Microarray-Based Transcriptional Profiling of Renieramycin M and Jorunnamycin C, Isolated from Thai Marine Organisms
Abstract
:1. Introduction
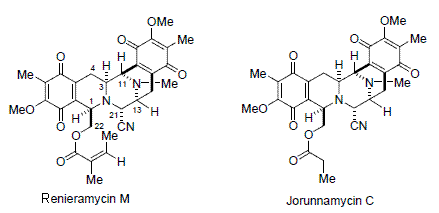
2. Results and Discussion
3. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
References and Notes
- Scott, JD; Williams, RM. Chemistry and Biology of the Tetrahydroisoquionoline Antitumor Antibiotics. Chem Rev 2002, 102, 1669–1730. [Google Scholar]
- Rinehart, KL. Antitumor Compounds from Tunicates. Med Drug Rev 2000, 20, 1–27. [Google Scholar]
- Friedman, D; Hu, Z; Kolb, EA; Gorfajn, B; Scotto, KW. Ecteinascidin 743 Inhibits Activated but not Constitutive Transcription. Cancer Res 2002, 62, 3377–3381. [Google Scholar]
- Takebayashi, Y; Pourquier, P; Zimonjic, DB; Nakayama, K; Emmert, S; Ueda, T; Urasaki, Y; Kanzaki, A; Akiyama, S; Popescu, N; Kraemer, KH. Pommier, Antiproliferativity of Ecteinascidin 743 Is Dependent upon Transcription-Coupled Nucleotide-Excision Repair. Nat Med 2001, 7, 961–966. [Google Scholar]
- Aune, GJ; Furuta, T; Pommier, Y. Ecteinascidin 743: A Novel Anticancer Drug with a Unique Mechanism of Action. Anticancer Drugs 2002, 13, 545–555. [Google Scholar]
- Gajate, C; An, F; Mollinedo, F. Differential Cytostatic and Apoptotic Effects of Ecteinascidin-743. J Biol Chem 2002, 277, 41580–41589. [Google Scholar]
- Jimeno, J; Faircloth, G; Fernandez Sousa-Faro, JM; Scheuer, PJ; Rinehart, K. New Marine Derived Anticancer Therapeutics – A Journey from the Sea to Clinical Trials. Mar Drugs 2004, 2, 14–29. [Google Scholar]
- Fayette, J; Coquard, IR; Alberti, L; Ranchère, D; Boyle, H; Blay, JY. ET-743: A Novel Agent with Activity in Soft Tissue Sarcomas. Oncologist 2005, 10, 827–832. [Google Scholar]
- David-Cordonnier, M-H; Gajate, C; Olmea, O; Laine, W; de la Iglesia-Vicente, J; Perez, C; Cuevas, C; Otero, G; Manzanares, I; Bailly, C; Mollinedo, F. DNA and Non-DNA Targets in the Mechanism of Action of the Antitumor Drug Trabectedin. Chem Biol 2005, 12, 1201–1210. [Google Scholar]
- Guirouin-Barbat, J; Antony, S; Pommier, Y. Zalypsis (PM00104) is a Potent Inducer of γ-H2AX foci and Reveals the Importance of the C Ring of Trabectedin for Transcription-coupled Repair Inhibition. Mol Cancer Ther. 2009, 8, pp. 2007–2014.
