Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae
Abstract
:1. Introduction
2. Conventional AMPs
2.1. Linear, α-helical peptides
2.1.1. Invertebrates
2.1.2. Fish
2.2. Cysteine-rich peptides
2.2.1. Invertebrates
2.2.2. Fish
2.3. Cationic peptides, amino acid enriched
2.4. Miscellaneous AMPs
3. Unconventional Anti-infectives
3.1. Antimicrobials derived from intracellular structures
3.1.1. Histones
3.1.2. Other intracellular proteins
3.2. Membrane-derived antimicrobial compounds
3.2.1. Free fatty acids
3.2.2. Oxylipins
3.3. Pigment or pigment-derived antimicrobials
3.3.1. Respiratory pigments
3.3.2. Other pigments
3.4. Pore-forming toxins
3.5. Neuropeptides
3.6. Regulatory binding and other molecules
3.7. Lectins
3.8. Binding molecules
4. Discussion
4.1. Diversity and evolution of antibacterial molecules
4.2. Potential value of marine eukaryotic anti-infectives
4.3. Promise for future medical or commercial exploitation
4.3.1. Synergy
4.3.2. Chimeric compounds
- Samples Availability: Not available from the authors.
References
- Brahmachary, M; Krishnan, SPT; Koh, JLY; Khan, AM; Seah, SH; Tan, TW; Brusic, TW; Bajic, TW. ANTIMIC: A database of antimicrobial sequences. Nucleic Acids Res 2004, 32, D586–589. [Google Scholar]
- Wang, Z; Wang, G. APD: The antimicrobial peptide database. Nucleic Acids Res 2004, 32, 590–592. [Google Scholar]
- Thomas, S; Karnik, S; Shanker, Barai R; Jayaraman, VK; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res 2010, 38, D774–780. [Google Scholar]
- Boman, HG. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol 1995, 13, 61–92. [Google Scholar]
- Hancock, REW; Scott, MG. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci USA 2000, 97, 8856–8861. [Google Scholar]
- Ovchinnikova, TV; Balandin, SV; Aleshina, GM; Tagaev, AA; Leonova, YF; Krasnodembsky, ED; Men’shenin, AV; Kokryakov, VN. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem Biophys Res Comm 2006, 348, 514–523. [Google Scholar]
- Ovchinnikova, TV; Aleshina, GM; Balandin, SV; Krasnosdembskaya, AD; Markelov, ML; Frolova, EI; Leonova, YF; Tagaev, AA; Krasnodembsky, EG; Kokryakov, VN. Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett 2004, 577, 209–214. [Google Scholar]
- Tasiemski, A; Schikorski, D; Le Marrec-Croq, F; Pontoire-Van Camp, C; Boidin-Wichlacz, C; Sautière, P-E. Hedistin: A novel antimicrobial peptide containing bromotryptophan constitutively expressed in the NK cells-like of the marine annelid Nereis diversicolor. Dev Comp Immunol 2007, 31, 749–762. [Google Scholar]
- Pan, W; Liu, X; Ge, F; Han, J; Zheng, T. Perinerin, a novel antimicrobial peptide purified from the clamworm Perinereis aibuhitensis Grube and its partial characterization. J Biochem 2004, 135, 297–304. [Google Scholar]
- Li, C-H; Zhoa, JM; Song, LS. A review of advances in research on marine molluscan antimicrobial peptides and their potential application in aquaculture. Mollusc Res 2009, 29, 17–26. [Google Scholar]
- Stensvåg, K; Haug, T; Sperstad, SV; Rekdal, O; Indrevoll, B; Styrvold, OB. Arasin 1, a proline-arginine-rich antimicrobial peptide isolated from the spider crab Hyas araneus. Dev Comp Immunol 2008, 32, 275–285. [Google Scholar]
- Schnapp, D; Kemp, GD; Smith, VJ. Purification and characterization of a proline-rich antibacterial peptide, with sequence similarity to bactenecin-7, from the haemocytes of the shore crab Carcinus maenas. Eur J Biochem 1996, 240, 532–539. [Google Scholar]
- Khoo, L; Robinette, DW; Noga, NJ. Callinectin, an antibacterial peptide from blue crab, Callinectes sapidus, hemocytes. Mar Biotechnol 1999, 1, 44–51. [Google Scholar]
- Smith, VJ; Fernandes, JMO; Kemp, GD; Hauton, C. Crustins: Enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev Comp Immunol 2008, 32, 758–772. [Google Scholar]
- Battison, AL; Summerfield, R; Patrzykat, A. Isolation and characterisation of two antimicrobial peptides from haemocytes of the American lobster Homarus americanus. Fish Shellfish Immunol 2008, 25, 181–187. [Google Scholar]
- Sperstad, SV; Haug, T; Vasskog, T; Stensvåg, K. Hyastatin, a glycine-rich multi-domain antimicrobial peptide isolated from the spider crab (Hyas araneus) hemocyes. Mol Immunol 2009, 46, 2604–2612. [Google Scholar]
- Cuthbertson, BJ; Deterding, LJ; Williams, JG; Tomer, KB; Etienne, K; Blackshear, PJ; Büllesbach, EE; Gross, PS. Diversity in penaeidin antimicrobial peptide form and function. Dev Comp Immunol 2008, 32, 167–181. [Google Scholar]
- Huang, WS; Wang, KJ; Yang, M; Cai, JJ; Li, SJ; Wang, GZ. Purification and part characterization of a novel antibacterial protein scygonadin, isolated from the seminal plasma of mud crab, Scylla serrata (Forskal). J Exp Mar Biol Ecol 2006, 339, 37–42. [Google Scholar]
- Yedery, RD; Reddy, KVR. Identification, cloning, characterization and recombinant expression of an anti-lipopolysaccharide factor from the hemocytes of Indian mud crab Scylla serrata. Fish Shellfish Immunol 2009, 27, 275–284. [Google Scholar]
- Saito, T; Kawabata, S; Shigenaga, T; Takayenoki, Y; Cho, J; Nakajima, H; Hirata, M; Iwanaga, S. A novel big defensin identified in horseshoe-crab hemocytes: Isolation, amino acid-sequence, and antibacterial activity. J Biochem 1995, 117, 1131–1137. [Google Scholar]
- Miyata, T; Tokunaga, F; Yoneya, T; Yoshikawa, K; Iwanaga, S; Niwa, M; Takao, T; Shimonishi, Y. Antimicrobial peptides, isolated from horseshoe-crab hemocytes, tachyplesin-II, and polyphemusin-I and polyphemusin-II: Chemical structures and biological activity. J Biochem 1989, 106, 663–668. [Google Scholar]
- Kawabata, SI; Nagayama, R; Hirata, M; Shigenaga, T; Agarwala, KL; Saito, T; Cho, J; Nakajima, H; Takagi, T; Iwanaga, S. Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem 1996, 120, 1253–1260. [Google Scholar]
- Osaki, T; Omotezako, M; Nagayama, R; Hirata, M; Iwanaga, S; Kasahara, J; Hattori, J; Ito, I; Sugiyama, H; Kawabata, SI. Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J Biol Chem 1999, 274, 26172–26178. [Google Scholar]
- Li, C; Haug, T; Styrvold, OB; Jørgensen, TØ; Stensvåg, K. Strongylocins, novel antimicrobial peptides from the green sea urchin Stronglyocentrotus droebachiensis. Dev Comp Immunol 2008, 32, 1430–1440. [Google Scholar]
- Lee, IH; Zhao, C; Cho, Y; Harwig, SS; Cooper, EL; Lehrer, RI. Clavanins, alpha-helical antimicrobial peptides from tunicate hemocytes. FEBS Lett 1997, 400, 158–162. [Google Scholar]
- Lee, IH; Lee, YS; Kim, CH; Kim, CR; Hong, T; Menzel, L; Boo, LM; Pohl, J; Sherman, MA; Waring, A; Lehrer, RI. Dicynthaurin: an antimicrobial peptide from hemocytes of the solitary tunicate Halocynthia aurantium. Biochim Biophys Acta 2001, 1527, 141–148. [Google Scholar]
- Jang, W; Kim, K; Lee, Y; Nam, M; Lee, I. Halocidin: a new antimicrobial peptide from hemocytes of the solitary tunicate Halocynthia aurantium. FEBS Lett 2002, 521, 81–86. [Google Scholar]
- Azumi, K; Yokosawa, H; Ishii, S. Halocyamines – novel antimicrobial tetrapeptide-like substances isolated from the hemocytes of the solitary ascidian Halocynthia roretzi. Biochemistry 1990, 29, 159–165. [Google Scholar]
- Galinier, R; Roger, E; Sautiere, PE; Aumelas, A; Banaigs, B; Mitta, G. Halocyntin and papillosin, two new antimicrobial peptides isolated from hemocytes of the solitary tunicate Halocynthia papillosa. J Pept Sci 2009, 15, 48–55. [Google Scholar]
- Lee, IH; Cho, Y; Lehrer, RI. Styelins, broad-spectrum antimicrobial peptides from the solitary tunicate Styela clava. Comp Biochem Physiol 1997, 118, 515–521. [Google Scholar]
- Smith, VJ; Fernandes, JMO. Zaccone, G, Masseguer, J, García-Ayala, A, Kapoor, BG, Eds.; Non-specific antimicrobial proteins of the innate system. In Fish Defences; Science Publishers: Enfield, NH, USA, 2009; Volume 1, pp. 241–275. [Google Scholar]
- Gueguen, Y; Herpin, A; Aumelas, A; Garnier, J; Fievet, J; Escoubas, JM; Bulet, P; Gonzalez, M; Lelong, C; Favrel, P; Bachère, E. Characterization of a defensin from the oyster Crassostrea gigas. Recombinant production, folding, solution structure, antimicrobial activities, and gene expression. J Biol Chem 2006, 281, 313–323. [Google Scholar]
- Gonzalez, M; Gueguen, Y; Desserre, G; de Lorgeril, J; Romestand, B; Bachère, E. Molecular characterization of two isoforms of defensin from hemocytes of the oyster Crassostrea gigas. Dev Comp Immunol 2007, 31, 332–339. [Google Scholar]
- Brogden, KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria. Nat Rev Microbiol 2005, 3, 238–250. [Google Scholar]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by K-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta 1999, 1462, 55–70. [Google Scholar]
- Huang, HW. Action of antimicrobial peptides: two-state model. Biochemistry 2000, 39, 8347–8352. [Google Scholar]
