Integrated Omics-Based Discovery of Bioactive Halogenated Metabolites from the Deep-Sea Streptomyces sp. B188M101
Abstract
1. Introduction
2. Results and Discussion
2.1. Non-Targeted Multivariate Analysis of OSMAC Extracts
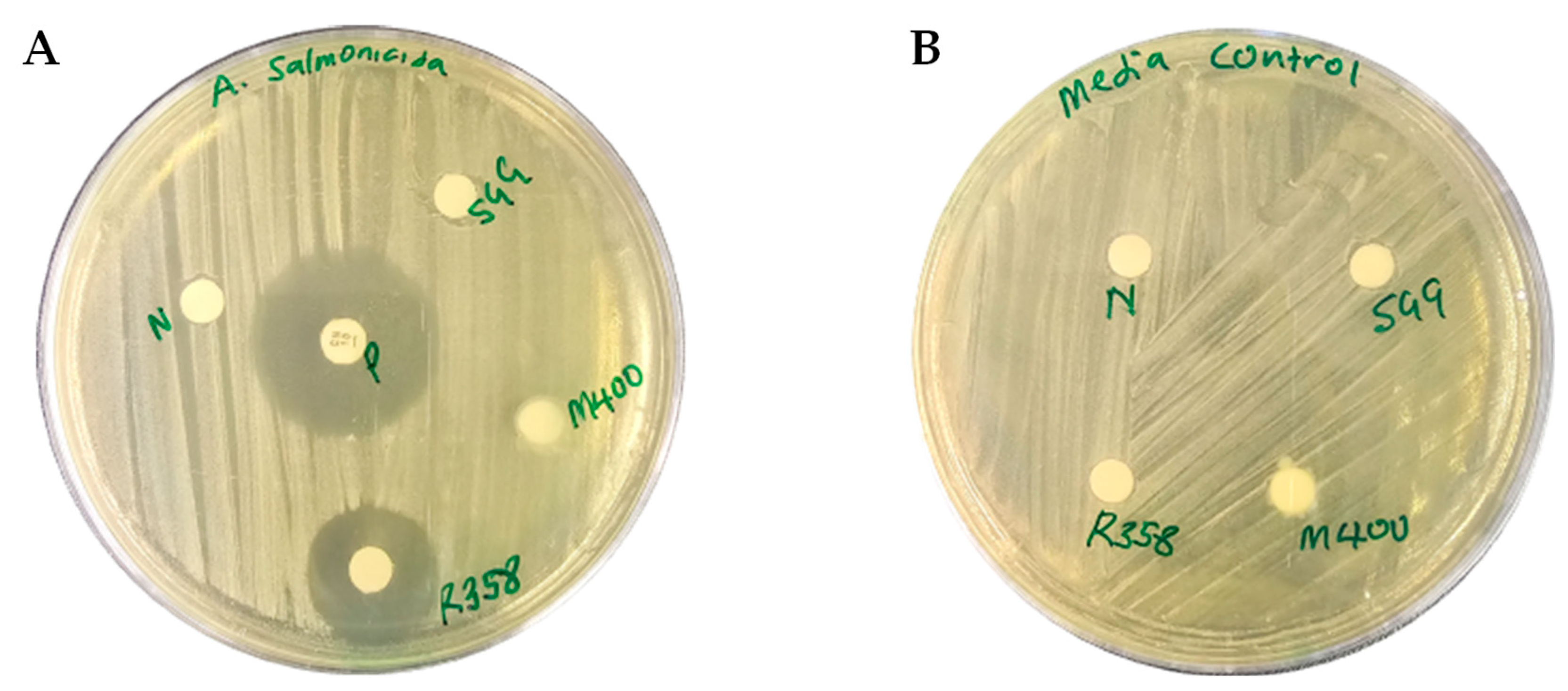
2.2. Preliminary Disc Diffusion Assays
2.3. Chemoinformatic Feature-Based Molecular Networking and Database Dereplication Analysis