- Martinez, EJ; Owa, T; Schreiber, SL; Corey, EJ. Phthalascidin, a Synthetic Antitumor Agent with Potency and Mode of Action Comparable to Ecteinascidin 743. Proc Natl Acad Sci USA 1999, 96, 3496–3501. [Google Scholar]
- Martinez, EJ; Corey, EJ; Owa, T. Antitumor Activity and Gene Expression-Based Profiling of Ecteinascidin 743 and Phthalascidin Pt 650. Chem Biol 2001, 8, 1151–1160. [Google Scholar]
- Plowright, AT; Schaus, SE; Myers, AG. Transcriptional Response Pathways in a Yeast Strain Sensitive Saframycin A and a More Potent Analog Evidence for a Common Based of Activity. Chem Biol 2002, 9, 607–618. [Google Scholar]
- He, H; Faulkner, DJ. Renieramycins E and F from the Sponge Reniera sp: Reassignment of the Stereochemistry of the Renieramycins. J Org Chem 1989, 54, 5822–5824. [Google Scholar]
- Frincke, JM; Faulkner, DJ. Antimicrobial Metabolites of the Sponge Reniera sp. J Am Chem Soc 1982, 104, 265–269. [Google Scholar]
- Fontana, A; Cavaliere, P; Wahidulla, S; Naik, CG; Cimino, G. A New Antitumor Isoquinoline Alkaloid from the Marine Nudibranch. Jorunna funebris Tetrahedron 2000, 56, 7305–7308. [Google Scholar]
- Suwanborirux, K; Amnuoypol, S; Plubrukarn, A; Pummangura, S; Kubo, A; Tanaka, C; Saito, N. Chemistry of Renieramycins. Part 3. Isolation and Structure of Stabilized Renieramycin Type Derivatives Possessing Antitumor Activity from Thai Sponge, Xestospongia Species Pretreated with Potassium Cyanide. J Nat Prod 2003, 66, 1441–1446. [Google Scholar]
- Amnuoypol, S; Suwanborirux, K; Pummangura, S; Kubo, A; Tanaka, C; Saito, N. Chemistry of Renieramycins. Part 5. Structure Elucidation of Minor Components of Renieramycins O and Q-S, from Thai Marine Sponge, Xestospongia Species, Pretreated with Potassium Cyanide. J Nat Prod 2004, 67, 1023–1028. [Google Scholar]
- Charupant, K; Daikuhara, N; Saito, E; Amnuoypol, S; Suwanborirux, K; Owa, T; Saito, N. Chemistry of Renieramycins. Part 8. Synthesis and Cytotoxicity Evaluation of Renieramycin M-Jorunnamycin A Analogues. Bioorg Med Chem 2009, 17, 4548–4558. [Google Scholar]
- Charupant, K; Suwanborirux, K; Amnuoypol, S; Saito, E; Kubo, A; Saito, N. Jorunnamycins A–C, New Stabilized Renieramycin-Type Bistetrahydroisoquinolines Isolated from the Thai Nudibranch Jorunna funebris. Chem. Pharm. Bull. 2007, 55, 81–86. [Google Scholar]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J Immunol Methods 1983, 65, 55–63. [Google Scholar]
- In vitro antiproliferative activity: Exponentially growing cells (1,500 cells per well for HCT116 and 3,000 cells per well for MDA-MB-435) were seeded into 96-well microtiter plates and pre-cultured for one day. Both compounds (renieramycin M and jorunnamycin C) were dissolved in dimethyl sulfoxide (DMSO) to make 20 mM and further diluted with the culture medium to prepare threefold serial dilutions with the maximum concentration being 100 nM after the addition into each well. The obtained dilutions were added to the plates and incubation was continued for an additional three days. The antiproliferative activity was measured in triplicate by the MTT colorimetric assay. Absorbance was measured with a TECAN microplate reader at a test wavelength of 540 nm and a reference wavelength of 660 nm to be taken as an index of the number of viable cells. The IC50 value (the concentration required to inhibit cell growth by 50%) was determined by the least squares method.
- Oligonucleotide microarray gene expression analysis: HCT116 and MDA-MB-435 cells were each plated at 20 × 106 cells per dish in 10-cm-diameter dishes with 10 mL of fresh RPMI 1640 medium After 24 h pre-incubation, HCT116 cells were treated with 2 × IC50 concentration of each test compound (33 nM renieramycin M or 55 nM jorunnamycin C) for 4 h and 12 h MDA-MB-453 cells were also treated with 2 × IC50 concentration of each test compound (13 nM renieramycin M or 33 nM jorunnamycin C) for 4 h and 12 h DMSO (02%) treatment was used as control Total RNA was extracted from the cells using Trizol (Invitrogen) The extracted RNA was purified using an RNeasy kit (Qiagen) Double-stranded cDNA was synthesized from 5 μg of total RNA by means of a SuperScript double-stranded cDNA synthesis kit (Invitrogen) with T7-d(T)24 primer The cDNA product was purified by phenol/chloroform/isoamyl alcohol extraction In vitro transcription was carried out by means of a GeneChip IVT Labeling kit (Affymetrix). The resulting biotin-labeled cRNA was purified using the RNeasy kit. cRNA was fragmented at 94 °C for 35 min, and then hybridized for 16 h onto an Affymetrix GeneChip Human Genome Focus array that is capable of probing approximately 8,500 transcripts. The probe arrays were washed and stained with streptavidin-phycoerythrin and biotinylated goat anti-streptavidin on an Affymetrix Fluidics Station. Fluorescence intensities were captured with a Hewlett-Packard confocal laser scanner. All quantitative data were processed using the robust multi-array average (RMA) method [27], and transcriptional signature was defined as the differences between the data for compound treatment and those for DMSO treatment (control) on a logarithmic scale. Hierarchical clustering of the obtained transcriptional signatures for all test samples was done using the unweighted pair grouping method with arithmetic mean (UPGMA) in GeneSpring software to afford a dendrogram (tree graph) based on the similarity (cosine correlation). Up- and down-regulated genes were selected according to the following criteria: i) at least twofold change compared with control data; and ii) statistical significance with p-value < 0.05 by the t-test in triplicate data. Gene Ontology (GO) analysis was used to illuminate compound-related biological processes, cellular components, and molecular functions. Enriched GO terms with Benjamini-Hochberg adjusted p-values < 0.05 were selected using the Bioconductor GOstats library [28].
