the demosponge Suberites domuncula. For example, fungi are recognized by specific d-glucan carbohydrate receptors [9] and subsequently eliminated by expression of a fibrinogen-like protein. Gram-positive bacteria are killed by an increased induction of lysozyme [10], resulting in lysis of the microorganisms and subsequent phagocytosis. Gram-negative bacteria are recognized by sponges through a lipopolysaccharide-interacting protein and eliminated by expression of perforins through activation of a MyD88-dependent signaling pathway [11]. It should be noted that most of the recognition and effector molecules display high sequence similarities to polypeptides identified in deuterostomians. However, sponges also produce molecules that are only found in some protostomian taxa, e.g., a GlcNAc/GalNAc-binding lectin (tachylectin; [12]). It is amazing that this rich arsenal exists in the phylum Porifera which is the oldest, still extant, animal phylum, originating from the common metazoan ancestor, the Urmetazoa [13].
Considering the large and diverse number of noxious prokaryotic and eukaryotic organisms passing through sponges, it appeared to be compelling to search for antimicrobial peptides displaying broad activity spectra. It is well established that cationic antimicrobial peptides are produced by higher metazoans as host defense against microorganisms [14]. Recently they have been recognized as promising novel antimicrobial and antitumor agents [15]. The defensins and the ASABF (Ascaris suum antibacterial factor) are grouped to those peptides [16]. By screening the S. domuncula database that comprises over 40,000 sequences [17] several ESTs (Expressed Sequence Tag) were identified that displayed defensin-like sequence similarity. More specifically, the deduced protein shared highest relationship with the antimicrobial peptide designated as ASABF. ASABF peptides were first identified in Ascaris suum [18] and subsequently also in Caenorhabditis elegans [19]. The ASABF peptides and the defensins are both cysteine-rich. While the defensins are widespread both in protostomians and in deuterostomians [20], the ASABF peptides are restricted, among the triploblastic animals, to the nematodes [21]. In contrast to the defensins which comprise two to three antiparallel β-sheets that are linked together by three to four intramolecular disulfide bridges, the characteristic ASABFs have eight cysteine residues that form four disulfide bridges [22]. In the present study we report on the first sponge cDNA encoding an ASABF peptide. The predicted, mature peptide from S. domuncula, comprising 64 aa (amino acid) residues, was shown to be cationic (isoelectric point >8), a characteristic that it shares with the other known ASABFs [23]. Furthermore, we also checked the database from the Baikalian freshwater sponge Lubomirskia baicalensis for the existence of an ASABF-encoding cDNA; a cDNA fragment was found and completed. However, this sequence shared only little sequence similarity with other ASABFs or defensins.
The recombinant sponge ASABF protein was shown to be biologically active, as microbicidal (as expected) peptide possessing also hemolytic activity. Furthermore, we report here for the first time that an ASABF peptide is toxic also for metazoans. It is shown that the ASABF peptide was toxic for the marine gastropod/molluscs Bittium sp. which settled on apoptotic patches on the sponge surface [24]. The gastropod was attracted by the sponge after induction of the 3-hydroxyanthranilate 3,4-dioxygenase (HAD) gene, through quinolinic acid, a metabolite of the kynurenine pathway. In parallel with the increase in the steady-state level of HAD, the expression of a gene encoding a caspase was induced [25] that is known to be one key enzyme induced during apoptosis in S. domuncula. Both effector limbs, grazing of the gastropod and the caspase-dependent apoptosis pathway, led to the removal of the apoptotic tissue. Temporarily delayed, the expression of the ASABF gene encoding the functionally active peptide occurred, a process that resulted in the elimination of the gastropods. The data summarized show that in the sponge system the evolutionary most ancient ASABF peptide is involved not only in the microbicidal defense but also in the elimination of eukaryotic invaders. Both functions contribute to the rich arsenal of the innate immunity in the most basal metazoan phylum.
2. Results and Discussion
2.1. Sponge (S. domuncula, L. baicalensis) ASABF Peptide: Sequence Analysis and Phylogenetic Relationships
By the technique of differential display of transcripts, in total RNA from a field specimen of S. domuncula 32 fragments were identified that were strongly upregulated, compared to the RNA from a specimen kept in the aquarium. After finding by this technique, the corresponding ESTs were also identified in the sponge database [17]. Then the cDNAs were completed. Blast similarity search of the deduced polypeptides revealed that two of those sequences shared highest sequence relationship with the A. suum ASABF-γ peptide from (BAC00498.1; [23]), and that the sequence from S. domuncula was complete; Figure 1A. Therefore, the sponge sequence was termed ASABF peptide (ASABF_SUBDO), deduced from the SDASABF cDNA. The similarity/identity score of the deduced peptide was 36%/27%. The open reading frame (ORF) of the 467 nt long cDNA ranges from nt47–49 to nt296–293(stop) and comprises 83 aa contributing to a theoretical size of 9029 Da and an isoelectric point (pI) of 9.28. The peptide possesses one distinct signal sequence at the N-terminal end from aa1 to aa19 (SignalP 3.0 Server; Technical University of Denmark); Figure 1A. The length of this segment is identical to that predicted for the A. suum ASABF-γ peptide [23]; taking this sequence as reference, the predicted mature region starts at aa20 and ranges to the C-terminus. Hence, the theoretical Mw (molecular weight) is 7026, and the corresponding pI is 9.24. Therefore, the S. domuncula ASABF is a cationic protein, with a pI almost identical to that calculated for the mature ASABF from A. suum with 8.9 [23]. The S. domuncula ASABF sequence shows the characteristic consensus of the nematode CSαβ motif reading C-X(3,18)-C-X(3)-C-X(7,9)-[GS]-X-CX(4-13)-CXC [14]. Like the nematode peptide the mature S. domuncula ASABF sequence includes 8 Cys moieties (aa23, aa30, aa34, aa39, aa44, aa49, aa51, aa54) that allow the formation of disulfide bridges as highlighted in Figure 1A, and as predicted [21]. Secondary structure prediction was performed according to Cole et al. [26]. The characteristic CSαβ (cysteine-stabilized α-helix and β-sheet) structural motif [14] has been predicted according to Chou and Fasman [27]; Figure 1A. The N-terminal α-helix includes the first two Cys residues while the second β-strand comprises the last two Cys residues. No other domain could be identified in the sponge peptide [28].
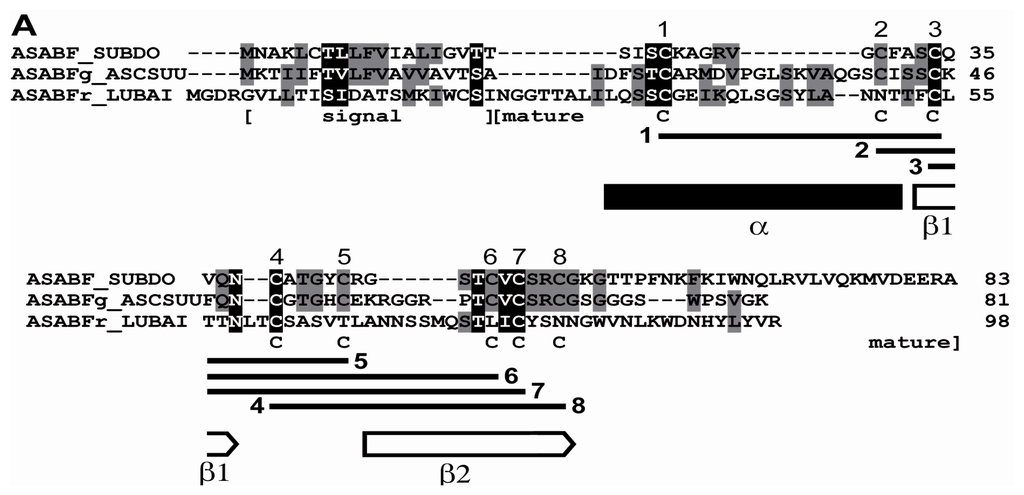
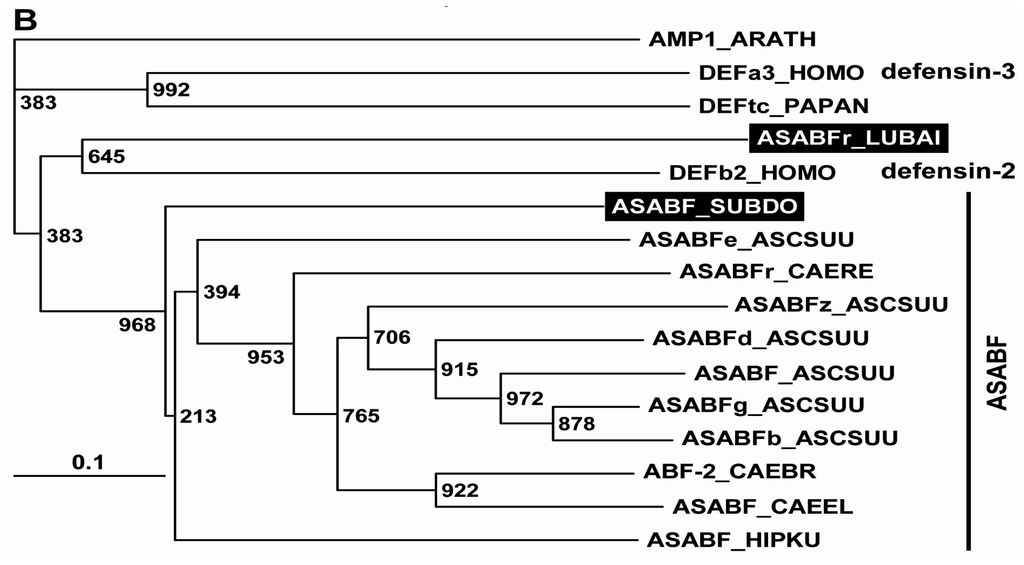
Figure 1.