- Yang, L; Harroun, TA; Weiss, TM; Ding, L; Huang, HW. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys J 2001, 81, 1475–1485. [Google Scholar]
- Campagna, S; Saint, N; Molle, G; Aumelas, A. Structure and mechanism of action of the antimicrobial peptide piscidin. Biochemistry 2007, 46, 1771–1778. [Google Scholar]
- Park, CB; Kim, HS; Kim, SC. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem Biophys Res Comm 1998, 244, 253–257. [Google Scholar]
- Yonezawa, A; Kuwahara, J; Fujii, N; Sugiura, Y. Binding of tachyplesin I to DNA revealed by footprinting analysis: significant contribution of secondary structure to DNA binding and implication for biological action. Biochemistry 1992, 31, 2998–3004. [Google Scholar]
- Patrzykat, A; Friedrich, CL; Zhang, L; Mendoza, V; Hancock, RE. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob Agents Chemother 2002, 46, 605–614. [Google Scholar]
- Gusman, H; Travis, J; Helmerhorst, EJ; Potempa, J; Troxler, RF; Oppenheim, FG. Salivary histatin 5 is an inhibitor of both host and bacterial enzymes implicated in periodontal disease. Infection and Immunity 2001, 69, 402–1408. [Google Scholar]
- Otvos, LOI, Jr; Rogers, ME; Consolvo, PJ; Condie, BA; Lovas, S; Bulet, P; Blaszczyk-Thurin, M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 2000, 39, 14150–14159. [Google Scholar]
- Lee, IH; Cho, Y; Lehrer, RI. Effects of pH and salinity on the antimicrobial properties of clavanins. Infect Immun 1997, 65, 2898–2903. [Google Scholar]
- van Kan, EJM; Demel, RA; Breukink, E; van der Bent, A; de Kruijff, B. Clavanin permeabilizes target membranes via two distinctly different pH-dependent mechanisms. Biochemistry 2002, 41, 7529–7539. [Google Scholar]
- Zhao, C; Liaw, L; Lee, IH; Lehrer, RI. cDNA cloning of three cecropin-like antimicrobial peptides (Styelins) from the tunicate Styela clava. FEBS Lett 1997, 412, 144–148. [Google Scholar]
- Taylor, SW; Craig, AG; Fischer, WH; Park, M; Lehrer, RI. Styelin D, an extensively modified antimicrobial peptide from ascidian hemocytes. J Biol Chem 2000, 275, 38417–38426. [Google Scholar]
- Lazarovici, P; Primor, N; Loew, LM. Purification and pore-forming activity of two polypeptides from the secretion of the Red Sea Moses sole (Pardachirus marmoratus). J Biol Chem 1986, 261, 16704–16713. [Google Scholar]
- Thompson, SA; Tachibana, K; Nakanishi, K; Kubota, I. Melittin-like peptides from the shark-repelling defense secretion of the sole Pardachirus pavoninus. Science 1986, 233, 341–343. [Google Scholar]
- Oren, Z; Shai, Y. A class of highly potent antibacterial peptides derived from pardaxin, a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur J Biochem 1996, 237, 303–310. [Google Scholar]
- Cole, AM; Weis, P; Diamond, G. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J Biol Chem 1997, 272, 12008–12013. [Google Scholar]
- Noga, EJ; Silphaduang, U. Piscidins, a novel family of peptide antibiotics from fish. Drug News & Perspect 2003, 16, 87–92. [Google Scholar]
- Mulero, I; Noga, EJ; Meseguer, J; García-Alya, A; Mulero, V. The antimicrobial peptides piscidins are stored in the granules of professional phagocytic granulocytes of fish and are delivered to the bacteria-containing phagosome upon phagocytosis. Dev Comp Immunol 2008, 32, 1531–1538. [Google Scholar]
- Salerno, G; Parrinello, N; Roch, P; Cammarata, M. cDNA sequence and tissue expression of an antimicrobial peptide, dicentracin; a new component of the moronecidin family isolated from head kidney leukocytes of sea bass Dicentrarchus labrax. Comp Biochem Physiol 2007, 146B, 521–529. [Google Scholar]
- Iijima, N; Tanimoto, N; Emoto, Y; Morita, Y; Uematsu, K; Murakami, T; Nakai, T. Purification and characterization of three isoforms of chrysophsin, a novel antimicrobial peptide in the gills of the red sea bream Chrysophrys major. Eur J Biochem 2003, 270, 675–686. [Google Scholar]
- Yin, ZX; Chen, WJ; Yan, JH; Yang, JN; Chan, SM; He, JG. Cloning, expression and antimicrobial activity of an antimicrobial peptide, epinecidin-1, from the orange-spotted grouper Epinephelus coioides. Aquaculture 2006, 253, 204–211. [Google Scholar]
- Silphaduang, U; Noga, EJ. Peptide antibiotics in mast cells of fish. Nature 2001, 414, 268–269. [Google Scholar]
- Lauth, X; Shike, H; Burns, JC; Westerman, ME; Ostland, VE; Carlberg, JM; Van Olst, JC; Nizet, V; Taylor, SW; Shimizu, C; Bulet, P. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J Biol Chem 2002, 277, 5030–5039. [Google Scholar]
- Noga, EJ; Silphaduang, U; Park, NG; Seo, JK; Stephenson, J; Kozlowicz, S. Piscidin 4, a novel member of the piscidin family of antimicrobial peptides. Comp Biochem Physiol 2009, 152B, 299–305. [Google Scholar]
- Lee, SA; Kim, YK; Lim, SS; Zhu, WL; Ko, H; Shin, SY; Hahm, KS; Kim, Y. Solution structure and cell selectivity of piscidin 1 and its analogues. Biochemistry 2007, 46, 3653–3663. [Google Scholar]
- Sung, WS; Lee, J; Lee, DG. Fungicidal effect and the mode of action of piscidin 2 derived from hybrid striped bass. Biochem Biophys Res Comm 2008, 371, 551–555. [Google Scholar]
- Ganz, T. The role of antimicrobial peptides in innate immunity. Integr Comp Biol 2003, 43, 300–304. [Google Scholar]
- Hubert, F; Noel, T; Roch, P. A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis). Eur J Biochem 1996, 240, 302–306. [Google Scholar]
- Iwanaga, S. The molecular basis of innate immunity in the horseshoe crab. Curr Opin Immunol 2002, 14, 87–95. [Google Scholar]
- Katsu, T; Nakao, S; Iwanaga, S. Mode of action of an antimicrobial peptide, tachyplesin I, on biomembranes. Biol Pharm Bull 1993, 16, 178–181. [Google Scholar]
- Fujitani, N; Kawabata, SI; Osaki, T; Kumaki, Y; Demura, M; Nitta, K; Kawano, K. Structure of the antimicrobial peptide tachystatin A. J Biol Chem 2002, 277, 23651–23657. [Google Scholar]
- Kawabata, S; Nagayama, R; Hirata, M; Shigenaga, T; Agarwala, K; Saito, T; Cho, J; Nakajima, H; Takagi, T; Iwanaga, S. Tachycitin, a small granular component in horseshoe crab hemocytes is an antimicrobial protein with chitin-binding activity. J Biochem 1996, 120, 1253–1260. [Google Scholar]
- Kouno, T; Fujitani, N; Mizuguchi, M; Osaki, T; Nishimura, SI; Kawabata, SI; Aizawa, T; Demura, M; Nitta, K; Kawano, K. A novel, β-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria. Biochemistry 2008, 47, 10611–10619. [Google Scholar]
- Zhao, J; Song, L; Li, C; Ni, D; Wu, L; Zhu, L; Wang, H; Xu, W. Molecular cloning, expression of a big defensin gene from bay scallop Argopecten irradians and the antimicrobial activity of its recombinant protein. Mol Immunol 2007, 44, 360–368. [Google Scholar]
- Charlet, M; Chernysh, S; Philippe, H; Hetru, C; Hoffmann, JA; Bulet, P. Innate immunity. Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusc Mytilus edulis. J Biol Chem 1996, 271, 21808–21813. [Google Scholar]
- Romestand, B; Molina, F; Richard, V; Roch, P; Granier, C. Key role of the loop connecting the two beta strands of mussel defensin in its antimicrobial activity. Eur J Biochem 2003, 270, 2805–2813. [Google Scholar]
- Yang, YS; Mitta, G; Chavanieu, A; Calas, B; Sanchez, JF; Roch, P; Aumelas, A. Solution structure and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1). Biochemistry 2000, 39, 14436–14447. [Google Scholar]
- Mitta, G; Vandenbulcke, F; Hubert, F; Roch, P. Mussel defensins are synthesised and processed in granulocytes then released into the plasma after bacterial challenge. J Cell Sci 1999, 112, 4233–4242. [Google Scholar]
- Mitta, G; Vandenbulcke, F; Roch, P. Original involvement of antimicrobial peptides in mussel innate immunity. FEBS Lett 2000, 486, 185–190. [Google Scholar]
- Mitta, G; Hubert, F; Noël, T; Roch, P. Myticin, a novel cysteine-rich antimicrobial peptide isolated from haemocytes and plasma of the mussel Mytilus galloprovincialis. Eur J Biochem 1999, 265, 71–78. [Google Scholar]
- Roch, P; Yang, Y; Toubiana, M; Aumelas, A. NMR structure of mussel mytilin, and antiviral-antibacterial activities of derived synthetic peptides. Dev Comp Immunol 2008, 32, 227–238. [Google Scholar]
- Li, H; Toubiana, M; Monfort, P; Roch, P. Influence of temperature, salinity and E. coli tissue content on immune gene expression in mussel: Results from a 2005–2008 survey. Dev Comp Immunol 2009, 33, 974–979. [Google Scholar]
- Mitta, G; Hubert, F; Dyrynda, EA; Boudry, P; Roch, P. Mytilin B and MGD2, two antimicrobial peptides of marine mussels: gene structure and expression analysis. Dev Comp Immunol 2000, 24, 381–393. [Google Scholar]
- Mitta, G; Vandenbulcke, F; Noel, T; Romestand, B; Beauvillain, JC; Salzet, M; Roch, P. Differential distribution and defence involvement of antimicrobial peptides in mussel. J Cell Sci 1999, 113, 2759–2769. [Google Scholar]