- Xu, Y; Tan, LJ; Grachtchouk, V; Voorhees, JJ; Fisher, GJ. Receptor-type Protein-tyrosine Phosphatse-K regulates Epidermal Growth Factor Receptor Function. J Biol Chem 2005, 280, 42694–42700. [Google Scholar]
- Martínez, N; Sánchez-Beato, M; Carnero, A; Moneo, V; Tercero, JC; Fernández, I; Navarrete, M; Jimeno, J; Piris, MA. Transcriptional Signature of Ecteinascidin 743 (Yondelis, Trabectedin) in Human Sarcoma Cells Explanted from Chemo-Naive Patients. Mol Cancer Ther 2005, 4, 814–823. [Google Scholar]
- Owa, T; Yokoi, A; Yamazaki, K; Yoshimatsu, K; Yamori, T; Nagasu, T. Array-Based Structure and Gene Expression Relationship Study of Antitumor Sulfonamide Including N-[2-(4-hydroxyphenyl)amino]3-pyridinyl]-4-methoxybenzenesulfonamide and N-(3-Chloro-7-indolyl)-1,4-benzenedisulfonamide. J Med Chem 2002, 45, 4913–4922. [Google Scholar]
- Irizarry, RA; Hobbs, B; Collin, F; Beazer-Barclay, YD; Antonellis, KJ; Scherf, U. Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe. Biostatistic 2003, 4, 249–264. [Google Scholar]
- Gentleman, R. Using GO for Statistical Analyses. Proceedings of COMPSTAT 2004 Symposium. 2004, pp. 171–180.








| Compound | Human cancer cell line, IC50 ± SD (nM) | |
|---|---|---|
| HCT116 (colon) | MDA-MB-435 (breast) | |
| Renieramycin M | 16.4 ± 0.3 | 6.3 ± 0.1 |
| Jorunnamycin C | 27.3 ± 1.0 | 16.3 ± 1.3 |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Charupant, K.; Suwanborirux, K.; Daikuhara, N.; Yokoya, M.; Ushijima-Sugano, R.; Kawai, T.; Owa, T.; Saito, N. Microarray-Based Transcriptional Profiling of Renieramycin M and Jorunnamycin C, Isolated from Thai Marine Organisms. Mar. Drugs 2009, 7, 483-494. https://doi.org/10.3390/md7040483
Charupant K, Suwanborirux K, Daikuhara N, Yokoya M, Ushijima-Sugano R, Kawai T, Owa T, Saito N. Microarray-Based Transcriptional Profiling of Renieramycin M and Jorunnamycin C, Isolated from Thai Marine Organisms. Marine Drugs. 2009; 7(4):483-494. https://doi.org/10.3390/md7040483
Chicago/Turabian StyleCharupant, Kornvika, Khanit Suwanborirux, Naomi Daikuhara, Masashi Yokoya, Rie Ushijima-Sugano, Takatoshi Kawai, Takashi Owa, and Naoki Saito. 2009. "Microarray-Based Transcriptional Profiling of Renieramycin M and Jorunnamycin C, Isolated from Thai Marine Organisms" Marine Drugs 7, no. 4: 483-494. https://doi.org/10.3390/md7040483
APA StyleCharupant, K., Suwanborirux, K., Daikuhara, N., Yokoya, M., Ushijima-Sugano, R., Kawai, T., Owa, T., & Saito, N. (2009). Microarray-Based Transcriptional Profiling of Renieramycin M and Jorunnamycin C, Isolated from Thai Marine Organisms. Marine Drugs, 7(4), 483-494. https://doi.org/10.3390/md7040483