The S. domuncula ASABF peptide (ASABF_SUBDO), deduced from its cDNA (SDASABF). (A) The deduced protein was aligned with the most related sequence from the database, the ASABF-γ peptide from A. suum (ASABFg_ASCSUU; BAC00498.1) and the distantly similar sequence, the ASABF-related peptide from the Baikalian sponge L. baicalensis (ASABFr_LUBAI). Residues conserved (identical or similar with respect to their physico-chemical properties) in all sequences are shown in white on black; those which share similarity to at least two residues are in black on grey. The borders of the signal peptide and the mature peptide are given. The 8 Cys moieties that are involved in disulfide bridge formation are numbered; the connectivities of the Cys are indicated. The α-helix and the two β-strands are marked with a black solid bar and two open arrows, respectively; (B) These three proteins were compared with the related ASABFs from A. suum: ASABF-beta (ASABFb_ASCSUU; BAC00497.1), ASABF-delta (ASABFd_ASCSUU; BAC00499.1), ASABF-zeta (ASABFz_ASCSUU; BAC57992.1), ASABF-epsilon (ASABFe_ASCSUU; BAC41495.1) and the genuine ASABF (ASABF_ASCSUU; BAA11943.1); the ASABF from C. remanei: the CRE_25171/ASABF protein (ASABFr_CAERE; XP_003113198.1), the ASABF from C. briggsae: CBR-ABF-2 protein (ABF-2_CAEBR; XP_002646276.1), the ASABF from C. elegans: ASABF/abf-2 (ASABF_CAEEL; NP_491252.1), as well as the ASABF-like antimicrobial protein from H. kuda (ASABF_HIPKU; AAX58116.2). In addition, the two human defensins, beta defensin-2 (DEFb2_HOMO; AAC69554.1) and the neutrophil defensin-3 preproprotein (DEFa3_HOMO; NP_005208.1) as well as the theta defensin precursor from P. anubis (DEFtc_PAPAN; NP_001135410.1) were aligned. The plant putative defensin AMP1 protein from A. thaliana (AMP1_ARATH; AAM45086.1) was used as outgroup to root the tree. The scale bar indicates an evolutionary distance of 0.1 aa substitutions per position in the sequence.
The complete cDNA (LBASABFr), encoding the L. baicalensis ASABF-related peptide (ASABFr_LUBAI), has a length of 558 nts and an ORF spanning the segment nt27–29 to nt321–323(stop), predicting a 98 aa moieties long peptide. The theoretical Mw is 10,665 and the pI was predicted with 7.61. Based on comparisons the signal peptide ranges from aa1 to aa22, followed by the mature peptide with an Mw of 8,172 and a theoretical pI of 7.86. This only slightly cationic predicted peptide has only 4 Cys residues.
In order to estimate the phylogenetic relationship of the sponge peptides to other metazoan sequences, for the alignment the highest related A. suum, Caenorhabditis remanei, Caenorhabditis briggsae, and the C. elegans sequences of the ASABF family, as well as the ASABF-related sequence from the seahorse Hippocampus kuda have been chosen [29]; additionally the two human defensins (the beta defensin-2 and the neutrophil alpha defensin-3 preproprotein) and the only metazoan theta defensin (from Papio anubis) were selected. The phylogenetic tree was established by using the plant ASABF-related putative defensin AMP1 protein from Arabidopsis thaliana as outgroup. The construction of the tree revealed that the ASABF sequences cluster together, including the sponge peptide (ASABF from S. domuncula) which forms the basis for that branch (Figure 1B). Separated from them are the human and baboon defensins. Interesting is the finding that the L. baicalensis ASABF-related peptide clusters together with the defensins that comprise likewise less Cys moieties in their mature peptide [23].
2.2. Preparation of Recombinant ASABF
Recombinant protein was prepared in the yeast Pichia pastoris. This eukaryotic expression system allows post-translational modifications such as disulfide bonding, which is required for proper folding of defensins. Accordingly, the complete ORF of SDASABF, without the putative signal sequence (i.e., aa20 to aa83), was expressed as fusion to a 6xHis tag. Following Ni2+-NTA metal-affinity purification (Figure 2Ab), the recombinant protein (8.0 kDa (minus His tag: 7.1 kDa)) was used for immunization of rabbits. Then, specificity of the polyclonal anti-ASABF serum (PoAb-rASABFSUBDO) was assessed on Western blots (Figure 2Ba), showing a band at the expected size of the recombinant protein. In control experiments, 100 μL of the antibodies were adsorbed to 20 μg of the recombinant protein prior to their use. Consequently, no band could be observed on the immunoblots after application of these blocked antibodies (Figure 2Bb).
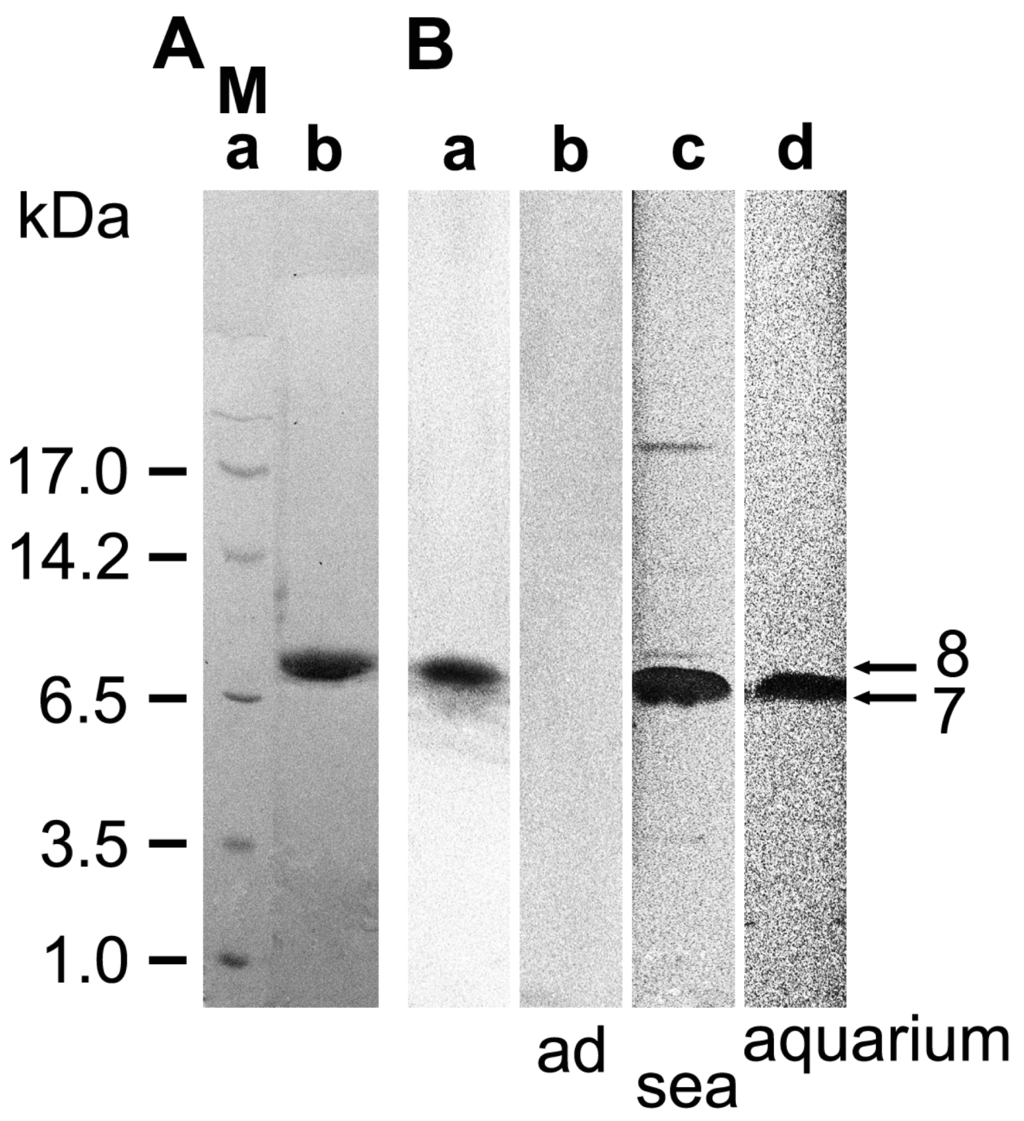
Figure 2.
Analysis of recombinant and naturally occurring ASABF by SDS-PAGE. (A) P. pastoris cells were transformed with the ASABF cDNA (SDASABF), as described under “Experimental Section”. After purification, the recombinant protein was analyzed by SDS-PAGE. (a) Size markers (M); (b) Purified recombinant ASABF. After size separation the gel was stained by Coomassie brilliant blue; (B) (a and b) Western blot analysis of recombinant ASABF. The blot-transferred filters were incubated with (a) untreated PoAb-rASABFSUBDO, or (b) with PoAb-rASABFSUBDO adsorbed to rASABF_SUBDO; (c and d) Protein extracts from animals (c) sampled from the sea, or (d) kept for over 1 year in the aquarium were used for gel electrophoresis; 5 μg of protein were applied per slot.
In a parallel set of experiments, the PoAb was applied to detect S. domuncula ASABF in protein extracts of sponge specimens that had been sampled from the sea (Figure 2Bc), or that had been cultured in aquaria for 12 months (Figure 2Bd). Thus, following SDS-PAGE and immunoblotting of identical protein concentrations, a band was detected with a size corresponding to the processed, mature variant of ASABF, lacking the signal sequence (i.e., 7.0 kDa). The band intensity was considerably stronger in extracts of field specimens than in those of aquarium specimens.
2.3. Microbicidal Assay
The recombinant ASABF caused strong anti-microbial activity in the in vitro assay system (Table 1). Especially strong was the effect towards Gram-positive bacteria (S. aureus, B. subtilis, and M. luteus) with minimum inhibitory concentrations (MIC) between 2 and 5 μg/mL, while the activity against Gram-negative bacteria (P. aeruginosa and E. coli) was considerably lower with concentrations between 12 and 17 μg/mL. Interestingly, ASABF also caused distinct inhibitory effects on growth of eukaryotic fungi (C. albicans and A. niger) with MIC concentrations between 9 and 13 μg/mL. In order to prove that the effect measured was specific, the microorganisms were exposed to recombinant ASABF that had been adsorbed with antibodies prior to the addition to the cultures; such samples were found to be inhibitory only at MIC above 20 μg/mL.

Table 1.
Antimicrobial activity of non-adsorbed or antibody-adsorbed recombinant ASABF against Gram-positive bacteria, Gram-negative bacteria and two fungi. The inhibition is given as minimum inhibitory concentrations (MIC) value in μg/mL.
2.4. Hemolytic Activity
As a convenient assay to study hemolysis human erythrocytes have been used and exposed to recombinant ASABF. The cells were exposed to the peptide and the percentage of the lysed erythrocytes was determined on the basis of the released hemoglobin. Under the assay conditions described under “Experimental Section” it was seen that the recombinant, non-adsorbed ASABF caused at a concentration of about 1 μg/mL 35% hemolysis. The degree of lysis increased to 76% at a concentration of 10 μg/mL (Figure 3). As a control, the ASABF peptide was adsorbed to anti-ASABF serum (PoAb-rASABFSUBDO), as described under “Experimental Section”. Using this ASABF sample the degree of lysis was very low and amounted to about 10% at the high concentration of 10 μg/mL.