- Costa, MM; Dios, S; Alonso-Gutierrez, J; Romero, A; Novoa, B; Figueras, A. Evidence of high individual diversity on myticin C in mussel (Mytilus galloprovincialis). Dev Comp Immunol 2009. [Google Scholar]
- Seo, JK; Crawford, JM; Stone, KL; Noga, EJ. Purification of a novel arthropod defensin from the American oyster Crassostrea virginica. Biochem Biophys Res Comm 2005, 338, 1998–2005. [Google Scholar]
- Relf, JM; Chisholm, JRS; Kemp, GD; Smith, VJ. Purification and characterisation of a cysteine-rich 11.5 kDa antibacterial peptide from the granular haemocytes of the shore crab Carcinus maenas. Eur J Biochem 1999, 264, 1–9. [Google Scholar]
- Smith, VJ; Chisholm, JRS. Beck, G, Sugumaran, M, Cooper, EL, Eds.; Antimicrobial proteins in crustaceans. In Phylogenetic Perspectives on the Vertebrate Immune System; Kluwer: London, UK, 2000; Volume 484, pp. 95–112. [Google Scholar]
- Destoumieux, D; Bulet, P; Loew, D; Van Dorsselaer, A; Rodriguez, J; Bachère, E. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp Penaeus vannamei (decapoda). J Biol Chem 1997, 272, 28398–28406. [Google Scholar]
- Destoumieux, D; Munoz, M; Cosseau, C; Rodriguez, J; Bulet, P; Comps, M; Bachère, E. Penaeidins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J Cell Sci 2000, 113, 461–469. [Google Scholar]
- Chang, CI; Pleguezuelos, O; Zhang, YA; Zou, J; Secombes, CJ. Identification of a novel cathelicidin gene in the rainbow trout Oncorhynchus mykiss. Infect Immun 2005, 73, 5053–5064. [Google Scholar]
- Chang, CI; Zhang, YA; Zou, J; Nie, P; Secombes, CJ. Two cathelicidin genes are present in both rainbow trout (Oncorhynchus mykiss) and atlantic salmon (Salmo salar). Antimicrob Agents Chemother 2006, 50, 185–195. [Google Scholar]
- Maier, VH; Dorn, KV; Gudmundsdottir, BK; Gudmundsson, GH. Characterisation of cathelicidin gene family members in divergent fish species. Mol Immunol 2008, 45, 3723–3730. [Google Scholar]
- Uzzell, T; Stolzenberg, ED; Shinnar, AE; Zasloff, M. Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 2003, 24, 1655–1667. [Google Scholar]
- Zou, J; Mercier, C; Koussounadis, A; Secombes, C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol Immunol 2007, 44, 638–647. [Google Scholar]
- Falco, A; Chico, V; Marroquí, L; Perez, L; Coll, JM; Estepa, A. Expression and antiviral activity of a β-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol Immunol 2008, 45, 757–765. [Google Scholar]
- Casadei, E; Wang, T; Zou, J; González Vecino, JL; Wadsworth, S; Secombes, CJ. Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol Immunol 2009, 46, 3358–3366. [Google Scholar]
- Nam, BH; Moon, J-Y; Kim, Y-O; Kong, HJ; Kim, W-J; Lee, S-J; Kim, K-K. Multiple β-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol 2010, 28, 267–274. [Google Scholar]
- Park, CH; Valore, EV; Waring, AJ; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 2001, 276, 7806–7810. [Google Scholar]
- Shike, H; Lauth, X; Westerman, ME; Ostland, VE; Carlberg, JM; Van Olst, JC; Shimizu, C; Bulet, P; Burns, JC. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur J Biochem 2002, 269, 2232–2237. [Google Scholar]
- Lauth, X; Babon, JJ; Stannard, JA; Singh, S; Nizet, V; Carlberg, JM; Ostland, VE; Pennington, MW; Norton, RS; Westerman, ME. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J Biol Chem 2005, 280, 9272–9282. [Google Scholar]
- Gueguen, Y; Romestand, B; Fievet, J; Schmitt, P; Destoumieux-Garzon, D; Vandenbulcke, F; Bulet, P; Bachère, E. Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol Immunol 2009, 46, 516–522. [Google Scholar]
- Andrä, J; Jakovkin, I; Grötzinger, J; Hecht, O; Krasnosdembskaya, AD; Goldmann, T; Gutsmann, T; Leippe, M. Structure and mode of action of the antimicrobial peptide arenicin. Biochem J 2008, 410, 113–122. [Google Scholar]
- Hirsch, JG. Bactericidal action of histone. J Exp Med 1958, 108, 925–944. [Google Scholar]
- Frohm, M; Gunne, H; Bergman, AC; Agerberth, B; Bergman, T; Boman, A; Liden, S; Jornvall, H; Boman, HG. Biochemical and antibacterial analysis of human wound and blister fluid. Eur J Biochem 1996, 237, 86–92. [Google Scholar]
- Park, IY; Park, CB; Kim, MS; Kim, SC. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish Parasilurus asotus. FEBS Lett 1998, 437, 258–262. [Google Scholar]
- Robinette, D; Wada, S; Arroll, T; Levy, MG; Miller, WL; Noga, EJ. Antimicrobial activity in the skin of the channel catfish, Ictalurus punctatus: characterization of broad-spectrum histone-like antimicrobial proteins. Cell Mol Life Sci 1998, 54, 467–475. [Google Scholar]
- Patrzykat, A; Zhang, L; Mendoza, V; Iwama, G; Hancock, R. Synergy of histone-derived peptides of coho salmon with lysozyme and flounder pleurocidin. Antimicrob Agents Chemother 2001, 45, 1337–1342. [Google Scholar]
- Richards, RC; O’Neil, DB; Thibault, P; Ewart, KV. Histone H1: an antimicrobial protein of Atlantic salmon (Salmo salar). Biochem Biophys Res Commun 2001, 284, 549–555. [Google Scholar]
- Fernandes, JMO; Kemp, GD; Molle, MG; Smith, VJ. Anti-microbial properties of histone H2A from skin secretions of rainbow trout Oncorhynchus mykiss. Biochem J 2002, 368, 611–620. [Google Scholar]
- Noga, EJ; Fan, Z; Silphaduang, U. Histone-like proteins from fish are lethal to the parasitic dinoflagellate Amyloodinium ocellatum. Parasitology 2001, 123, 57–65. [Google Scholar]
- Fernandes, JMO; Molle, GM; Kemp, GD; Smith, VJ. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout Oncorhynchus mykiss. Dev Comp Immunol 2004, 28, 127–138. [Google Scholar]
- Patat, SA; Carnegie, RB; Kingsbury, C; Gross, PS; Chapman, RB; Schey, KL. Antimicrobial activity of histones from hemocytes of the Pacific white shrimp. Eur J Biochem 2004, 271, 4825–4833. [Google Scholar]
- Birkemo, GA; Luders, T; Andersen, O; Nes, IF; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim Biophys Acta 2003, 1646, 207–215. [Google Scholar]
- De Zoysa, M; Nikapitiya, C; Whang, I; Lee, JS; Lee, J. Abhisin: A potential antimicrobial peptide derived from histone H2A of disk abalone (Haliotis discus discus). Fish Shellfish Immunol 2009, 27, 639–646. [Google Scholar]
- Bergsson, G; Agerberth, B; Jörnvell, H; Gudmundsson, GH. Isolation and identification of antimicrobial components from epidermal mucus of Atlantic cod Gadus morhua. FEBS J 2005, 272, 4960–4969. [Google Scholar]
- Fernandes, JMO; Saint, N; Kemp, GD; Smith, VJ. Oncorhyncin III: a potent antimicrobial peptide derived from the non-histone chromosomal protein H6 of rainbow trout Oncorhynchus mykiss. Biochem J 2003, 373, 621–628. [Google Scholar]
- Fernandes, JMO; Smith, VJ. A novel antimicrobial function for a ribosomal peptide from rainbow trout skin. Biochem Biophys Res Commun 2002, 296, 167–171. [Google Scholar]
- Cho, JH; Sung, BH; Kim, SC. Buforins: H2A-derived antimicrobial peptides from toad stomach. Biochim. Biophys. Acta 2009, 1788, 1564–1569. [Google Scholar]
- Li, CH; Song, LS; Zhou, JM; Zhu, L; Zhang, H; Wang, H; Cai, ZH. Preliminary study of potential antibacterial peptide derived from histone H2A in hemocytes of scallop Chlamys farreri. Fish Shellfish Immunol 2007, 22, 663–672. [Google Scholar]
- Cho, JH; Park, IY; Kim, HS; Lee, WT; Kim, MS; Kim, SC. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J 2002. [Google Scholar]
- Cho, JH; Park, IY; Kim, MS; Kim, SC. Matrix metalloproteinase 2 is involved in the regulation of the antimicrobial peptide parasin I production in catfish skin mucosa. FEBS Lett 2002, 531, 459–463. [Google Scholar]
- Hiemstra, PS; Eisenhauer, PB; Harwig, SS; van den Barselaar, MT; van Furth, R; Lehrer, RI. Antimicrobial proteins of murine macrophages. Infect Immun 1993, 61, 3038–3046. [Google Scholar]
- Kawasaki, H; Isaacson, T; Iwamuro, S; Conlon, JM. A protein with antimicrobial activity in the skin of Schlegel’s green frog, Rhacophorus schlegelii, (Rhacophoridae) identified as histone H2B. Biochem Biophys Res Comm 2003, 312, 1082–1086. [Google Scholar]
- Lee, DY; Huang, CM; Nakatsuji, T; Thiboutat, D; Kang, SA; Monestier, M; Gallo, RL. Histone H4 is a major component of the antimicrobial action of human sebocytes. J Invest Dermatol 2009, 129, 2489–2496. [Google Scholar]
- Tsao, HT; Spinella, SA; Lee, AT; Elmore, DE. Design of histone-derived antimicrobial peptides. Peptides 2009, 30, 2168–2173. [Google Scholar]
- Subramanian, S; Ross, NW; McKinnon, SL. Comparison of the biochemical composition of normal epidermal mucus and extruded slime of hagfish, (Myxine glutinosa L). Fish Shellfish Immunol 2008, 25, 625–632. [Google Scholar]
- Kashima, M. H1 histones contribute to candidacidal activities of human epidermal extract. J Dermatol 1991, 18, 695–706. [Google Scholar]