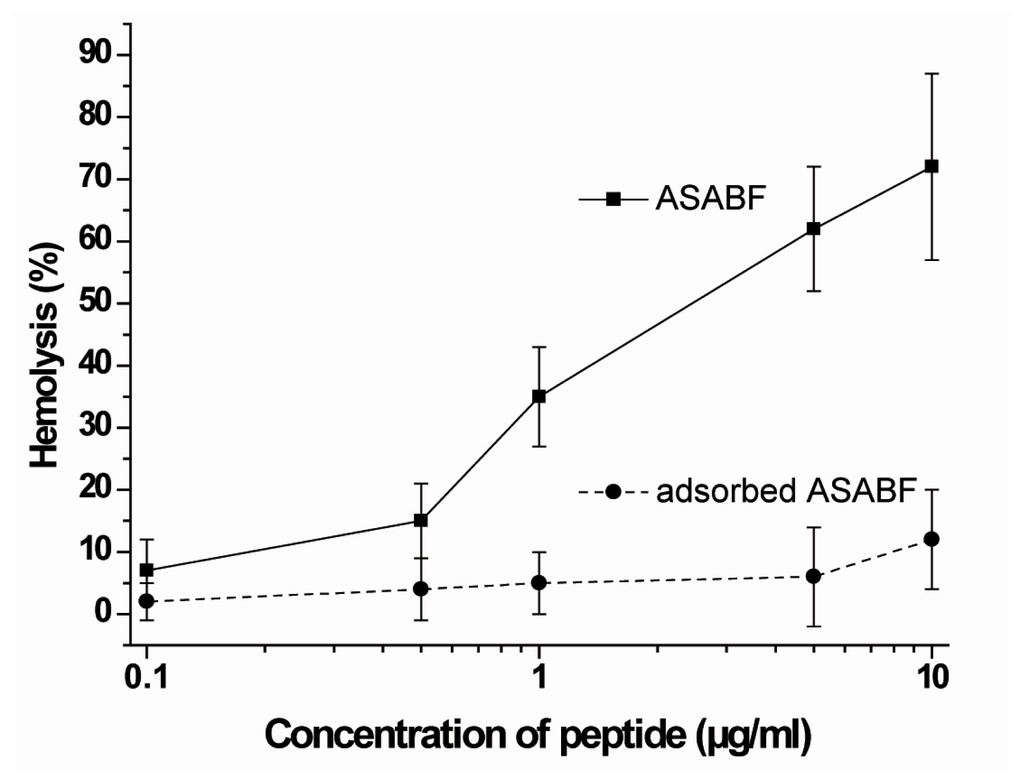
Figure 3.
Hemolytic activity of ASABF, assayed in the human erythrocyte lysis assay. Red blood cells were incubated for 1 h (37 °C) at increasing concentrations of recombinant ASABF (—▪—). In a parallel series of experiments the recombinant ASABF was adsorbed to anti-ASABF antiserum (PoAb-rASABFSUBDO) (- - • - -). Subsequently, the lysed erythrocytes were determined on the basis of the released hemoglobin. The values represent the means ± SEM of ten experiments each.
2.5. Induction of Apoptosis in S. domuncula after Exposure of the Animals to Dip
If sponge specimens were kept in aquaria at lower aeration rate or in the presence of Fe++ chelator Dip the animals started to develop apoptotic spots/areas on their surface, as described [24]. Those regions were initially (after 1 day) marked by a deeper-red color. At a later stage the apoptotic surface regions became unstructured to meander-like. Previously, those regions had been identified to undergo apoptosis, as had been proven by the “Cell Death Detection ELISA test system” [24]. At day 2 these apoptotic regions were colonized by the gastropod Bittium sp. that had been attracted by quinolinic acid, a metabolite that is formed after induction of the 3-hydroxyanthranilate 3,4-dioxygenase (HAD) gene [24]. At days 2 to 3 the specimens of Bittium sp. (1 to 6 per 5 mm2) grazed on the apoptotic areas (Figure 4A); no gastropods were seen on non-apoptotic surfaces (not shown). As observed before, the gastropods ate (Figure 4B,C) and, by that, removed the apoptotic tissue (Figure 4D). After day 7 the previously apoptotic regions had been removed.
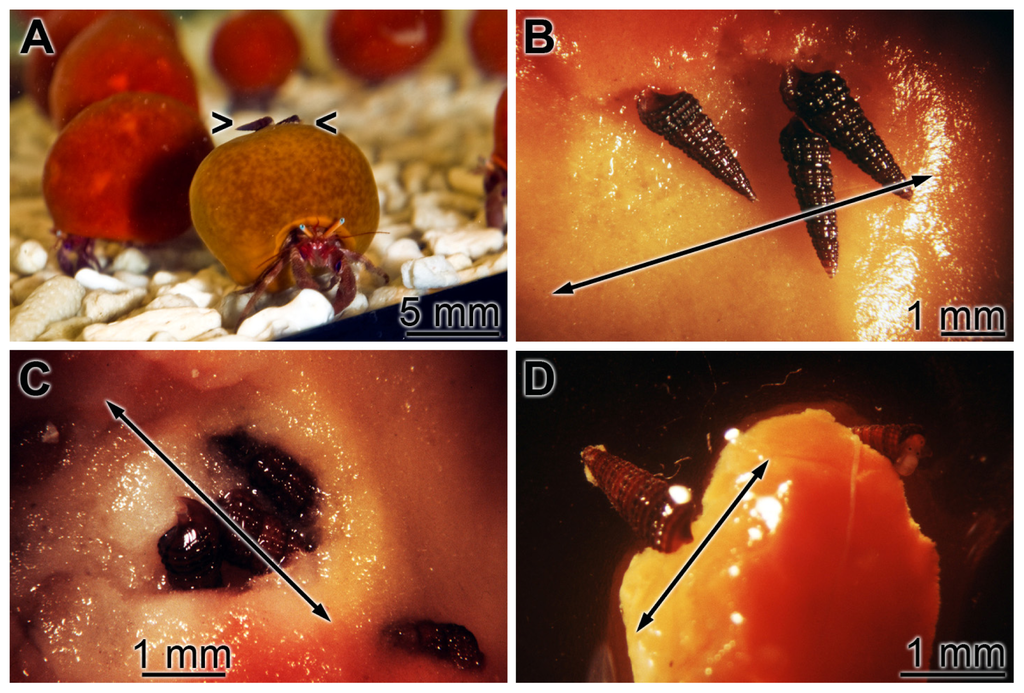
Figure 4.
Colonialization of S. domuncula specimens by the gastropod Bittium sp. Specimens were kept in seawater in the presence of the chelator Dip. After 1 to 2 days in the aquarium the surfaces of the specimens developed speckled-type darker-colored areas which became apoptotic. (A) In an earlier stage one and soon after up to six gastropods colonized those apoptotic areas (> <); (B and C) Grazing gastropods on apoptotic areas (double-headed arrows); resulting in (D) a removal of those apoptotic areas (double-headed arrow).
2.6. Expression of the ASABF Gene in S. domuncula Specimens: in Situ Hybridization
Samples from a control specimen and from an animal kept for 1 day in Dip were analyzed by in situ hybridization. Using the DIG-labeled anti-sense ssDNA (SDASABF) probe the extent of reaction to the tissue structures was low in control sections (Figure 5A,B). In contrast, the reaction of the labeled probe with tissue slices from Dip-exposed animals was intense (Figure 5D,E); at a higher magnification the reaction with cell structures was evident (Figure 5F). One control was performed by hybridizing a sample from the Dip-treated animal with a labeled sense ssDNA probe; there, only background signals were recorded (Figure 5C).
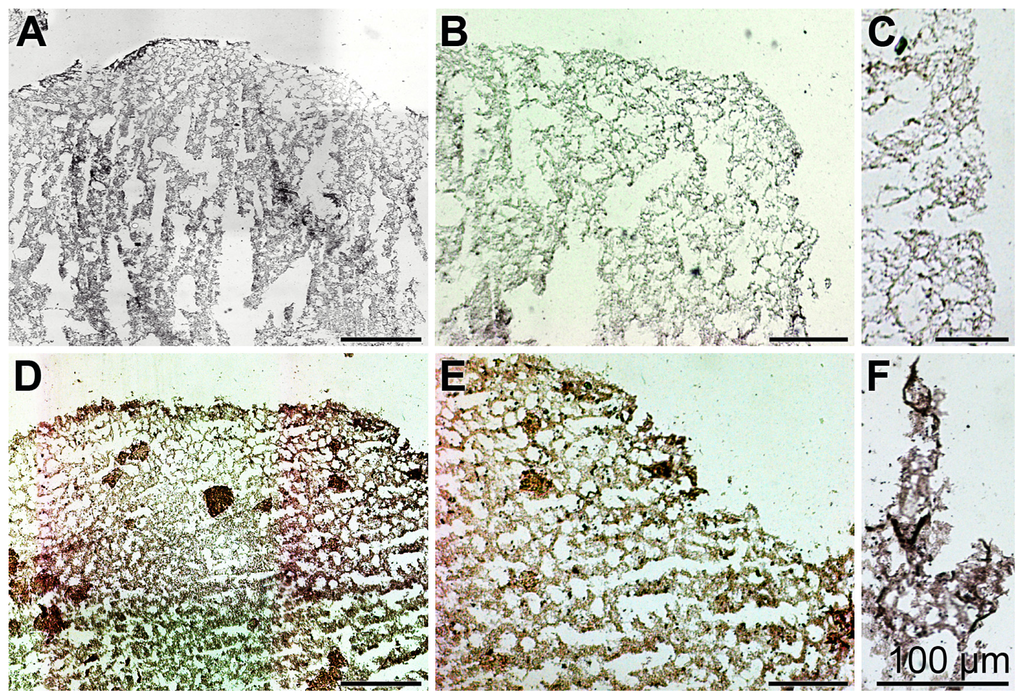
Figure 5.
Expression studies of ASABF gene by application of the in situ hybridization technique. (A,B) Control sponge tissue of non-treated sponge specimens or (C–F) from animals kept for 1 day in the presence of the chelator Dip and subsequently for an additional 1 day in the absence of the chemical, was sliced and the sections were analyzed for the expression of the ASABF gene using the SDASABF DIG-labeled probe. In (A,B,D–F) hybridization was performed with the anti-sense ssDNA probe; (C) had been hybridized with the labeled sense probe. All size bars represent 100 μm.
2.7. Expressions of the HAD, the ASABF as well as the Caspase Genes: Determination by qRT-PCR
Tissue samples from apoptotic areas on the surface of Dip-treated animals, as well as from control animals were taken and subjected to qRT-PCR analysis. For a quantitative assessment steady-state gene expressions of HAD (marker for quinolinic acid synthesis), the caspase (formation of apoptotic tissue [30]) as well as of ASABF (ASABF synthesis) were chosen. The tissue samples for the determination of the respective transcript levels in apoptotic areas were taken from the rim region of those regions. The determinations were performed at the beginning of the exposure (day 0), as well as after 1 to 9 days. The results showed that the expression level of HAD at the beginning of the experiments was approximately 2.1 × 10−3 with respect to the expression of the house-keeping gene tubulin (Table 2). While in the controls the expression level did not significantly change during the 9-day incubation period, a strong increase in the steady-state expression of HAD was measured in the Dip-treated animals. The maximum of expression was seen already at day 2 (27.1 × 10−3); during the end of the experiment (after 9 days) the expression level returned to normal (3.1 × 10−3). Focusing on ASABF expression, likewise no change of the steady-state expression was observed in the controls (around 1.3 × 10−2). In the Dip-treated specimens a strong and significant increase was found already after day 1 (2.8 × 10−2), while the maximum was seen 4 days after exposure to Dip (9.8 × 10−2). Subsequently, the level of expression decreased. Finally, the expression of the caspase gene likewise increased in the Dip-treated animals from 1.5 × 10−4 (day 0) to 12.4 × 10−4 at day 3 (Table 2). In the control tissue no significant changes were measured. A diagrammatic sketch, summarizing the expression levels in apoptotic tissue from the surface of Dip-treated specimens, as well as from controls, is given in Figure 6. There, also the surface texture of a 3-day-old apoptotic area with a grazing gastropod is shown with its uneven appearance in contrast to the smooth/plain surface in the control.