- Lüders, T; Birkemo, GA; Nissen-Meyer, J; Andersen, Ø; Nes, IF. Proline conformation-dependent antimicrobial activity of a proline-rich histone H1 N-terminal peptide fragment isolated from the skin mucus of Atlantic salmon. Antimicrob Agents Chemother 2005, 49, 2399–2406. [Google Scholar]
- Stollar, BD; Ward, M. Rabbit antibodies to histone fractions as specific reagents for preparative and comparative studies. J Biol Chem 1969, 245, 1261–1266. [Google Scholar]
- Jüttner, F. Liberation of 5,8,11,14,17-eicosapentaenoic acid and other polyunsaturated fatty acids from lipids as a grazer defense reaction in epilithic diatom biofilms. J Phycol 2001, 37, 744–755. [Google Scholar]
- Pohnert, G. Phospholipase A2 activity triggers the wound-activated chemical defense in the diatom Thalassiosira rotula. Plant Physiol 2002, 129, 103–111. [Google Scholar]
- Küpper, FC; Gaquerel, E; Bonenerg, E-M; Morath, S; Salaün, J-P; Potin, P. Early events in the perception of lipopolysaccharides in the brown alga Laminaria digitata include an oxidative burst and activation of fatty acid oxidation cascades. J Exp Bot 2006, 57, 1991–1999. [Google Scholar]
- Lion, U; Wiesemeier, T; Weinberger, F; Beltran, J; Flores, V; Faugeron, S; Correa, J; Pohnert, G. Phospholipases and galactolipases trigger oxylipin-mediated wound-activated defence in the red alga Gracilaria chilensis against epiphytes. ChemBioChem 2006, 7, 457–462. [Google Scholar]
- Bouarab, K; Adas, F; Gaquerel, E; Kloareg, B; Salaün, JP; Potin, P. The innate immunity of a marine red alga involves oxylipins from both the eicosanoid and octadecanoid pathways. Plant Physiol 2004, 135, 1838–1848. [Google Scholar]
- Desbois, AP; Smith, VJ. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 2010, 85, 1629–1642. [Google Scholar]
- Feldlaufer, MF; Knox, DA; Lusby, WR; Shimanuki, H. Antimicrobial activity of fatty acids against Bacillus larvae, the causative agent of American foulbrood disease. Apidologie 1993, 24, 95–99. [Google Scholar]
- Kabara, JJ; Swieczkowski, DM; Conley, AJ; Truant, JP. Fatty acids and derivatives as antimicrobial agents. Antimicrob Agents Chemother 1972, 2, 23–28. [Google Scholar]
- Bergsson, G; Arnfinnsson, J; Steingrímsson, Ó; Thormar, H. Killing of Gram-positive cocci by fatty acids and monoglycerides. APMIS 2001, 109, 670–678. [Google Scholar]
- Saito, H; Tomioka, H; Yoneyama, T. Growth of group IV mycobacteria on medium containing various saturated and unsaturated fatty acids. Antimicrob Agents Chemother 1984, 26, 164–169. [Google Scholar]
- Knapp, HR; Melly, MA. Bactericidal effects of polyunsaturated fatty acids. J Infect Dis 1986, 154, 84–94. [Google Scholar]
- Galbraith, H; Miller, TB; Paton, AM; Thompson, JK. Antibacterial activity of long chain fatty acids and the reversal with calcium, magnesium, ergocalciferol and cholesterol. J Appl Bacteriol 1971, 34, 803–813. [Google Scholar]
- d’Ippolito, G; Tucci, S; Cutignano, A; Romano, G; Cimino, G; Miralto, A; Fontana, A. The role of complex lipids in the synthesis of bioactive aldehydes of the marine diatom Skeletonema costatum. Biochim Biophys Acta 2004, 1686, 100–107. [Google Scholar]
- Wichard, T; Gerecht, A; Boersma, M; Poulet, SA; Wiltshire, K; Pohnert, G. Lipid and fatty acid composition of diatoms revisited: rapid wound-activated change of food quality parameters influences herbivorous copepod reproductive success. Chem Bio Chem 2007, 8, 1146–1153. [Google Scholar]
- Pohnert, G. Wound-activated chemical defense in unicellular planktonic algae. Agnew Chem Int Ed 2000, 39, 4352–4354. [Google Scholar]
- Leflaive, J; Ten-Hage, L. Chemical interactions in diatoms: role of polyunsaturated aldehydes and precursors. New Phytol 2009, 184, 794–805. [Google Scholar]
- Pohnert, G; Steinke, M; Tollrian, R. Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol Evol 2007, 22, 198–204. [Google Scholar]
- Pohnert, G. Chemical noise in the silent ocean. J Plankton Res 2010, 32, 141–144. [Google Scholar]
- Imai, I; Ishida, Y; Hata, Y. Killing of marine phytoplankton by a gliding bacterium Cytophaga sp., isolated from the coastal sea of Japan. Mar Biol 1993, 116, 527–532. [Google Scholar]
- Brussaard, CPD. Viral control of phytoplankton populations – a review. J Eukaryot Microbiol 2004, 51, 125–138. [Google Scholar]
- Mayali, X; Azam, F. Algicidal bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol 2004, 51, 139–144. [Google Scholar]
- Park, MG; Yih, W; Coats, DW. Parasites and phytoplankton, with special emphasis on dinoflagellate infections. J Eukaryot Microbiol 2004, 51, 145–155. [Google Scholar]
- Desbois, AP. Antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Ph.D. thesis, University of St Andrews, UK, 2007. [Google Scholar]
- Desbois, AP; Mearns-Spragg, A; Smith, VJ. A fatty acid from the diatom Phaeodactylum tricornutum is antibacterial against diverse bacteria including multi-resistant Staphylococcus aureus (MRSA). Mar Biotechnol 2009, 11, 45–52. [Google Scholar]
- Desbois, AP; Yan, L; Lebl, T; Smith, VJ. Isolation and structural characterisation of two antibacterial free fatty acids from the marine diatom Phaeodactylum tricornutum. Appl Microbiol Biotechnol 2008, 81, 755–764. [Google Scholar]
- Findlay, JA; Patil, AD. Antibacterial constituents of the diatom Navicula delognei. J Nat Prod 1984, 47, 815–818. [Google Scholar]
- Ohta, S; Chang, T; Kawashima, A; Nagate, T; Murase, M; Nakanishi, H; Miyata, H; Kondo, M. Anti methicillin-resistant Staphylococcus aureus (MRSA) activity by linolenic acid isolated from the marine microalga Chlorococcum HS-101. Bull Environ Contam Toxicol 1994, 52, 673–680. [Google Scholar]
- Benkendorff, K; Davis, AR; Rogers, CN; Bremner, JB. Free fatty acids and sterols in the benthic spawn of aquatic molluscs, and their associated antimicrobial properties. J Exp Mar Biol Ecol 2005, 316, 29–44. [Google Scholar]
- Clément, M; Tremblay, J; Lange, M; Thibodeau, J; Belhumeur, P. Whey-derived free fatty acids suppress the germination of Candida albicans in vitro. FEMS Yeast Res 2007, 7, 276–285. [Google Scholar]
- Desbois, AP; Walton, MJ; Smith, VJ. Differential antibacterial activities of fusiform and oval morphotypes of Phaeodactylum tricornutum (Bacillariophyceae). J. Mar. Biol. Assoc. UK. in press. [CrossRef]
- Willett, NP; Morse, GE. Long-chain fatty acid inhibition of growth of Streptococcus agalactiae in a chemically defined medium. J Bacteriol 1966, 91, 2245–2250. [Google Scholar]
- Thompson, L; Cockayne, A; Spiller, RC. Inhibitory effect of polyunsaturated fatty acids on the growth of Helicobacter pylori: a possible explanation of the effect of diet on peptic ulceration. Gut 1994, 35, 1557–1561. [Google Scholar]
- Wille, JJ; Kydonieus, A. Palmitoleic acid isomer (C16:1Δ6) in human skin sebum is effective against Gram-positive bacteria. Skin Pharmacol Appl Skin Physiol 2003, 16, 176–187. [Google Scholar]
- Ko, HL; Heczko, PB; Pulverer, G. Differential susceptibily of Propionibacterium acnes, P. granulosum and P. avidum to free fatty acids. J Invest Dermatol 1978, 71, 363–365. [Google Scholar]
- Sheu, CW; Freese, E. Effects of fatty acids on growth and envelope proteins of Bacillus subtilis. J Bacteriol 1972, 111, 516–524. [Google Scholar]
- Leekumjorn, S; Cho, HJ; Wu, Y; Wright, NTAKAK; Chan, C. The role of fatty acid unsaturation in minimizing biophysical changes on the structure and local effects of bilayer membranes. Biochim Biophys Acta 2009, 1788, 1508–1516. [Google Scholar]
- Cutignano, A; d’Ippolito, G; Romano, G; Lamari, N; Cimino, G; Febbraio, F; Nucci, R; Fontana, A. Chloroplastic glycolipids fuel aldehyde biosynthesis in the marine diatom Thalassiosira rotula. ChemBioChem 2006, 7, 450–456. [Google Scholar]
- d’Ippolito, G; Cutignano, A; Briante, R; Febbraio, F; Cimino, G; Fontana, A. New C16 fatty-acid-based oxylipin pathway in the marine diatom Thalassiosira rotula. Org Biomol Chem 2005, 3, 4065–4070. [Google Scholar]
- d’Ippolito, G; Cutignano, A; Tucci, S; Romano, G; Cimino, G; Fontana, A. Biosynthetic intermediates and stereochemical aspects of aldehyde biosynthesis in the marine diatom Thalassiosira rotula. Phytochemistry 2006, 67, 314–322. [Google Scholar]
- Blée, E. Phytooxylipins and plant defense reactions. Prog Lipid Res 1998, 37, 33–72. [Google Scholar]
- Barofsky, A; Pohnert, G. Biosynthesis of polyunsaturated short chain aldehydes in the diatom Thalassiosira rotula. Org Lett 2007, 9, 1017–1020. [Google Scholar]
- Fontana, A; d’Ippolito, G; Cutignano, A; Miralto, A; Ianora, A; Romano, G; Cimino, G. Chemistry of oxylipin pathways in marine diatoms. Pure Appl Chem 2007, 79, 481–490. [Google Scholar]
- Christie, WW. Plant oxylipins: Chemistry and biology [online]. Available from: http://lipidlibrary.aocs.org/Lipids/eicplant/file.pdf (Accessed 14 December 2009).
- Wichard, T; Poulet, SA; Halsband-Lenk, C; Albaina, A; Harris, R; Liu, D; Pohnert, G. Survey of the chemical defence potential of diatoms: screening of fifty one species for α,β,γ,δ-unsaturated aldehydes. J Chem Ecol 2005, 31, 949–958. [Google Scholar]
- Prost, I; Dhondt, S; Rothe, G; Vicente, J; Rodriguez, MJ; Kift, N; Carbonne, F; Griffiths, G; Esquerré-Tugayé, MT; Rosahl, S; Castresana, C; Hamberg, M; Fournier, J. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 2005, 139, 1902–1913. [Google Scholar]
- Bisignano, G; Grazia Laganà, M; Trombetta, D; Arena, S; Nostro, A; Uccella, N; Mazzanti, G; Saija, A. In vitro antibacterial activity of some aliphatic aldehydes from Olea europaea L. FEMS Microbiol Lett 2001, 198, 9–13. [Google Scholar]
- Adolph, S; Bach, S; Blondel, M; Cueff, A; Moreau, M; Pohnert, G; Poulet, SA; Wichard, T; Zuccaro, A. Cytotoxicity of diatom-derived oxylipins in organisms belonging to different phyla. J Exp Biol 2004, 207, 2935–2946. [Google Scholar]
- Ribalet, F; Intertaglia, L; Lebaron, P; Casotti, R. Differential effect of three polyunsaturated aldehydes on marine bacterial isolates. Aquat Toxicol 2008, 86, 249–255. [Google Scholar]
- Fernandes, JMO; Smith, VJ. Partial purification of antibacterial proteinaceous factors from erythrocytes of Oncorhynchus mykiss. Fish Shellfish Immunol 2004, 16, 1–9. [Google Scholar]
- Destoumieux-Garzón, D; Saulnier, D; Garnier, J; Jouffrey, C; Bulet, P; Bachère, E. Crustacean immunity. Antifungal peptides are generated from the C-terminus of shrimp hemocyanin in response to microbial challenge. J Biol Chem 2001, 276, 47070–47077. [Google Scholar]
- Service, M; Wardlaw, AC. Echinochrome-A as a bactericidal substance in the coelomic fluid of Echinus esculentus (L.). Comp Biochem Physiol 1984, 79B, 161–165. [Google Scholar]
- Söderhäll, K; Ajaxon, R. Effects of quinines and melanin on mycelial growth of Aphanomyces spp. and extracellular protease of Aphanomyces astaci, a parasite of crayfish. J Invertebr Pathol 1982, 39, 105–109. [Google Scholar]
- Nappi, AJ; Ottaviani, E. Cytotoxicity and cytotoxic molecules in invertebrates. Bioessays 2000, 22, 469–480. [Google Scholar]
- Lin, LC; Chen, WT. Antimicrobial and photoactive properties of melanin from various sources of animal. Taiwanese J Agric Chem Food Sci 2004, 42, 315–320. [Google Scholar]
- Burkhart, CG; Burkhart, CN. The mole theory: primary function of melanocytes and melanin may be antimicrobial defense and immune modulation (not solar protection). Int J Dermatol 2005, 44, 340–342. [Google Scholar]
- Prota, G; Ortonne, JP; Voulot, C; Khatchadourian, C; Nardi, G; Palumbo, A. Occurrence and properties of tyrosinase in the ejected ink of cephalopods. Comp Biochem Physiol 1981, 68B, 415–419. [Google Scholar]
- Hata, S; Azumi, K; Yokosawa, H. Ascidian phenoloxidase: its release from hemocytes, isolation, characterisation and physiological roles. Comp Biochem Physiol 1998, 119B, 769–776. [Google Scholar]
- Li, G; Zhang, S; Li, H. Ultrastructural localization and antibacterial activity of phenoloxidase in amphioxus Branchiostoma belcheri tsingtauense. J Mar Biol Assoc UK 2001, 81, 705–706. [Google Scholar]
- Yamazaki, M; Ohye, H; Kisugi, J; Kamiya, H. Bacteriostatic and cytolytic activity of purple fluid from the sea hare. Dev Comp Immunol 1990, 14, 379–383. [Google Scholar]
- Yamazaki, M. Antitumour and antimicrobial glycoproteins from sea hares. Comp Biochem Physiol 1993, 105C, 141–146. [Google Scholar]
- Derby, CD. Escape by inking and secreting: marine molluscs avoid predators through a rich array of chemicals and mechanisms. Biol Bull 2007, 213, 274–289. [Google Scholar]
- Kitani, Y; Tsukamoto, C; Zhang, C; Nagai, H; Ishida, M; Ishizaki, S; Shimakura, K; Shiomi, S; Nagashima, S. Identification of an antibacterial protein as L-amino acid oxidase in the skin mucus of rockfish Sebastes schlegeli. FEBS J 2007, 274, 125–136. [Google Scholar]
- Jørgensen, EG. Antibiotic substances from cells and culture solutions of unicellular algae with special reference to some chlorophyll derivatives. Physiol Plant 1962, 15, 530–545. [Google Scholar]
- Hansen, JA. Antibiotic activity of the chrysophyte Ochromonas malhamensis. Physiol Plant 1973, 29, 234–238. [Google Scholar]
- Bruce, DL; Duff, DCB; Antia, NJ. The identification of two antibacterial products of the marine planktonic alga Isochrysis galbana. J Gen Microbiol 1967, 48, 293–298. [Google Scholar]
- Kristan, KC; Viero, G; Dallo Serra, M; Macěk, P; Anderluh, G. Molecular mechanism of pore formation by actinoporins. Toxicon 2009, 54, 1125–1134. [Google Scholar]
- Álvarez, C; Mancheño, JM; Martínez, D; Tejuca, M; Pazos, F; Lanio, ME. Sticholysins, two pore-forming toxins produced by the Caribbean Sea anemone Stichodactyla helianthus: Their interaction with membranes. Toxicon 2009, 54, 1135–1147. [Google Scholar]
- Yokota, H; Nagashima, Y; Shiomi, K. Interaction of grammistins with lipids and their antibacterial activity. Fish Sci 2001, 67, 928–933. [Google Scholar]
- Sugiyama, N; Araki, M; Ishida, M; Nagashima, Y; Shiomi, K. Further isolation and characterization of grammistins from the skin secretion of the soapfish Grammistes sexlineatus. Toxicon 2005, 45, 595–601. [Google Scholar]
- Tasiemski, A; Verger-Bocquet, M; Cadet, M; Goumon, Y; Metz-Boutigue, MH; Aunis, D; Stefano, GB; Salzet, M. Proenkephalin A-derived peptides in invertebrate innate immune processes. Mol Brain Res 2000, 76, 237–252. [Google Scholar]
- Concha, MI; Smith, VJ; Castro, K; Bastias, A; Romero, A; Amthauer, R; Apolipoproteins, A-I. A-II are potentially important effectors of innate immunity in the teleost fish Cyprinus carpio. Eur J Biochem 2004, 271, 2984–2990. [Google Scholar]
- Villaroel, F; Bastías, A; Casandro, A; Amthauer, R; Concha, MI. Apolipoprotein A-I, an antimicrobial protein in Oncorhynchus mykiss: evaluation of its expression in primary defence barriers and plasma levels in sick and healthy fish. Fish Shellfish Immunol 2007, 23, 197–209. [Google Scholar]
- Liao, WR; Lin, JY; Shieh, WY; Jeng, WL; Huang, R. Antibiotic activity of lectins from marine algae against marine vibrios. J Ind Microbiol Biotech 2003, 30, 433–439. [Google Scholar]
- Schröder, HC; Ushijima, H; Krasko, A; Gamulin, V; Thakur, NL; Diehl-Seifert, B; Müller, IM; Müller, WEG. Emergence and disappearance of an immune molecule, an antimicrobial lectin, in basal metazoa. J Biol Chem 2003, 278, 32810–32817. [Google Scholar]
- Moura, RM; Queiroz, AFS; Fook, JMSLL; Dias, ASF; Monteiro, NKV; Ribeiro, JKC; Moura, GEDD; Macedo, LLP; Santos, EA; Sales, MP. CvL, a lectin from the marine sponge Cliona varians: Isolation, characterization and its effects on pathogenic bacteria and Leishmania promastigotes. Comp Biochem Physiol 2006, 145A, 517–523. [Google Scholar]
- Tunkijjanukij, S; Olafsen, JA. Sialic acid-binding lectin with antibacterial activity from the horse mussel: Further characterization and immunolocalization. Dev Comp Immunol 1998, 22, 139–150. [Google Scholar]
- Chattopadhyay, T; Chatterjee, BP. A low molecular weight lectin from the edible crab Scylla serrata hemolymph: purification and partial characterization. Biochem Arch 1993, 9, 65–72. [Google Scholar]
- Saito, T; Kawabata, SI; Hirata, M; Iwanaga, S. A novel type of Limulus lectin-L6 – Purification, primary structure, and antibacterial activity. J Biol Chem 1995, 270, 14493–14499. [Google Scholar]
- Gowda, NM; Goswami, U; Khan, MI. T-antigen binding lectin with antibacterial activity from marine invertebrate, sea cucumber (Holothuria scabra): Possible involvement in differential recognition of bacteria. J Invertebr Pathol 2008, 99, 141–145. [Google Scholar]
- Morita, T; Ohtsubo, S; Nakamura, T; Tanaka, S; Iwanaga, S; Ohashi, K; Niwa, M. Isolation and biological activities of Limulus anticoagulant (anti-LPS factor) which interacts with lipopolysaccharide (LPS). J Biochem 1985, 97, 1611–1620. [Google Scholar]
- Hobson, G; Hirsch, JG. The antibacterial activity of hemoglobin. J Exp Med 1958, 107, 167–183. [Google Scholar]
- Parish, CA; Jiang, H; Tokiwa, Y; Berova, N; Nakanishi, K; McCabe, D; Zuckerman, W; Xia, MM; Gabay, JE. Broad spectrum antimicrobial activity of haemoglobin. Bioorg Med Chem 2001, 9, 377–382. [Google Scholar]