Table 2.
Expression levels of S. domuncula HAD, ASABF as well as of the caspase genes on the surfaces of control animals, not treated with the Fe++ chelator, or treated with Dip as described under “Experimental Section”. Tissue samples from the Dip-treated animals were taken from the rim of apoptotic region, or after day 5, from the area which previously had been apoptotic and was then cleaned by the gastropod. Then, RNA was extracted and expression levels were quantified through qRT-PCR. Each data point represents the mRNA level of the respective expressed gene (x: HAD, ASABF or caspase) normalized to the amount of β-tubulin transcripts, as means ± SEM (n = five experiments per time point).
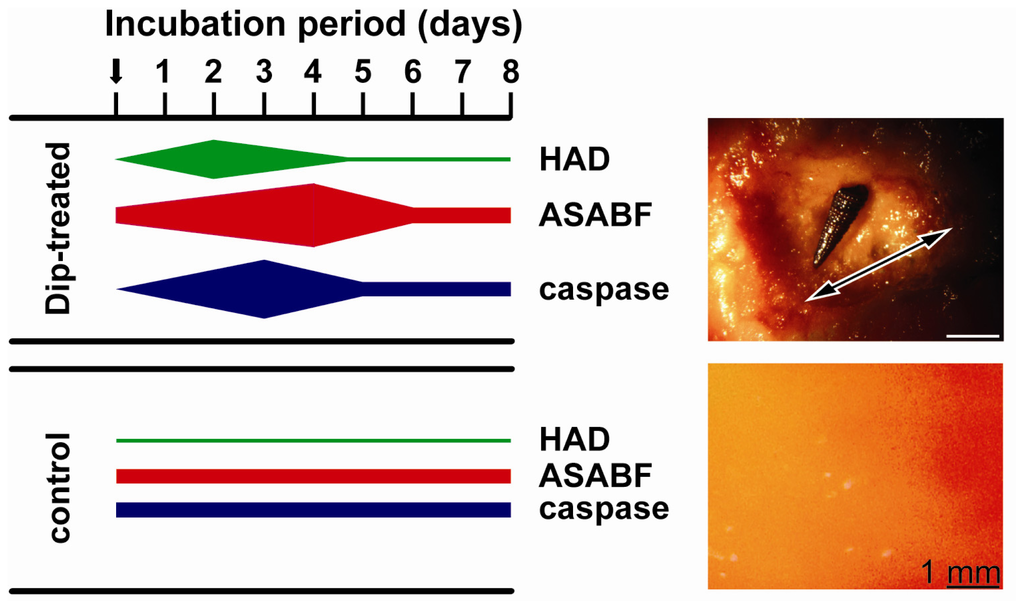
Figure 6.
Diagrammatic sketch of the expression levels of HAD, ASABF as well as of the caspase gene in Dip-treated and in control specimens (left). The widths of the bars symbolize the level of expression. At the upper right an image of the surface of a 3-day-old apoptotic area (double-headed arrow) with a gastropod is shown; lower right shows the plain surface of a non-treated specimen.
For the expression studies in the above described series of experiments tissue from the rim regions had been selected. To obtain a more detailed view of the expression levels of HAD, ASABF and caspase tissue samples were taken from within the apoptotic area. Such a spatially resolved study was only possible after a more progressed stage of apoptosis, 3 days after Dip exposure. In comparison, tissue samples from apoptotic regions after their ablation through the gastropods were analyzed (Figure 7). The data revealed that at day 3 the increase in the steady-state level of expression of the HAD gene and the ASABF gene was 4.8-fold and 5.9-fold, respectively, while the increase of the expression level for the caspase gene was much higher with 12.2-fold.
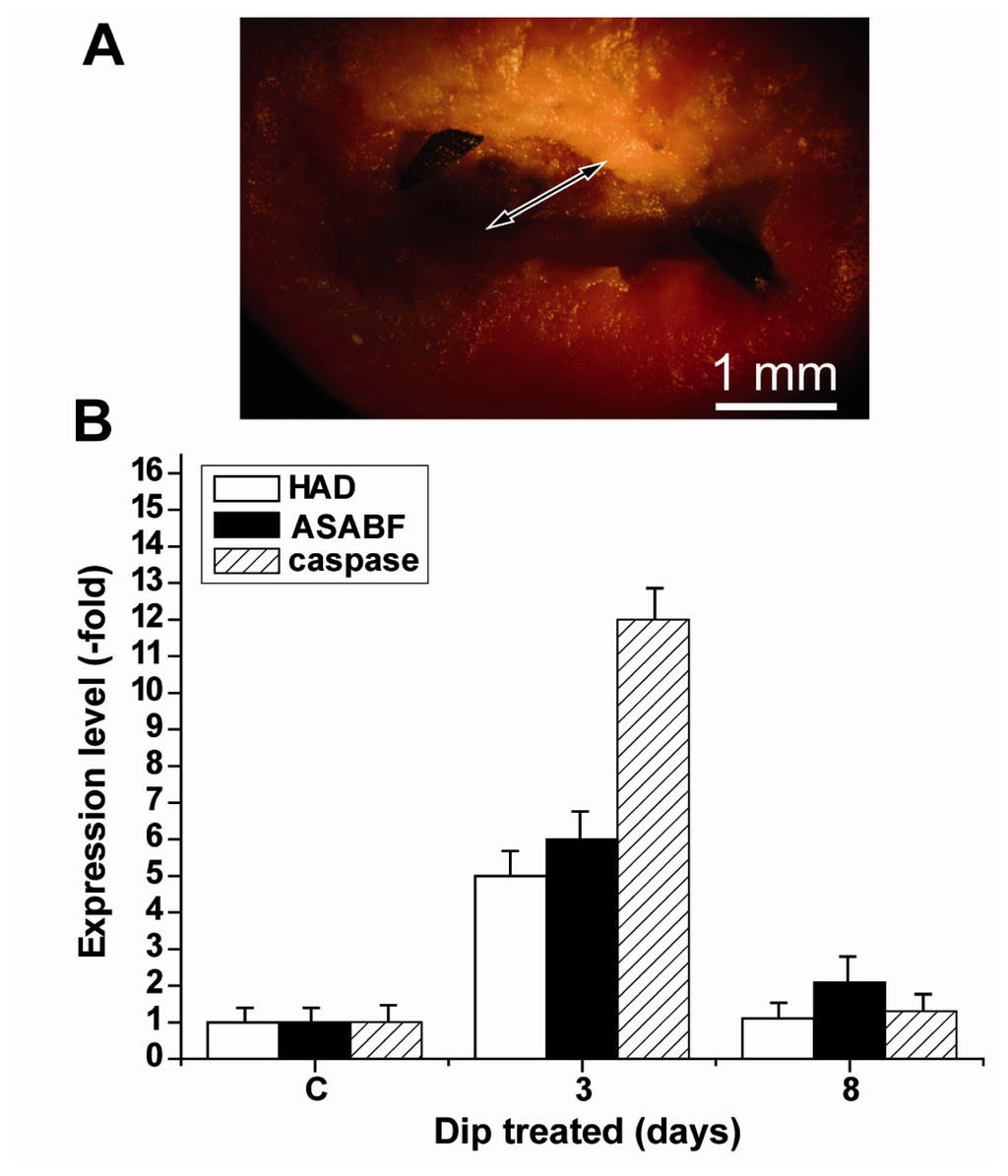
Figure 7.
Spatially resolved analysis of the expression level of HAD, ASABF as well as of the caspase gene. Tissue samples from controls as well as samples from within the apoptotic areas (the diameter of the area is given by a double-headed arrow) of Dip-treated specimens were analyzed. The expression is given in arbitrary units as multiple-fold of the transcript levels seen in the controls (non-treated animals; day 0). The analyses were performed by qRT-PCR 3 days and 8 days after exposure of the specimens to Dip using the expression of the tubulin gene as a reference house-keeping gene; the controls remained untreated. The means ± SEM (n = 5) are given.
2.8. Acute/Subchronic Toxicity Testing of the Gastropod
The above outlined results indicate that the drop of the HAD expression coincided (after about 4 days after exposure to Dip) with the maximum of the expression of the ASABF gene, encoding the antimicrobial peptide (Figure 6). In addition, it is seen that after about 5 days the gastropods disappeared from the previously apoptotic areas. The reason for the elimination of the gastropods is not quinolinic acid, since this dicarboxylic acid is not toxic for those epibionts. Therefore, it was intriguing to check if it was the ASABF peptide which may have caused a toxic effect. Accordingly it was necessary to test if the recombinant peptide affected the viability of the gastropods.
As outlined in the assay conditions described under “Experimental Section”, the gastropods were exposed for 96 h to ASABF in a serial concentration range. After incubation the mortality in the assays with ASABF were determined to be; at 3 μg/mL: 3.5 ± 2%, 10 μg/mL: 26.4 ± 8% and 30 μg/mL: 75.7 ± 12%. In the controls (untreated animals) 100% of the animals survived. According to the standards the toxicity of the ASABF towards the gastropods should be assessed as “highly toxic” (US EPA 2011); Figure 8.