- Liepke, C; Baxmann, S; Heine, C; Breithaupt, N; Ständker, L; Forssmann, W-G. Human haemoglobin-derived peptides exhibit antimicrobial activity: a class of defense peptides. J Chromatogr B 2003. [Google Scholar]
- Ullal, AJ; Litaker, RW; Noga, EJ. Antimicrobial peptides derived from haemoglobin are expressed in epithelium of channel catfish, Ictalurus punctata Rafinesque. Dev Comp Immunol 2008, 32, 1301–1312. [Google Scholar]
- Pan, J-Y; Zhang, Y-L; Wang, S-Y; Peng, X-X. Dodecamer is required for agglutination of Litopenaeus vannamei hemocyanin with bacterial cells and red blood cells. Mar Biotechnol 2008, 10, 645–652. [Google Scholar]
- Zhang, XB; Huang, CH; Qin, QW. Antiviral properties of hemocyanin isolated from shrimp Penaeus monodon. Antiviral Res 2004, 61, 93–99. [Google Scholar]
- Lee, SY; Lee, BL; Söderhäll, K. Processing of an antibacterial peptide from haemocyanin of the freshwater crayfish Pacifastacus leniusculus. J Biol Chem 2003, 278, 7927–7933. [Google Scholar]
- Lebedev, AV; Ivanova, M; Levitsky, DO. Echinochrome, a naturally occurring iron chelator and free radical scavenger in artificial and natural membrane systems. Life Sci 2005, 76, 863–875. [Google Scholar]
- Smith, VJ. Ratcliffe, NA, Rowley, AF, Eds.; Echinoderms. In Invertebrate Blood Cells; Academic Press: London, UK, 1981; Volume 2, pp. 513–561. [Google Scholar]
- Nosanchuk, JD; Casadevall, A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Chemother 2006, 50, 3519–3528. [Google Scholar]
- Nagai, T; Osaki, T; Kawabata, S. Functional conversion of hemocyanin by horseshoe crab antimicrobial peptides. J Biol Chem 2001, 276, 27166–27170. [Google Scholar]
- Fagutao, FF; Koyama, T; Kaizu, A; Saito-Taki, T; Kordo, H; Aoki, T; Hirono, I. Increased bacterial load in shrimp hemolymph in the absence of prophenoloxidase. FEBS J 2009, 276, 5298–5306. [Google Scholar]
- Lee, SY; Lee, BL; Söderhäll, K. Processing of hemocyanin subunits into prophenoloxidase. Biochem Biophys Res Comm 2004, 322, 490–496. [Google Scholar]
- Mochizuki, A. An antiseptic effect of cuttlefish ink. Nippon Suisan Gakkaishi 1979, 45, 1401–1403. [Google Scholar]
- Shen, TY; Chou, CC. Antimicrobial activity of squid ink. J Chin Agric Chem Soc 1990, 28, 59–68. [Google Scholar]
- Kamiya, H; Muramoto, K; Yamazaki, M. Aplysianin-A, an antibacterial and antineoplastic glycoprotein in the albumen gland of sea hare Aplysia kurodai. Cell Mol Life Sci 1986. [Google Scholar]
- Iijima, R; Kisugi, J; Yamazaki, M. Antifungal activity of aplysianin E, a cytotoxic protein of sea hare (Aplysia kuodai) eggs. Dev Comp Immunol 1995, 19, 13–19. [Google Scholar]
- Zuliani, JP; Kayano, AM; Zaqueo, KD; Neto, AC; Sampaio, SV; Soares, AM; Stabeli, RG. Snake venom L-amino acid oxidases: some considerations about their functional characterization. Protein Pept Lett 2009, 16, 908–912. [Google Scholar]
- Yang, H; Johnson, PM; Ko, KC; Kamio, M; Germann, MW; Derby, CD; Tai, PC. Cloning, characterisation and expression of escapin, a broadly antimicrobial FAD-containing L-amino acid oxidase from the ink of sea hare Aplysia californica. J Exp Biol 2005, 208, 3609–3622. [Google Scholar]
- Anderluh, G; Dalla Serra, M; Viero, G; Guella, G; Macěk, P; Menestrina, G. Pore formation by equinatoxin II, a eukaryotic protein toxin, occurs by induction of nonlamellar lipid structures. J Biol Chem 2003, 278, 45216–45223. [Google Scholar]
- Anderluh, G; Lakey, JH. Disparate proteins use similar architectures to damage membranes. Trends Biochem Sci 2008, 33, 482–490. [Google Scholar]
- Brogden, KA; Guthmiller, JM; Salzet, M; Zasloff, M. The nervous system and innate immunity: the neuropeptide connection. Nat Immunol 2005, 6, 558–564. [Google Scholar]
- Salzet, M; Tasiemski, A. Involvement of pro-enkephalin-derived peptides in immunity. Dev Comp Immunol 2001, 25, 177–185. [Google Scholar]
- Tada, N; Sakamoto, T; Kagami, A; Mochizuki, K; Kurosaka, K. Antimicrobial activity of lipoprotein particles containing apolipoprotein AI. Mol Cell Biochem 1993, 119, 171–178. [Google Scholar]
- Motizuki, M; Itoh, T; Yamada, M; Shimamura, S; Tsurugi, K. Purification, primary structure, and antimicrobial activities of bovine apolipoprotein A-II. J Biochem 1998, 123, 675–679. [Google Scholar]
- Motizuki, M; Itoh, T; Satoh, T; Yokoto, S; Yamada, M; Shimamura, S; Samejima, T; Tsurugi, K. Lipid-binding and antimicrobial properties of synthetic peptides of bovine apolipoprotein A-II. Biochem J 1999, 342, 215–221. [Google Scholar]
- Shiflett, AM; Bishop, JR; Pahwa, A; Hajduk, SL. Human high-density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem 2005, 280, 32578–32585. [Google Scholar]
- Ando, S; Mori, Y. Characteristics of serum lipoprotein features associated with lipid levels of muscle and liver from five species of fish. Nippon Suisan Gakkaishi 1993, 59, 1565–1571. [Google Scholar]
- Concha, MI; Molina, S; Oyarzun, C; Villanueva, J; Amthauer, R. Local expression of apolipoprotein A-I gene and possible role for HDL in primary defence in carp skin. Fish Shellfish Immunol 2003, 14, 259–275. [Google Scholar]
- Johnston, LD; Brown, G; Gauthier, D; Reece, K; Kator, H; van Veld, P. Apolipoprotein A-I from striped bass (Morone saxatilis) demonstrates antibacterial activity in vitro. Comp Biochem Physiol 2008, 151B, 167–175. [Google Scholar]
- Tunkijjanukij, S; Giaever, H; Chin, CC; Olafsen, JA. Sialic acid in hemolymph and affinity purified lectins from two marine bivalves. Comp Biochem Physiol 1998, 119B, 705–713. [Google Scholar]
- Takayama, S; Saito, E; Kimizuka, R; Yamada, S; Kato, T. Effect of eel galectin AJL-1 on periodontopathic bacterial biofilm formation and their lipopolysaccharide-mediated inflammatory cytokine induction. Int J Antimicrob Agents 2009, 34, 355–359. [Google Scholar]
- Ngai, PHK; Ng, TB. A mannose-specific tetrameric lectin with mitogenic and antibacterial activities from the ovary of a teleost, the cobia (Rachycentron canadum). Appl Microbiol Biotechnol 2007, 74, 433–438. [Google Scholar]
- Tanaka, S; Nakamura, T; Morita, T; Iwanaga, S. Limulus anti-LPS factor; an anticoagulant which inhibits endotoxin-mediated activation of Limulus coagulation system. Biochem Biophys Res Comm 1982. [Google Scholar]
- Hoess, A; Watson, S; Siber, GR; Liddington, R. Crystal structure of an endotoxin neutralizing protein from horseshoe crab, Limulus anti-LPS factor, at 1.5 A resolution. EMBO J 1993, 12, 3351–3356. [Google Scholar]
- Aketagawa, J; Miyata, T; Ohtsubo, S; Nakamura, T; Morita, T; Hayashida, H; Miyata, T; Iwanaga, S; Takao, T; Shimonishi, Y. Primary structure of Limulus anticoagulant and anti-lipopolysaccharide factor. J Biol Chem 1986, 261, 7357–7365. [Google Scholar]
- Muta, T; Miyata, T; Tokunaga, F; Nakamura, T; Iwanaga, S. Primary structure of anti-lipopolysaccharide factor from American horseshoe crab, Limulus Polyphemus. J Biol Chem 1987, 101, 1321–1330. [Google Scholar]
- Iwanaga, S; Kawabata, S; Muta, T. New types of clotting factors and defense molecules found in the horseshoe crab hemolymph: Their structures and functions. J Biochem 1998, 123, 1–15. [Google Scholar]
- Liu, FS; Liu, YC; Li, FH; Dong, B; Xiang, JH. Molecular cloning and expression profile of putative antilipopolysaccharide factor (ALF) in Chinese shrimp (Fenneropenaeus chinensis). Mar Biotechnol 2005, 7, 600–608. [Google Scholar]
- De la Vega, E; O’Leary, NA; Shockey, JE; Robalino, J; Payne, C; Browdy, CL; Warr, GW; Gross, PS. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): A broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection. Mol Immunol 2008, 45, 1916–1925. [Google Scholar]
- Rosa, RD; Stoco, PH; Barracco, MA. Cloning and characterisation of cDNA sequences encoding for anti-lipopolysaccharide factors (ALFs) in Brazilian palaemonid and penaeid shrimps. Fish Shellfish Immunol 2008, 25, 693–696. [Google Scholar]
- Beale, KM; Towle, DW; Jayasundara, N; Smith, CM; Shields, JD; Small, HJ; Greenwood, SJ. Anti-lipopolysaccharide factors in the American lobster Homarus americanus: Molecular characterization and transcriptional response to Vibrio fluvialis challenge. Comp Biochem Physiol 2008, 3D, 263–269. [Google Scholar]
- Li, CH; Zhao, JM; Song, LS; Mu, CK; Zhang, H; Gai, YC; Qiu, LM; Yu, YD; Ni, DJ; Xing, KZ. Molecular cloning, genomic organization and functional analysis of an anti-lipopolysaccharide factor from Chinese mitten crab Eriocheir sinensis. Dev Comp Immunol 2008, 32, 784–794. [Google Scholar]
- Vallespi, MG; Glaria, LA; Reyes, O; Garay, HA; Ferreo, J; Araña, MJ. A Limulus antilipopolysaccharide factor-derived peptide exhibits a new immunological activity with potential application in infectious diseases. Clin Diagn Lab Immunol 2000, 7, 669–675. [Google Scholar]
- Vallespi, MG; Alvarez-Obregon, JC; Rodriguez-Alonso, I; Montero, T; Garay, HA; Reyes, O; Araña, MJ. A Limulus anti-LPS factor-derived peptide modulates cytokine gene expression and promotes resolution of bacterial acute infection in mice. Int Immunopharmacol 2003, 3, 247–256. [Google Scholar]
- Brinkmann, V; Reichard, U; Goosmann, C; Fauler, B; Uhlemann, Y; Weiss, DS; Weinrauch, Y; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar]
- Fuchs, TA; Abed, U; Goosmann, C; Hurwitz, R; Schulze, I; Wahn, V; Weinrauch, Y; Brinkmann, V; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 2007, 176, 231–241. [Google Scholar]
- Köckritz-Blickwede, M; Nizet, V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med 2009, 87, 775–783. [Google Scholar]
- Von Köckritz-Blickede, M; Goldmann, O; Thulin, P; Heinemann, K; Norrby-Teglund, A; Rohde, M; Medina, E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 2008, 111, 3070–3080. [Google Scholar]
- Yousefi, S; Gold, JA; Andina, N; Lee, JJ; Kelly, AM; Kozlowski, E; Schmid, I; Straumann, A; Reichenbach, J; Gleich, GJ; Simon, H-U. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med 2008, 14, 949–953. [Google Scholar]
- Palić, D; Andreasen, CB; Ostojić, J; Tell, RM; Roth, JA. Zebrafish (Danio rerio) whole kidney assays to measure neutrophil extracellular trap release and degranulation of primary granules. J Immunol Methods 2007, 319, 87–97. [Google Scholar]
- Palić, D; Ostojić, J; Andreasen, CB; Roth, JA. Fish cast NETs: neutrohil extracellular traps are released from fish neutrophils. Dev Comp Immunol 2007, 31, 805–816. [Google Scholar]
- Altincicek, B; Stotzelm, S; Wygrecka, M; Preissner, KT; Vilcinskas, A. Host-derived extracellular nucleic acids enhance innate immune responses, induce coagulation and prolong survival upon insects. J Immunol 2008, 181, 2705–2712. [Google Scholar]
- Class, R; Zeppezauer, M. Antimicrobial histone H1 compositions, kits, and methods of use thereof. Official Gazette of the United States Patent and Trademark Office Patents. US 06,884,423, 2005. [Google Scholar]
- Pérez-Playá, E; Houghton, RA; Blondelle, SE. The role of amphipathicity in the folding self-association and biological activity of multiple subunit proteins. J Biol Chem 1995, 270, 1048–1056. [Google Scholar]
- Tam, JP; Lu, YA; Yang, JL. Design of salt-insensitive glycine-rich antimicrobial peptides with cyclic tricystine structures. Biochemistry 2000, 39, 7159–7169. [Google Scholar]
- Park, IY; Cho, JH; Kim, S; Kim, YB; Kim, MS; Kim, SC. Helix stability confers salt resistance upon helical antimicrobial peptides. J Biol Chem 2004, 279, 13896–13901. [Google Scholar]
- Uyterhoeven, ET; Butler, CH; Ko, D; Elmore, DE. Investigating the nucleic acid interactions and antimicrobial mechanism of buforin II. FEBS Lett 2008, 582, 1715–1718. [Google Scholar]
- Kawabata, SI; Nagayama, R; Hirata, M; Shigenaga, T; Agarwala, KL; Saito, T; Cho, J; Nakajima, H; Takagi, T; Iwanaga, S. Tachysin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem 1996, 120, 1253–1260. [Google Scholar]
- Fernandes, JMO; Kemp, GD; Smith, VJ. Two novel muramidases from skin mucosa of rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol 2004, 138B, 53–64. [Google Scholar]
- Thormar, H; Hilmarsson, H. The role of microbicidal lipids in host defense against pathogens and their potential as therapeutic agents. Chem Phys Lipids 2007, 150, 1–11. [Google Scholar]
- Drake, DR; Brogden, KA; Dawson, DV; Wertz, PW. Antimicrobial lipids at the skin surface. J Lipid Res 2008, 49, 4–11. [Google Scholar]
- United States Food and Drug Administration. Substances affirmed as generally recognized as safe: menhaden oil. Federal Register 1997, 62, 30751–30757.
- Lacey, RW; Lord, VL. Sensitivity of staphylococci to fatty acids: novel inactivation of linolenic acid by serum. J Med Microbiol 1981, 14, 41–49. [Google Scholar]
- Petschow, BW; Batema, RP; Ford, LL. Susceptibility of Helicobacter pylori to bactericidal properties of medium-chain monoglycerides and free fatty acids. Antimicrob Agents Chemother 1996, 40, 302–306. [Google Scholar]
- Lukowski, G; Lindequist, U; Mundt, S; Kramer, A; Jülich, WD. Inhibition of dermal MRSA colonisation by microalgal micro- and nanoparticles. Skin Pharmacol Physiol 2008, 21, 98–105. [Google Scholar]
- Sun, CQ; O’Connor, CJ; Roberton, AM. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol Med Microbiol 2003, 36, 9–17. [Google Scholar]
- Neyts, J; Kristmundsdóttir, T; De Clercq, E; Thormar, H. Hydrogels containing monocaprin prevent intravaginal and intracutaneous infections with HSV-2 in mice: impact on the search for vaginal microbicides. J Med Virol 2000, 61, 107–110. [Google Scholar]
- Clarke, SR; Mohamed, R; Bian, L; Routh, AF; Kokai-Kun, JF; Mond, JJ; Tarkowski, A; Foster, SJ. The Staphylococcus aureus surface protein isdA mediates resistance to innate defenses of human skin. Cell Host Microb 2007, 1, 1–14. [Google Scholar]
- Bachère, E; Gueguen, Y; Gonzalez, M; de Lorgeril, J; Garnier, J; Romestand, B. Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunol Rev 2004, 198, 149–168. [Google Scholar]
- Destoumieux, D; Munoz, M; Bulet, P; Bachère, E. Penaeidins, a new family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda). Cell Mol Life Sci 2000, 57, 1260–1271. [Google Scholar]
- Yang, Y; Poncet, J; Garnier, J; Zatylny, C; Bachère, E; Aumelas, A. Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide. J Biol Chem 2003, 278, 36859–36867. [Google Scholar]
- Cuthbertson, BJ; Yang, Y; Bachère, E; Bullesbach, EE; Gross, PS; Aumelas, A. Solution structure of synthetic penaeidin-4 with structural and functional comparisons with penaeidin-3. J Biol Chem 2005, 280, 16009–16018. [Google Scholar]
| Taxon | Peptide/peptide family | Key reference |
|---|---|---|
| Cnidaria (Scyphozoa) | Aurelin | Ovchinnikova et al. [6] |
| Annelida (Polychaeta) | Arenicin | Ovchinnikova et al. [7] |
| Hedistin | Tasiemski et al. [8] | |
| Perinerin | Pan et al. [9] | |
| Mollusca (Bivalvia) | Big defensins 1 | Li et al. [10] |
| Cg-prp | Li et al. [10] | |
| Defensins 1 | Li et al. [10] | |
| Myticins 1 | Li et al. [10] | |
| Mytilins 1 | Li et al. [10] | |
| Mytimycin | Li et al. [10] | |
| Crustacea (Decapoda) | Arasin-1 | Stensvåg et al. [11] |
| Bac-like | Schnapp et al. [12] | |
| Callinectin | Khoo et al. [13] | |
| Crustins 1 | Smith et al. [14] | |
| Homarin | Battison et al. [15] | |
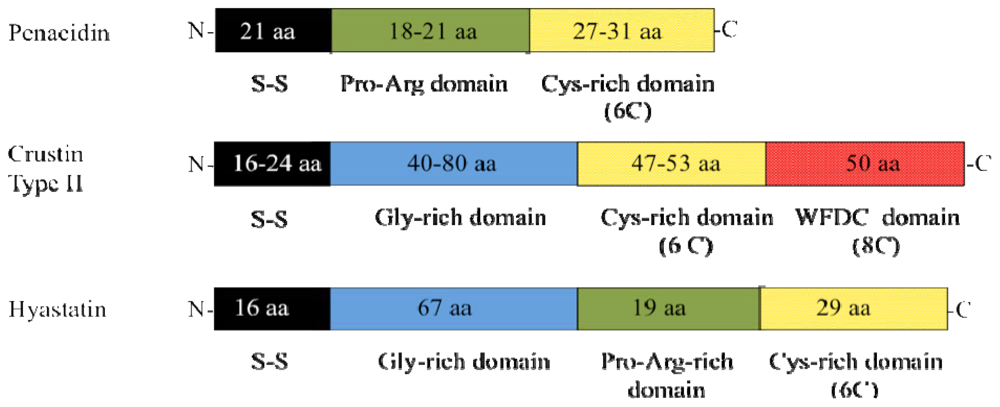
| Hyastatin | Spersted et al. [16] | |
| Penaeidins 1 | Cuthbertson et al. [17] | |
| Scygonadin | Huang et al. [18] | |
| Scylla serrata antimicrobial protein | Yedery and Reddy [19] | |
| Chelicerata | Big defensin | Saito et al. [20] |
| Polyphemusins 1 | Miyata et al. [21] | |
| Tachycitin | Kawabata et al. [22] | |
| Tachyplesins 1 | Miyata et al. [21] | |
| Tachystatins 1 | Osaki et al. [23] | |
| Echinodermata (Echinoidea) | Strongylocins 1 | Li et al. [24] |
| Urochordata (Ascidiacea) | Clavanins 1 | Lee et al. [25] |
| Dicynthaurin | Lee et al. [26] | |
| Halocidin | Jang et al. [27] | |
| Halocyamines 1 | Azumi et al. [28] | |
| Halycyntin | Galinier et al. [29] | |
| Papillosin | Galinier et al. [29] | |
| Styelins 1 | Lee et al. [30] | |
| Chordata (Pisces) | Cathelicidins 1 | Smith and Fernandes [31] |
| Defensins 1 | Smith and Fernandes [31] | |
| Hepcidins | Smith and Fernandes [31] | |
| Liver-expressed antimicrobial peptides (LEAPs) 1 | Smith and Fernandes [31] | |
| Piscidins 1 | Smith and Fernandes [31] | |
| Categories of AMPs | Examples | Activity 1 | |
|---|---|---|---|
| Linear, α-helical | Clavanins (Ascidians) | G+, G−, F | |
| Dicynthaurin (Ascidians) | G+, G−, H | ||
| Halocyntin (Ascidians) | G+, G− | ||