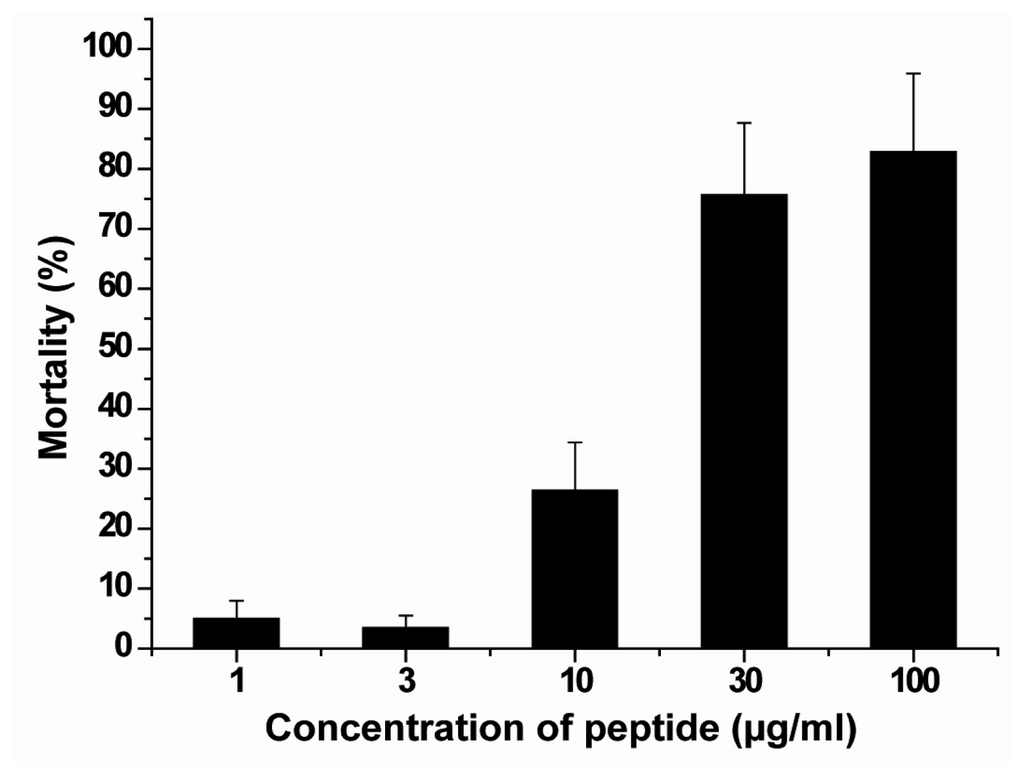
Figure 8.
Acute/subchronic toxicity of recombinant ASABF peptide on the gastropod Bittium sp. The gastropods were incubated for 4 days with the recombinant peptide. Subsequently cultivation of the animals continued for 4 days in seawater without the peptide; the mortality was determined as described under “Experimental Section”.
3. Discussion
Antimicrobial agents that usually represent peptides with a length of less than 50 amino acid residues are potent effector molecules of the innate immune system. They interact with both microbial and eukaryotic cell membranes and, in turn, cause cell lysis. Besides direct interactions with biomembranes, they function in vertebrates also as mediators of inflammation, cause cytokine release, affect cell proliferation, and interfere with angiogenesis, wound healing, or chemotaxis [31,32]. Most of those peptides are classified to the group of cationic peptides [33], comprising characteristic domains, e.g., the cysteine-rich CSαβ-type motif [14]. Already 20 years ago it had been proposed that those “antimicrobial peptides” act not only on cell membranes but also affect intracellular targets, as has been shown e.g., for the small tachyplesin peptides from horseshoe crabs [34]. The remarkable feature of those antimicrobial peptides is their property to act in a broad spectrum against Gram-positive and Gram-negative bacteria as well as fungi [35].
Even though it is known that sponges are rich sources of bioactive compounds of low molecular weight [3], no screening for genes encoding antimicrobial peptides from the sponges themselves has yet been reported. Unlike the antibiotic peptides, synthesized by the sponge symbionts and encoded by them, e.g., by synthases [36], the amino acid sequences of the antimicrobial peptides are encoded by the host organism, the sponge itself. Hence, such antimicrobial peptides can be considered to be components of the metazoan innate or natural immune system which protects them against prokaryotic and eukaryotic invading organisms. The present contribution reports on a gene from S. domuncula, encoding an ASABF-type antimicrobial peptide that acts against microorganisms as well as against eukaryotic cells. The S. domuncula ASABF is a cationic peptide that comprises eight cysteines allowing the formation of four intramolecular disulfide bridges. From the ORF of the full-length cDNA sequence an 83-residue precursor peptide can be predicted. From this polypeptide a signal peptide of 19 amino acids in length is cleaved off, converting the precursor into a 64 aa long, mature peptide. The four disulfide bridges follow the prediction based on the nematode ASABFs [21]. The sequence also shares the CSαβ motif with the nematode ASABFs. The sponge polypeptide is, like the other ASABFs, related to the defensins which comprise 3–4 intramolecular disulfide bridges [22]. The phylogenetic analysis likewise indicated a separation of the ASABFs, including the sponge sequence, from the defensins, regardless of whether they come from vertebrates or invertebrates. Like the ASABFs the defensins are key elements of the immune system and provide a broad-range defense against eukaryotic and protozoan pathogens [16]. It is important to mention that in metazoans the defensins are expressed cell-type/tissue specifically and often show modulatory effects on immune cells, e.g., macrophages, lymphocytes or mast cells [37]. Even more, an interference of the defensins with host cell recognition proteins has been described [38]. These findings on the sequence and function polymorphism of the ASABFs/defensins underscore their important role in the innate immune system of metazoans.
The sponge ASABF has been successfully expressed in the P. pastoris system and the biologically active peptide was obtained. The recombinant peptide turned out to be highly active against Gram-positive bacteria with a MIC concentration between 2 and 5 μg/mL (0.3 to 0.7 μM), and is, hence, similarly active as the antimicrobial nematode ASABF peptides, abf-1 and abf-2 [19]. Less effective is the sponge peptide against Gram-negative bacteria with >12 μg/mL. In addition, microbicidal activity was seen in eukaryotic fungal systems (C. albicans and A. niger) with an inhibitory activity at 9–13 μg/mL. Considering the screening range of Zhang et al. [39] we also tested the sponge peptide in the human hemolysis system with erythrocytes. Interestingly, even at the low concentration of 1 μg/mL (0.15 μM) the sponge peptide caused a significant hemolytic activity; about 30–40% of the erythrocytes were found to be lysed at that concentration. This efficiency is considerably higher than that known for the nematode A. suum ASABFs [39], which varies around a MIC of 10–30 μg/mL.
The potent hemolytic activity prompted us to determine the effect of the recombinant sponge ASABF on the gastropod Bittium sp. an eukaryotic epibiont that occasionally grazes on S. domuncula. Consequently we had to answer two questions. First, the determination of the gene expression status of ASABF in a control region and in the areas where the gastropod settled, and second, of the potential toxic effect of the recombinant ASABF peptide on the gastropod directly. Previously it had been shown that this gastropod settles on the surface of S. domuncula after induction of apoptosis in response to a lower oxygen partial pressure [24]. In this previous report apoptosis was measured with the “Cell Death Detection ELISA test system” [40] that bases on the quantification of the degree of chromatin fragmentation to mono- and oligonucleosomes. During the process of apoptosis the expression of the gene for the key enzyme of the quinolinic acid pathway is induced, the 3-hydroxyanthranilate 3,4-dioxygenase (HAD) that converts 3-hydroxyanthranilate to 2-amino-3-carboxymuconate 6-semialdehyde under consumption of oxygen. Since the latter metabolite is not stable, it non-enzymatically converts to quinolinic acid [41]. This dicarboxylic acid is not only in sponges [24] but also in human cells a selective inducer of apoptosis [42]. In sponges the initiating enzyme, HAD, is rapidly induced during 2 days after impairing the environment of the animals. After formation of quinolinic acid in the apoptotic tissue, the gastropod Bittium sp. colonizes that region, very likely attracted by the dicarboxylic acid. Subsequently, as implicated [24], the gastropods ablate the apoptotic tissue during a period of 2–3 days. In parallel with the induction of HAD, a significant upregulation of the steady-state expression of caspase occurs. Increased transcript levels of caspase in apoptotic tissue are also known from earlier studies [30]. After a lag period of 2 days after the induction of HAD, the gene encoding the ASABF peptide is induced in the tissue, surrounding the apoptotic area. The expression of ASABF in apoptotic tissue is low, almost not existing, while it is reasonably expressed in control, non-apoptotic tissue (data not shown). However, if samples are taken from regions bordering the apoptotic area that has been eliminated by the gastropods, a strong upregulation is seen. The induction of the gene for the ASABF peptide has been described previously in the nematode A. suum, after exposure of the worm to heat-killed bacteria [23].
The toxicity of the ASABF peptide on the gastropod Bittium sp. has been determined under controlled, closed conditions. Our toxicity studies revealed that ASABF is already lethal for the animals at a concentration of the recombinant peptide as low as 10–30 μg/mL during an incubation period of 4 days. This first description of an antimicrobial peptide from sponges which is toxic for animal cells and is likely used as a defense against attacking predators, widens the view of the activity spectra of the “antimicrobial” peptides. Consequently, the sponge ASABF, a member of the innate immune system, most likely acts as a broad-spectrum defense molecule. The perhaps ubiquitous presence of antimicrobial peptides of the ASABF type in sponges is supported by the identification of the ASABF-related gene in the freshwater sponge L. baicalensis.
4. Experimental Section
4.1. Chemicals, Materials and Enzymes
The sources of the materials used for this study were given previously [43]. In addition, Dip (2,2′-dipyridyl) was purchased from Sigma/Aldrich (St. Louis, MO; USA), and PCR DIG (digoxigenin) Probe Synthesis Kit, anti-DIG AP Fab fragments, as well as disodium 2-chloro-5-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)-1-phenyl phosphate (CDP-Star) were obtained from Roche (Mannheim; Germany).
4.2. Sponges and Gastropod/Mollusc Colonization
Specimens of the marine sponge S. domuncula (Porifera, Demospongiae, Hadromerida) were sampled in the Northern Adriatic near Rovinj (Croatia), and then kept in aquaria in Mainz (Germany) at a temperature of 17 °C for more than 12 months. Extracts were prepared from tissue samples by treatment with four parts of a 0.4 M Tris-HCl buffer (pH 8.0; supplemented with 50 mM NaCl, 0.5 mM EDTA and 5 mM MgCl2). After grinding, and stirring of the suspension at 2 °C for 1 h a clear supernatant was obtained after centrifugation (20,000 × g, 30 min, 2 °C); this was collected and contained between 3 and 7 mg of protein per mL. This extract was diluted down to 2 mg/mL and used for the experiments.
The experiments were performed in 5 L (artificial sea water [24]) aquaria, containing 3 sponges each. Apoptosis was induced in sponges by keeping the specimens in the presence of 50 μg/mL of the Fe++ chelator Dip [44] for 1 day. Under these conditions the specimens responded with the formation of speckled patches on their surfaces, as described earlier [24]. By application of the “Cell Death Detection ELISA test system” (Roche) apoptosis was proven in these regions [24]. To each aquarium, both controls and Dip (2,2′-dipyridyl) supplemented ones, approximately 100 specimens of the gastropod/mollusc Bittium sp. (Mollusca, Gastropoda, Cerithiidae, Bittium) were added. Already 1 day later the apoptotic patches on the sponge surfaces were covered by 1 to 6 specimens of Bittium sp. per 5 mm2 (Dip-treated), while on the surfaces of specimens kept in absence of Dip (controls) no gastropods were seen. As described before, the gastropods ablated the apoptotic tissue within 3 days. Seven days after the beginning of the experiments no signs of apoptotic tissue could be seen. Tissue samples were taken at the beginning of the experiments as well as after 1 day, 2 days, 3 days, 4 days, 5 days, 7/8 days, and 9 days from Dip-treated (speckled-type regions) and from control regions, and analyzed for gene expression.