| Papillosin (Ascidians) | G+, G− | ||
| Piscidins (Fish) | G+, G−, F, H | ||
| Styelins (Ascidians) | G+, G−, H | ||
| Cysteine-rich | No. disulphide bonds | ||
| 2 | Cathelicidins (Fish) | G+, G− | |
| LEAPs (Fish) | G−, F | ||
| Tachyplesins (Horseshoe crabs) | G+, G−, F | ||
| Polyphemusins (Horseshoe crabs) | G+, G−, F | ||
| 3 | Aurelin (Jellyfish) | G+, G− | |
| Big defensins (Horseshoe crabs) | G+, G−, F | ||
| Penaeidins (Shrimp) | G+, G−, F, Cb | ||
| Strongylocins (Sea urchins) | G+, G− | ||
| Tachystatin (Horseshoe crabs) | G+, G−, F, H 2, Cb | ||
| 4 | Defensins (Molluscs) | G+, G−3, F | |
| LEAPs (Fish) | G−, F | ||
| Myticins (Molluscs) | G+, G−4, F 4 | ||
| Mytilins (Molluscs) | G+, G−, F 5 | ||
| Crustins (WAP domain; crabs) | G+ | ||
| 5 | Tachycitin (Horseshoe crabs) | G+, G−, F, Cb | |
| Cationic peptides: specific amino acid enriched | Arasin-1 (Spider crab) (proline and arginine rich) Bac-like (Crab) (proline rich) Cg-prp (Oyster) (proline rich) Hyastatin (Spider crab) (glycine rich) Penaeidins (Shrimp) (proline and cysteine rich) | G+, G− G+, G− Synergises with defensin G+, G−, F, Cb G+, F, Cb | |
| Miscellaneous | Arenicin (Polychaete) Hedistin (Polychaete) Perinerin (Polychaete) | G+, G−, F G+, G− G+, G−, F | |
| Protein/peptide | Location | Active factor | Reported activity 1 | Source | Key reference |
|---|---|---|---|---|---|
| Histone H1 | Nucleus | Whole protein (20.7 kDa) | G− | Salmon | Richards et al. [104] |
| N-terminus (26 aa) (HSDF-1) | G− | Salmon | Patrzykat et al. [103] | ||
| C-terminus (69 aa) (oncorhyncin II) | G+, G− | Rainbow trout | Fernandes et al. [107] | ||
| Fragment (not specified) | G+ | Shrimp | Patat et al. [108] | ||
| Histone H2A | Nucleus | Whole protein (13.5 kDa) | G+, F | Channel catfish | Robinette et al. [102] |
| Whole protein (13.5 kDa) | G+, H | Rainbow trout | Fernandes et al. [105] | ||
| Whole protein (13.5 kDa) | G+, G− | Shrimp | Patat et al. [108] | ||
| N-terminus (51 aa) (hipposin) | G+, G− | Halibut | Birkemo et al. [109] | ||
| N-terminus (19 aa) (parasin-1) | G+, G−, F | Catfish | Park et al. [101] | ||
| N-terminus (40 aa) (abhisin) | G+, F, Cy | Abalone | De Zoysa et al. [110] | ||
| Histone H2B | Nucleus | Whole protein (13.8 kDa) | G− | Cod | Bergsson et al. [111] |
| Whole protein (15.5 kDa) | G−, F | Channel catfish | Robinette et al. [102] | ||
| Whole protein (13.5 kDa) | G+ | Shrimp | Patat et al. [108] | ||
| Histone H3 | Nucleus | Whole protein (15.3 kDa) | G+ | Shrimp | Patat et al. [108] |
| Histone H4 | Nucleus | Whole protein (11.3 kDa) | G+ | Shrimp | Patat et al. [108] |
| HMG H6 | Nucleus | Whole protein (6.7 kDa) (oncorhyncin III) | G+, G− | Fish | Fernandes et al. [112] |
| 40Rsp30 | Ribosomes | Whole protein (6.7 kDa) | G+ | Rainbow trout | Fernandes and Smith [113] |
| 60RspL40 | Whole protein (6.4 kDa) | G+, G− | Cod | Bergsson et al. [111] | |
| 60RspL36A | Whole protein (12.3 kDa) | G+ G− | Cod | Bergsson et al. [111] | |
| 60RspL35 | Whole protein (14.2 kDa) | G+ | Cod | Bergsson et al. [111] | |
| Structural feature | Fatty acid | Clear zone area (mm2) |
|---|---|---|
| Carbon chain length: saturated fatty acids | C6:0 | 0 |
| C8:0 | 223 | |
| C9:0 | 1230 | |
| C10:0 | 2260 | |
| C11:0 | 2800 | |
| C12:0 | 5000 | |
| C13:0 | 1230 | |
| C14:0 | 46.9 | |
| C15:0 | 0 | |
| C16:0 | 0 | |
| C17:0 | 0 | |
| C18:0 | 0 | |
| Carbon chain length: monounsaturated fatty acids | C14:1 n-5 | 5000 |
| C16:1 n-7 | 4040 | |
| C18:1 n-9 | 0 | |
| C20:1 n-9t | 584 | |
| C22:1 n-9 | 0 | |
| Degree of unsaturation | C22:1 n-9 | 0 |
| C22:2 n-6 | 584 | |
| C22:3 n-3 | 1230 | |
| C22:4 n-6 | 1930 | |
| C22:6 n-3 | 2090 | |
| Bond orientation(s) | C16:1 n-7 | 4040 |
| C16:1 n-7t | 675 | |
| C18:2 n-9 | 3600 | |
| C18:2 n-9t1 | 1230 | |
| Species, strain and Gram’s stain 1 | Clear zone area (mm2) | |
|---|---|---|
| DD | EPA | |
| Marine isolates | ||
| Aeromonas hydrophila NCIMB 1108(G−) | 483 | 0.0 |
| Alteromonas haloplanktis NCIMB 19 (G−) | 22.0 | 0.0 |
| Listonella anguillarum MT 1637 (G−) | 566 | 15.9 |
| Photobacteriumphosphoreum NCIMB 64 (G−) | 22.0 | 22.0 |
| Psychrobacter immobilisNCIMB 308 (G−) | 84.8 | 0.0 |
| Micrococcus luteus NCIMB 9278 (G+) | 42.6 | 50.3 |
| Planococcus citreus NCIMB 1493 (G+) | 1600 | 50.3 |
| Opportunistic human and animal pathogens | ||
| Escherichia coli B (G−) | 18.9 | 0.0 |
| Pseudomonas aeroginosa NCIMB 10775 (G−) | 10.2 | 0.0 |
| Staphylococcus aureus SH 1000 (G+) | 50.3 | 173 |
| Staphylococcus epidermidis ATCC 10145 (G+) | 22.0 | 105 |
| Name | Main function | Size | Activities 1 | Organism(s) | Key references |
|---|---|---|---|---|---|
| Respiratory pigments | |||||
| Haemoglobin fragments | Respiratory pigment | 28, 41 kDa | G+, G− | Rainbow trout | Fernandes and Smith [174] |
| Haemocyanin fragments | Respiratory pigment | 7.9, 8.3 kDa | F | Shrimp | Destoumieux et al. [175] |
| Other pigments | |||||
| Echinochrome A | Blood pigment | 266 Da | G+, G− | Sea urchins | Service and Wardlaw [176] |
| Melanin | Blood pigment | ~318 kDa | G+, G−, F | Crustaceans | Söderhäll and Ajaxon [177]; Nappi and Ottaviani [178]; Lin and Chen [179]; Burkhart and Burkhart [180] |
| Melanin | Ink pigment | ~318 kDa | G+, G−, F | Octopus | Prota et al. [181] |
| Prophenoloxidase | Enzyme | 60–77 kDa | F | Solitary ascidian | Hata et al. [182] |
| Prophenoloxidase | Enzyme | 60–77 kDa | G+, G− | Amphioxus | Li et al. [183] |
| Aplysianins | Ink component | 60–320 kDa | G+, G−, F, Cy | Sea hares | Yamazaki et al. [184] |
| Dolabellin | Ink component | 60 kDa | G+, G−, Cy | Sea hares | Yamazaki et al. [185] |
| l-amino acid oxidases | Ink component | 340 kDa | G+, G− | Many organisms | Derby [186] |
| Sebastes schlegeli antibacterial protein (l-amino acid oxidase) | Skin mucus | 120 kDa | G− | Fish | Kitani et al. [187] |
| Chlorophyll derivatives | Photosynthetic pigment | ~0.6 kDa | G+, G− | Various micro-algae | Jorgensen [188]; Hansen [189]; Bruce et al. [190] |
| Pore-forming toxins | |||||
| Pardaxin | Skin toxin | 3–4 kDa | G+, G−, H | Flatfish | Lazarovici et al. [48] |
| Actinoporins | Skin toxin | 20 kDa | H, Cy | Sea anemones | Kristan et al. [191] |
| Sticholysins | Skin toxin | 20 kDa | H, Cy | Sea anemones | Alvarez et al. [192] |
| Grammistins | Skin toxin | ~1–3 kDa | G+, G− | Soapfish | Yokota et al. [193]; Sugiyama et al. [194] |
| Neuropeptides | |||||
| Peptide B | Neuropeptide fragment | 2.5–3.5 kDa | G+ | Mussel | Tasiemski et al. [195] |
| Regulatory factors | |||||
| HDL/ApoA-1 | Various functions | 29.5 kDa | G+, G− | Carp | Concha et al. [196] |
| HDL/ApoA-1 | Various functions | 29.5 kDa | G+, G− | Rainbow trout | Villaroel et al. [197] |
| Lectins | |||||
| ESA | Lectin | Not specified | G− | Red alga | Liao et al. [198] |
| GMA | Lectin | Not specified | G− | Red alga | Liao et al. [198] |
| LEC_SUBDO | Lectin | 27 kDa | G+, G− | Sponge | Schroeder et al. [199] |
| CvL | Lectin | 106 kDa | G+ | Sponge | Moura et al. [200] |
| Sialic-acid binding lectin | Lectin | ~51 kDa | G− | Bivalve mollusc | Tunkijjanukij and Olafsen [201] |
| Scyllin | Lectin | 5 kDa | G+, G− | Mud crab | Chattopadhyay and Chatterjee [202] |
| Tachylectin-1 | Lectin | 27 kDa | G− | Horseshoe crabs | Saito et al. [203] |
| HSL | Lectin | 182 kDa | G+, G− | Holothurian | Gowda et al. [204] |
| Binding molecules | |||||
| Anti-lipopolysaccharide factor (ALF) | Endotoxin-binding protein | ~15 kDa | G− | Horseshoe crabs | Morita et al. [205] |
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae. Mar. Drugs 2010, 8, 1213-1262. https://doi.org/10.3390/md8041213
Smith VJ, Desbois AP, Dyrynda EA. Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae. Marine Drugs. 2010; 8(4):1213-1262. https://doi.org/10.3390/md8041213
Chicago/Turabian StyleSmith, Valerie J., Andrew P. Desbois, and Elisabeth A. Dyrynda. 2010. "Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae" Marine Drugs 8, no. 4: 1213-1262. https://doi.org/10.3390/md8041213
APA StyleSmith, V. J., Desbois, A. P., & Dyrynda, E. A. (2010). Conventional and Unconventional Antimicrobials from Fish, Marine Invertebrates and Micro-algae. Marine Drugs, 8(4), 1213-1262. https://doi.org/10.3390/md8041213