4.3. Maintenance of the Gastropod Bittium sp
Bittium sp. (Gastropoda: Cerithiidae) are common grazers, epizoons [45], abundantly seen on the surfaces of S. domuncula apoptotic specimens. The gastropods were collected, transferred to nets (mesh size 44 μm) and could be kept in aquaria, for over 30 days, without losing more than 10% of the specimens. Routinely, the gastropods were used for the experiments 6 days after collection from the apoptotic specimens.
4.4. Identification and Cloning of the Poriferan ASABF Peptides
S. domuncula ASABF: For the identification of the cDNAs encoding putative defensin-like molecules the technique of differential display was employed according to Müller et al. [4]. Total RNA was extracted from S. domuncula specimens that had been sampled from the sea or had been cultivated in aquaria for more than one year. Specimens that had been taken from nature showed a higher load of bacteria and, in turn, were more likely to express those polypeptides in a wider array of antibacterial defense molecules and at higher expression levels than aquarium animals [46]. The cloning procedure, given in details before [4] is outlined in brief: 1.8 μg of total RNA of aquarium animals or of field specimens was reverse transcribed using the anchored oligo(dT) primer T11CC and SuperScript III Reverse Transcriptase (Invitrogen, Darmstadt; Germany). Following first-strand cDNA synthesis, the samples were diluted 1:10 with H2O and, then, subjected to polymerase chain reaction (PCR). For this reaction, Platinum Taq DNA polymerase (Invitrogen) was employed in combination with the IR-labeled arbitrary primer 5′-GTGATCGCAG-3′ and an oligo(dT) primer. After initial denaturation at 94 °C for 2 min, cDNA was amplified during 38 cycles each, at 94 °C for 30 s, 42 °C for 120 s, and 72 °C for 30 s, followed by a final incubation of 10 min at 72 °C. Subsequently, amplicons were size-separated during polyacrylamide gel electrophoresis, using an automated DNA sequencer (LI-COR 4300 DNA Analysis System). Then, cDNAs between 250 and 400 nucleotides (nt) of differentially expressed transcripts were excised using the Odyssey (LI-COR) infrared imaging system and subsequently reamplified by PCR. Finally, the products were sub-cloned in the pGEM-T vector (Promega) and sequenced. Complete cDNAs were obtained by primer walking [47]. Thus, the clone SDASABF was identified (among 32 differentially expressed transcripts found within field animals), comprising 467 nts (excluding the poly(A) tail) with an ORF encoding the putative protein ASABF_SUBDO.
L. baicalensis ASABF: Following the identification of S. domuncula ASABF, >2500 L. baicalensis ESTs [28] were screened for related sequences. One clone was discovered, comprising 558 nts with considerable sequence homology to SDASABF, which was termed LBASABFr. The ORF was found to be complete.
4.5. Sequence Analyses
Homology searches were conducted through servers at the European Bioinformatics Institute, Hinxton, United Kingdom and the National Center for Biotechnology Information (NCBI), Bethesda, MD [29]. Multiple alignments were carried out with ClustalW version 1.6 [48]. Phylogenetic trees were constructed on the basis of aa sequence alignments applying the Neighbor-Joining method to distance matrices that were calculated using the Dayhoff PAM matrix model [49,50]. The degree of support for internal branches was further assessed by bootstrapping [51]. The graphical output of the bootstrap figures was produced through the “Treeview” software [52] and GeneDoc [53]. Potential domains, subunits, patterns, and transmembrane regions were predicted searching the Pfam [54] or the SMART database [55].
4.6. Preparation of Recombinant S. domuncula ASABF Peptide
cDNA of S. domuncula ASABF, coding for the complete mature protein (aa20 to aa83), was cloned as gene of interest into the pPIC3.5 yeast expression vector (Invitrogen) via SnaB I and EcoR I. At the 5′ terminus, an ATG initiation codon was placed within the context of a yeast translation initiation sequence ([56] and according to the manufacturer’s protocol). Additionally, the cDNA contained a 3′ 6xhistidine (His) tag coding sequence (to facilitate metal-affinity chromatography purification of the recombinant protein), followed by a stop codon. After digestion with Sal I, chemically competent Pichia pastoris GS115 cells were transformed with the linearized construct. Subsequently, transformants were selected for their ability to grow on histidine-deficient medium. For recombinant protein expression, transformants were cultured for 84–96 h at 30 °C in appropriate medium with 0.5% (v/v) methanol. Then, cells were harvested, transferred into Breaking Buffer (50 mM sodium phosphate pH 7.4, 1 mM PMSF (phenylmethylsulfonyl fluoride), 1 mM EDTA, 5% glycerol), and homogenized (Precellys; PeqLab, Erlangen; Germany). Finally, the recombinant protein (termed rASABF_SUBDO) was purified through nickel-nitrilotriacetic acid (Ni2+-NTA) metal-affinity chromatography and checked through SDS-PAGE.
4.7. Preparation of Antibodies
The purified, recombinant ASABF protein (rASABF_SUBDO) was used for the production of polyclonal antibodies (PoAb), as described [8,57]. Thus, female rabbits (White New Zealand) were immunized thrice with 20 μg/boost. Following collection of the PoAb (termed PoAb-rASABFSUBDO) the titer was determined to be 1:5,000. For control experiments, 100 μL of PoAb were incubated with 20 μg of the antigen during an incubation period of 1 h (4 °C). Only then (adsorbed antibody preparation) these blocked antibodies were applied to immunodetection assays.
4.8. SDS-PAGE and Immunodetection Analysis
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was routinely performed as follows. Samples of 6 μg of protein were mixed with loading buffer (Roti-Load; Carl Roth, Karlsruhe; Germany), boiled for 8 min, and subjected to SDS-PAGE (15% acrylamide and 0.1% SDS) as described [58]. The gels were stained with Coomassie brilliant blue. The protein size standard “Dual Color” (Carl Roth) was used to estimate protein sizes. For Western blot analyses, size-separated proteins were transferred to PVDF-Immobilon membranes. Then, the membranes were blocked at room temperature with blocking solution (Roche; 1% (v/v) in TBS-T buffer; 20 mM Tris-HCl pH 7.6, 137 mM NaCl, 0.1% (v/v) Tween-20). Following consecutive incubations with PoAb-rASABFSUBDO (1:3,000 dilution in TBS-T, supplemented with 0.1% (v/v) Blocking solution), alkaline phosphatase (AP)-conjugated species-specific secondary antibodies (1:4000 dilution), and 4-nitro blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Invitrogen), proteins were detected colorimetrically. The “Ultra-Low Range Marker” (Sigma; M3546) was used to determine the apparent size of the peptide.
4.9. Quantitative Real-Time RT-PCR (qRT-PCR)
Quantitative real-time RT-PCR (qRT-PCR) was applied to determine the steady-state expression of the gene encoding ASABF (SDASABF), as recently described in detail [59,60]. Following extraction of tissue RNA was treated with DNAse to eliminate contaminating genomic DNA. First strand cDNA synthesis was performed by SuperScript III Reverse Transcriptase in a reaction mixture containing 5 μg of total RNA, dNTPs, oligo(dT)18, and reverse transcriptase buffer (Invitrogen) at 42 °C for 1 h. After enzyme inactivation (65 °C, 15 min), qPCR experiments were performed in an iCycler (Bio-Rad), using 1/10 serial dilutions in triplicate as described [61]. After appropriate dilution, 2 μL of the reaction mixture were employed as a template for 30 μL qPCR assays. Each reaction contained “Absolute Blue SYBR Green” master mixture (ABgene, Hamburg; Germany) and 5 pmol of each primer. All reactions were run with an initial denaturation at 95 °C for 3 min, followed by 40 cycles each, of 95 °C for 15 s, 62 °C for 30 s, 72 °C for 35 s, and 80 °C for 20 s. The following primers were used for amplification of SDASABF: Fwd: 5′-GCAAAGCTGGCAGAGTTGGAT-3′ (nt114 to nt134) and Rev: 5′-AGATTTTGAACTTGTTGAAAGG-3′ (nt245 to nt224); the size of the fragment obtained was 132 bp. For the amplification of the S. domuncula caspase gene SDCASPL (NCBI accession number AM392430.1; [8]) Fwd: 5′-GGACAAACAGATGTACACAGAG-3′ (nt597 to nt618) and Rev: 5′-GATGTAGTTTCCTTCTGGAGGT-3′ (nt732 to nt711) were applied, resulting in a fragment of 136 bp. Finally, the expression level of the S. domuncula 3-hydroxyanthranilate 3,4-dioxygenase (HAD; AJ298053.1) was quantified using the primer pair Fwd: 5′-CTCAGGTACTATGTTGATGGTA-3′ (nt369 to nt390) and Rev: 5′-TGGACTTCCATTCTTGTGTTCC-3′ (nt300 to nt279); size of the fragment, 132 bp. The expression of the house-keeping gene β-tubulin (AJ550806; [62]) was determined with the following primers, Fwd: 5′-AACCGCTGTTTGCGACATCC-3′ (nt1,126 to nt1,145) and Rev: 5′-CAATGCAAGAAAGCCTTTCGCC-3′ (nt1266 to nt1245); fragment size, 141 bp. The threshold position was set to 50.0 RFU above PCR subtracted baseline for all runs. Mean Ct values and efficiencies were calculated by the iCycler software. Expression levels of the respective transcripts were correlated with the one for β-tubulin to assess the relative expression levels (ETub CtTub/EASABF CtASABF; ETub CtTub/ECasp CtCasp; ETub CtTub/EHAD CtHAD); whereby “E” describes the PCR efficiency and “Ct” represents the threshold cycle.
4.10. In Situ Hybridization
In situ hybridization assays of S. domuncula tissue slices were performed according to Schröder et al. [62]. Briefly, labeling of ssDNA probes (SDASABF) was carried out with the PCR DIG Probe Synthesis Kit (Roche), using either forward primer 5′-ATGAATGCAAAGTTGTGCACTC-3′ (nt47 to nt68; for the sense probe) or reverse primer 5′-CAAGCGCGTTCTTCATCGAC-3′ (nt297 to nt278; for the anti-sense probe) in combination with a linearized template. The resulting labeled ssDNA probes spanned nt47 to nt297 of the SDASABF cDNA. Then, cryosections (8 μm thickness) were fixed with paraformaldehyde, washed with PBS (10 mM sodium phosphate pH 7.4, in 150 mM NaCl) at room temperature, and incubated with Proteinase K. For rehydration, the sections were hybridized with the anti-sense or sense probe (negative control) overnight at 40°C. After blocking, the sections were incubated for 1 h at 37 °C with anti-DIG Fab fragments (conjugated to alkaline phosphatase) and, then, washed twice. For visualization of the signals, the dye reagents BCIP/NBT (5-bromo-4-chloro-3′-indolyphosphate/nitro-blue tetrazolium) were used in a Tris-buffer (100 mM Tris-HCl pH 9.5, 100 mM NaCl, and 50 mM MgCl2).
4.11. Microbicidal Assay
Microbicidal/antibacterial assay was performed as earlier described [63,64] using the Mueller Hinton Medium (DIFCO Invitrogen, Karlsruhe; Germany) at pH 7.0. The following microorganisms were tested: the Gram-positive bacteria Staphylococcus aureus ATCC 25293, Bacillus subtilis ATCC 6633, and Micrococcus luteus ATCC 9341, the Gram-negative bacteria Pseudomonas aeruginosa ATCC 9027, and Escherichia coli ATCC 25922), and the fungi Candida albicans ATCC 10231 and Aspergillus niger ATCC 16404. The minimal inhibitory concentration (MIC) was determined and the values were given in μg/mL.
4.12. Hemolytic Assay
Human type A erythrocytes were used for the hemolytic assay; they came from healthy individuals. The detailed assay has been described earlier [39,65]. PBS-washed erythrocytes were used to prepare a 10% (v/v) suspension by using an isotonic glucose phosphate buffer (1 mM K-phosphate buffer, pH 7.0; supplemented with 287 mM glucose). Serial dilutions of recombinant non-adsorbed or adsorbed ASABF were prepared to a final volume of 200 μL in 96-well V-bottomed microtiter plates (Nunc/Thermo Electron, Langenselbold; Germany). To reach total hemolysis the samples were subjected to 1% (v/v) Triton X-100. After incubation for 1 h (37 °C) the plates were centrifuged (10 min at 2000 × g at 20 °C) and a 150 μL aliquot from the supernatant was taken and used for absorbance measurements at 450 nm. The percentage of hemolysis was calculated as described [65]
.
4.13. Acute/Subchronic Toxicity Testing
The gastropods, collected from one affected specimen of S. domuncula, were exposed at 17°C to 3 μg/mL, 10 μg/mL, or 30 μg/mL of recombinant ASABF in seawater, or remained without this peptide. The animals, 10 specimens per assay, were kept in parallel in aerated aquaria (volume of 200 mL) for 96 h. Subsequently the gastropods were transferred to seawater without the peptide and culture continued for 96 more hours. Then the living gastropods were counted. The means ± standard deviations of three series of experiments were calculated by using the paired Student’s t-test [66].
4.14. Further Methods
For the quantification of protein, the Bradford method ([67]; Roti-Quant solution-Roth) was used.
5. Conclusion
In the present study we describe, for the first time, a sponge ASABF peptide and show that the polypeptide, deduced from the complete cDNA, comprises the characteristic features of ASABF peptides so far identified especially in nematodes. The induction of the ASABF gene was monitored in tissue undergoing apoptosis. In those apoptotic regions the expression of ASABF is preceded by the expression of HAD, which encodes a key enzyme involved in the formation of quinolinic acid, and in parallel by the expression of the caspase gene that initiates and regulates the onset and the completion of the apoptotic process. The elimination of the apoptotic tissue is supported by the gastropod Bittium sp. which is attracted by the initially synthesized quinolinic acid and finally eliminated by ASABF. In parallel to the elimination of the gastropod, also potentially attacking microorganisms are killed by ASABF. Our data demonstrate that the innate immune network, based on antimicrobial peptides, in sponges is complex and together with the host-encoded immune defense arm, ASABF, involves a transient grazing of symbiotic gastropods that accelerate the elimination of apoptotic tissue (Figure 9).
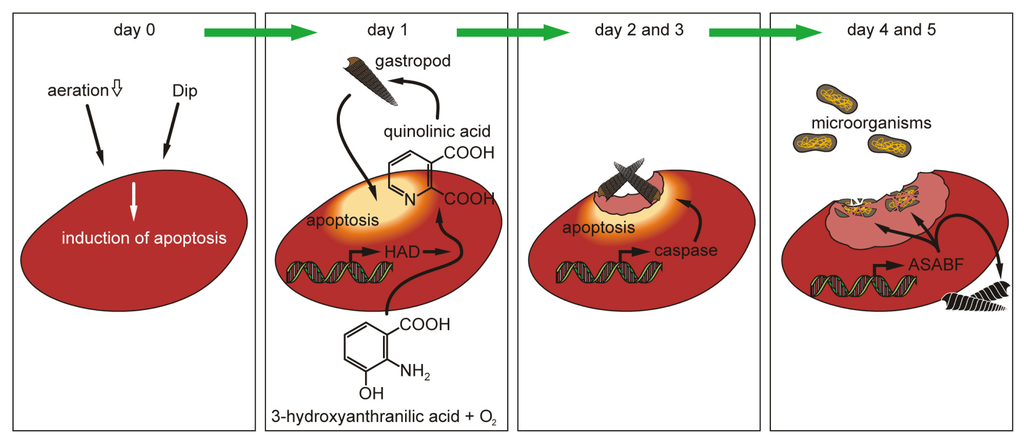
Figure 9.
Schematic outline of the elimination of apoptotic tissue from the sponge S. domuncula, after induction with the Fe++ chelator Dip. Initially the expression of the HAD gene is induced as a response to Dip. After initiation of Dip-induced apoptosis the caspase gene is upregulated and, sequentially, the expression of the ASABF gene sets on. While quinolinic acid, a metabolite synthesized via the HAD/kynurenine pathway, attracts the gastropod Bittium sp. the terminally expressed ASABF causes, after removal of the apoptotic tissue, an repelling of the gastropods as well as a killing of the microorganisms.
Acknowledgements
W.E.G., M. is holder of an ERC Advanced Investigator Grant (no. 268476–BIOSILICA). We wish to thank Sanja Perović-Ottstadt and Anatoli Krasko for their help during the in situ hybridization analyses and the isolation of the cDNA sequence data. This work was supported by grants from the German Bundesministerium für Bildung und Forschung (project “Center of Excellence BIOTECmarin”), the European Commission/EUREKA (EUROSTARS, no. 4289-SILIBACTS), the International Human Frontier Science Program, the European Commission (project no. 244967-Mem-S), and the Deutsche Forschungsgemeinschaft (WI 2116/2-2).
The sequences have been deposited (EMBL/GenBank); the cDNA for the Suberites domuncula ASABF-related peptide (SDASABF gene) under the accession number HE573029 and the cDNA from Lubomirskia baicalensis ASABF-related peptide (LBASABFr gene) under the accession number HE573030.
- †This contribution is dedicated to Dr. Peter Murphy (Townsville, Australia) in memory of his comprehensive studies in worldwide marine biodiversity.
References
- Sponges (Porifera) Marine Molecular Biotechnology; Müller, WEG (Ed.) Springer-Press: Berlin, Germany, 2003.
- Osinga, R; Belarbi, EH; Grima, EM; Tramper, J; Wijffels, RH. Progress towards a controlled culture of the marine sponge Pseudosuberites andrewsi in a bioreactor. J. Biotechnol 2003, 100, 141–144. [Google Scholar]
- Fattorusso, E; Gerwick, WH; Taglialatela-Scafati, O. Handbook of Marine Natural Products; Springer-Press: Berlin, Germany, 2011; in press. [Google Scholar]
- Müller, WEG; Klemt, M; Thakur, NL; Schröder, HC; Aiello, A; D’Esposito, M; Menna, M; Fattorusso, E. Molecular/chemical ecology in sponges: Evidence for an adaptive antibacterial response in Suberites domuncula. Mar. Biol 2003, 144, 19–29. [Google Scholar]
- Thakur, NL; Müller, WEG. Biotechnological potential of marine sponges. Curr. Sci 2004, 86, 1506–1512. [Google Scholar]
- Müller, WEG; Blumbach, B; Müller, IM. Evolution of the innate and adaptive immune systems: Relationships between potential immune molecules in the lowest metazoan phylum [Porifera] and those in vertebrates. Transplantation 1999, 68, 1215–1227. [Google Scholar]
- Blumbach, B; Diehl-Seifert, B; Seack, J; Steffen, R; Müller, IM; Müller, WEG. Cloning and expression of new receptors belonging to the immunoglobulin superfamily from the marine sponge Geodia cydonium. Immunogenetics 1999, 49, 751–763. [Google Scholar]
- Wiens, M; Korzhev, M; Perovic-Ottstadt, S; Luthringer, B; Brandt, D; Klein, S; Müller, WEG. Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol 2007, 24, 792–804. [Google Scholar]
- Perović-Ottstadt, S; Adell, T; Proksch, P; Wiens, M; Korzhev, M; Gamulin, V; Müller, IM; Müller, WEG. A (1→3)-β-d-glucan recognition protein from the sponge Suberites domuncula: Mediated activation of fibrinogen-like protein and epidermal growth factor gene expression. Eur. J. Biochem 2004, 271, 1924–1937. [Google Scholar]
- Thakur, NL; Perović-Ottstadt, S; Batel, R; Korzhev, M; Diehl-Seifert, B; Müller, IM; Müller, WEG. Innate immune defense of the sponge Suberites domuncula against gram-positive bacteria: Induction of lysozyme and AdaPTin. Mar. Biol 2005, 146, 271–282. [Google Scholar]
- Wiens, M; Korzhev, M; Krasko, A; Thakur, NL; Perović-Ottstadt, S; Breter, HJ; Ushijima, H; Diehl-Seifert, B; Müller, IM; Müller, WEG. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway: Induction of a perforin-like molecule. J. Biol. Chem 2005, 280, 27949–27959. [Google Scholar]
- Schröder, HC; Ushijima, H; Krasko, A; Gamulin, V; Schütze, J; Müller, IM; Müller, WEG. Emergence and disappearance of an immune molecule, an antimicrobial lectin, in basal Metazoa: The tachylectin family. J. Biol. Chem 2003, 278, 32810–32817. [Google Scholar]
- Müller, WEG; Wiens, M; Adell, T; Gamulin, V; Schröder, HC; Müller, IM. Bauplan of Urmetazoa: Basis for genetic complexity of Metazoa. Int. Rev. Cytol 2004, 235, 53–92. [Google Scholar]
- Zhua, S; Gao, B; Tytgat, J. Phylogenetic distribution, functional epitopes and evolution of the CSαβ superfamily. Cell. Mol. Life Sci 2005, 62, 2257–2269. [Google Scholar]
- Ueno, S; Masaomi, M; Yuji, N; Misako, T; Yasushi, T; Toshimasa, Y; Kato, Y. Generation of novel cationic antimicrobial peptides from natural non-antimicrobial sequences by acid-amide substitution. Ann. Clin. Microbiol. Antimicrobials 2011, 10, 1–7. [Google Scholar]
- Nijnik, A; Hancock, REW. Host defence peptides: Antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg Health Threats J 2009, 2, e1. [Google Scholar] [CrossRef]
- Poriferan EST database. Available online: https://octavia.vk.medizin.uni-mainz.de/login.cgi accessed on 15 January 2011.
- Kato, Y; Komatsu, S. ASABF, a novel cysteine-rich antibacterial peptide isolated from the nematode Ascaris suum: Purification, primary structure, and molecular cloning of cDNA. J. Biol. Chem 1996, 271, 30493–30498. [Google Scholar]
- Kato, Y; Aizawa, T; Hoshino, H; Kawano, K; Nitta, K; Zhang, H. Abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J 2002, 361, 221–230. [Google Scholar]
- Selsted, ME; Ouellette, AJ. Mammalian defensins in the antimicrobial immune response. Nature Immunol 2005, 6, 551–557. [Google Scholar]
- Minaba, M; Ueno, S; Pillai, A; Kato, Y. Evolution of ASABF (Ascaris suum antibacterial factor)-type antimicrobial peptides in nematodes: Putative rearrangement of disulfide bonding patterns. Develop. Comp. Immunol 2009, 33, 1147–1150. [Google Scholar]
- Kato, Y. Nematode antimicrobial peptides. Curr. Topics Pept. Prot. Res 2007, 8, 1–10. [Google Scholar]
- Pillai, A; Ueno, S; Zhang, H; Kato, Y. Induction of ASABF (Ascaris suum antibacterial factor)-type antimicrobial peptides by bacterial injection: Novel members of ASABF in the nematode Ascaris suum. Biochem. J 2003, 371, 663–668. [Google Scholar]
- Schröder, HC; Sudek, S; de Caro, S; de Rosa, S; Perović, S; Steffen, R; Müller, IM; Müller, WEG. Synthesis of the neurotoxin quinolinic acid in apoptotic tissue from Suberites domuncula: Cell biological, molecular biological and chemical analyses. Mar. Biotechnol 2002, 4, 546–558. [Google Scholar]
- Wiens, M; Krasko, A; Perović, S; Müller, WEG. Caspase-mediated apoptosis in sponges: Cloning and function of the phylogenetic oldest apoptotic proteases from Metazoa. Biochim. Biophys. Acta 2003, 1593, 179–189. [Google Scholar]
- Cole, C; Barber, JD; Barton, GJ. The Jpred 3 secondary structure prediction server. Nucl. Acids Res 2008, 35, W197–W201. [Google Scholar]
- Chou, PY; Fasman, GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol 1978, 47, 45–148. [Google Scholar]
- InterProScan Sequence Search. Available online: http://www.ebi.ac.uk/Tools/pfa/iprscan/ accessed on 27 January 2011.
- BLAST Assembled RefSeq Genomes. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 31 January 2011.
- Wiens, M; Luckas, B; Brümmer, F; Ammar, MSA; Steffen, R; Batel, R; Diehl-Seifert, B; Schröder, HC; Müller, WEG. Okadaic acid: A potential defense molecule for the sponge Suberites domuncula. Mar. Biol 2003, 142, 213–223. [Google Scholar]
- Yeaman, MR; Yount, NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev 2003, 55, 27–55. [Google Scholar]
- Toke, O. Antimicrobial peptides: New candidates in the fight against bacterial infections. Biopolymers 2005, 80, 717–735. [Google Scholar]
- Brogden, KA; Ackermann, M; McCray, PB, Jr; Tack, BF. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob Agents 2003, 22, 465–478. [Google Scholar]
- Yonezawa, A; Kuwahara, J; Fujii, N; Sugiura, Y. Binding of tachyplesin I to DNA revealed by footprinting analysis: Significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 1992, 31, 2998–3004. [Google Scholar]
- Beisswenger, C; Bals, R. Functions of antimicrobial peptides in host defense and immunity. Curr. Protein Pept. Sci 2005, 6, 255–264. [Google Scholar]
- Hochmuth, T; Piel, J. Polyketide synthases of bacterial symbionts in sponges—Evolution-based applications in natural products research. Phytochemistry 2009, 70, 1841–1849. [Google Scholar]
- Soruri, A; Grigat, J; Forssmann, U; Riggert, J; Zwirner, J. β-Defensins chemoattract macrophages and mast cells but not lymphocytes and dendritic cells: CCR6 is not involved. Eur. J. Immunol 2007, 37, 2474–2486. [Google Scholar]
- Klotman, ME; Chang, TL. Defensins in innate antiviral immunity. Nat. Rev. Immunol 2006, 6, 447–456. [Google Scholar]
- Zhang, H; Yoshida, S; Aizawa, T; Murakami, R; Suzuki, M; Koganezawa, N; Matsuura, A; Miyazawa, M; Kawano, K; Nitta, K; et al. In vitro antimicrobial properties of recombinant ASABF, an antimicrobial peptide isolated from the nematode Ascaris suum. Antimicrob. Agents Chemother 2000, 44, 2701–2705. [Google Scholar]
- Roche Applied Science, Apoptosis, Cell Death and Cell Proliferation; Roche Diagnostics: Mannheim, Germany, 2011.
- Pucci, L; Perozzi, S; Cimadamore, F; Orsomando, G; Raffaelli, N. Tissue expression and biochemical characterization of human 2-amino 3-carboxymuconate 6-semialdehyde decarboxylase, a key enzyme in tryptophan catabolism. FEBS J 2007, 274, 827–840. [Google Scholar]
- Guillemin, GJ; Wang, L; Brew, BJ. Quinolinic acid selectively induces apoptosis of human astrocytes: Potential role in AIDS dementia complex. J Neuroinflammation 2005, 2. [Google Scholar] [CrossRef] [Green Version]
- Müller, WEG; Wang, XH; Wiens, M; Schloßmacher, U; Jochum, KP; Schröder, HC. Hardening of bio-silica in sponge spicules involves an aging process after its enzymatic polycondensation: Evidence for an aquaporin-mediated water absorption. Biochim. Biophys. Acta 2011, 1810, 713–726. [Google Scholar]
- Actis, LA; Tolmasky, ME; Crosa, LM; Crosa, JH. Effect of iron-limiting conditions on growth of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol 1993, 31, 2812–2815. [Google Scholar]
- Wright, RA; Crowder, LB; Martin, TH. Selective predation by blue crabs on the gastropod, Bittium varium: Confirmation from opercula found in the sediments. Estuaries 1996, 19, 75–81. [Google Scholar]
- LePennec, G; Perović, S; Ammar, MSA; Grebenjuk, VA; Steffen, R; Müller, WEG. Cultivation of primmorphs from the marine sponge Suberites domuncula: Morphogenetic potential of silicon and iron. J. Biotechnol 2003, 100, 93–108. [Google Scholar]
- Müller, WEG; Krasko, A; Skorokhod, A; Bünz, C; Grebenjuk, VA; Steffen, R; Batel, R; Schröder, HC. Histocompatibility reaction in tissue and cells of the marine sponge Suberites domuncula in vitro and in vivo: Central role of the allograft inflammatory factor 1. Immunogenetics 2002, 54, 48–58. [Google Scholar]
- Thompson, JD; Higgins, DG; Gibson, TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl. Acids Res 1994, 22, 4673–4680. [Google Scholar]
- Dayhoff, MO; Schwartz, RM; Orcutt, BC. A Model of Evolutionary Change in Protein. In Atlas of Protein Sequence and Structure; Dayhoff, MO, Ed.; Natural Biomedical Research Foundation: Washington, DC, USA, 1978; pp. 345–352. [Google Scholar]
- Saitou, N; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. PHYLIP, Version 35; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Treeview. Available online: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html accessed on 17 February 2011.
- Nicholas, KB; Nicholas, HB, Jr. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments, Version 1.1.004; 1997. [Google Scholar]
- Finn, RD; Mistry, J; Schuster-Böckler, B; Griffiths-Jones, S; Hollich, V; Lassmann, T; Moxon, S; Marshall, M; Khanna, A; Durbin, R; et al. Pfam: Clans, web tools and services. Nucl. Acids Res 2006, 34, D247–D251. [Google Scholar]
- Letunic, I; Copley, RR; Pils, B; Pinkert, S; Schultz, J; Bork, P. SMART 5: Domains in the context of genomes and networks. Nucl. Acids Res 2006, 34, D257–D260. [Google Scholar]
- Romanos, MA; Scorer, CA; Clare, JJ. Foreign gene expression in yeast: A review. Yeast 1992, 8, 423–488. [Google Scholar]
- Müller, WEG; Rothenberger, M; Boreiko, A; Tremel, W; Reiber, A; Schröder, HC. Formation of siliceous spicules in the marine demosponge Suberites domuncula. Cell Tissue Res 2005, 321, 285–297. [Google Scholar]
- Laemmli, UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
- Wiens, M; Wang, XH; Unger, A; Schröder, HC; Grebenjuk, VA; Pisignano, D; Jochum, KP; Müller, WEG. Flashing light signaling circuit in sponges: Endogenous light generation after tissue ablation in Suberites domuncula. J. Cell Biochem 2010, 111, 1377–1389. [Google Scholar]
- Müller, WEG; Wang, XH; Schröder, HC; Korzhev, M; Grebenjuk, VA; Markl, JS; Jochum, KP; Pisignano, D; Wiens, M. A cryptochrome-based photosensory system in the siliceous sponge Suberites domuncula (Demospongiae). FEBS J 2010, 277, 1182–1201. [Google Scholar]
- Livak, KJ; Schmittgen, TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 D DCT method. Methods 2001, 25, 402–408. [Google Scholar]
- Schröder, HC; Perović-Ottstadt, S; Rothenberger, M; Wiens, M; Schwertner, H; Batel, R; Korzhev, M; Müller, IM; Müller, WEG. Silica transport in the demosponge Suberites domuncula: Fluorescence emission analysis using the PDMPO probe and cloning of a potential transporter. Biochem. J 2004, 381, 665–673. [Google Scholar]
- Seibert, G; Raether, W; Dogovic, N; Gasic, MJ; Zahn, RK; Müller, WEG. Antibacterial and antifungal activity of avarone and avarol. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 1985, 260, 379–386. [Google Scholar]
- Thakur, NL; Hentschel, U; Krasko, A; Pabel, CT; Anil, AC; Müller, WEG. Antibacterial activity of the sponge Suberites domuncula and its primmorphs: Potential basis for the defense. Aquat. Microbiol. Ecol 2003, 31, 77–83. [Google Scholar]
- Wei, GX; Bobek, LA. In vitro synergic antifungal effect of MUC7 12-mer with histatin-5 12-mer or miconazole. J. Antimicr. Chemother 2004, 53, 750–758. [Google Scholar]
- Sachs, L. Angewandte Statistik; Springer: Berlin, Germany, 1984; p. 242. [Google Scholar]
- Compton, S; Jones, CG. Mechanism of dye response and interference in the Bradford protein assay. Anal. Biochem 1985, 151, 369–374. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).