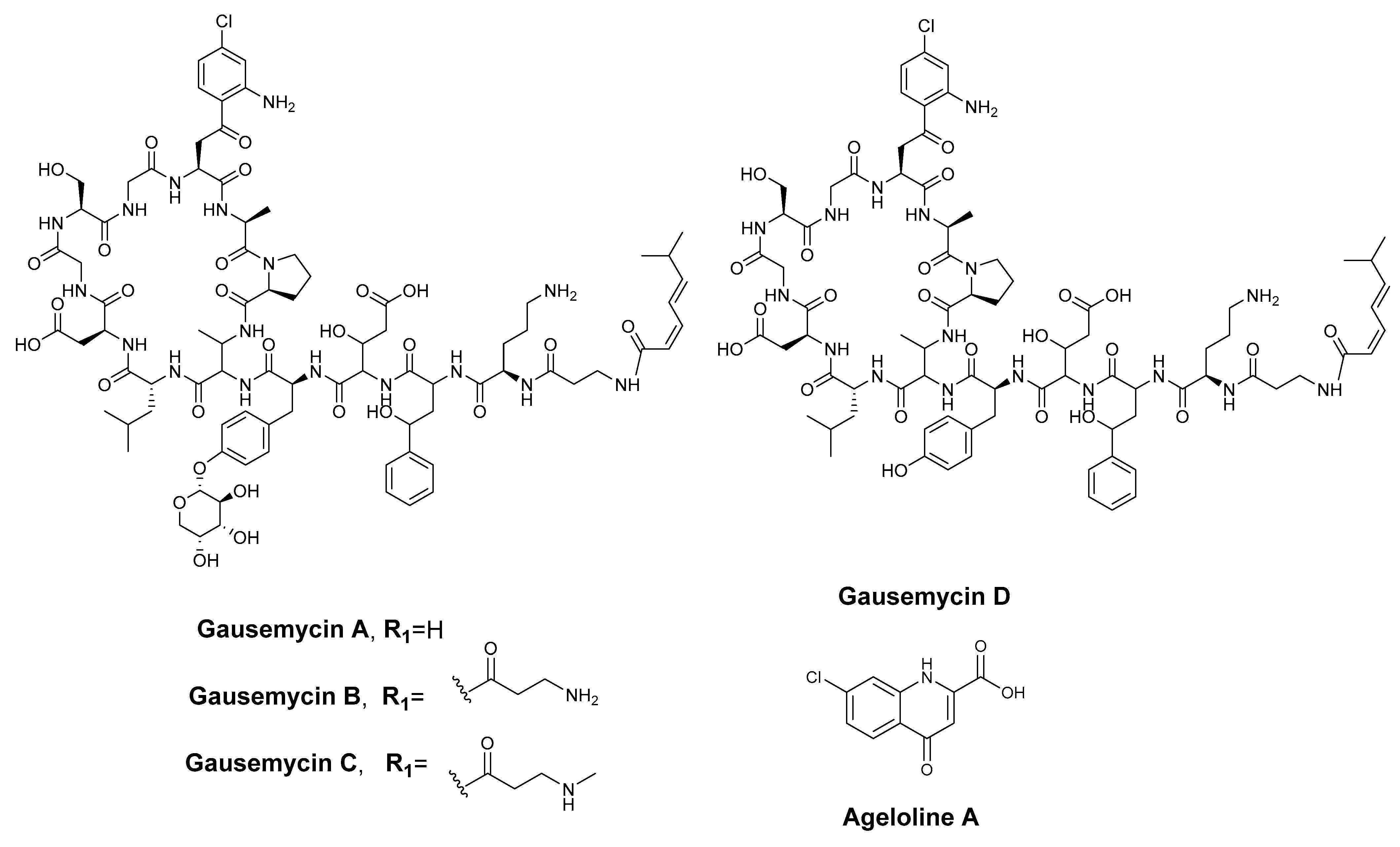
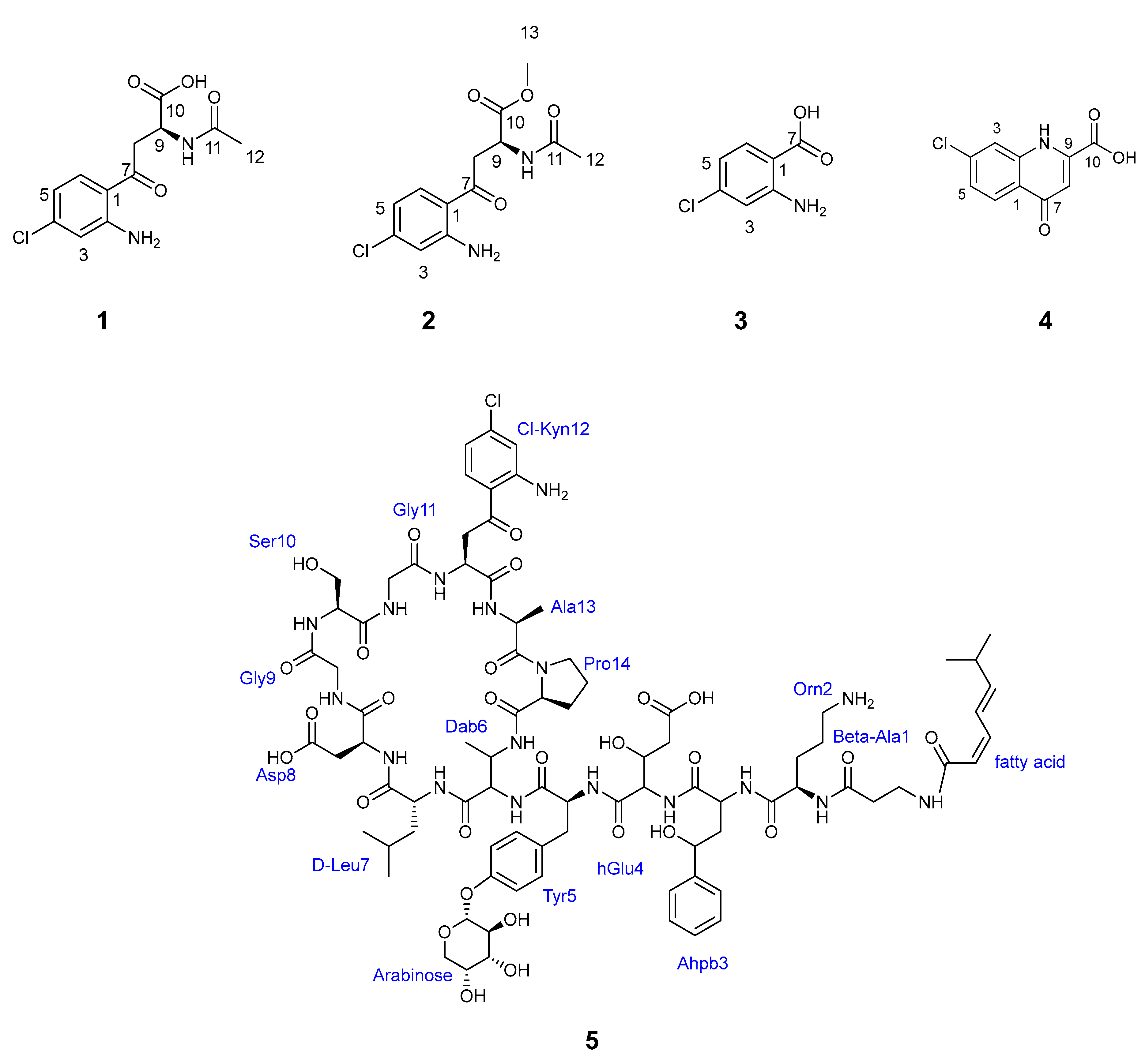
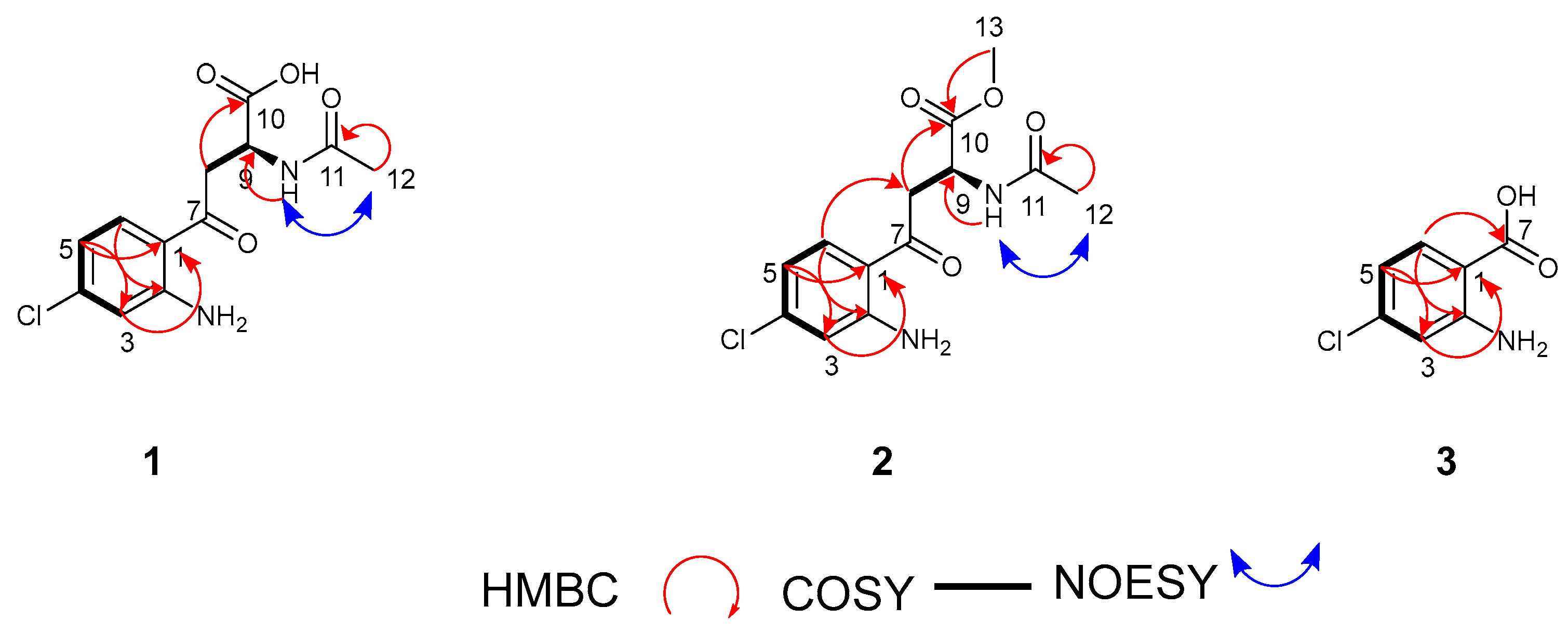
2.4. Isolation and Structure Elucidation of New Chlorinated Compounds
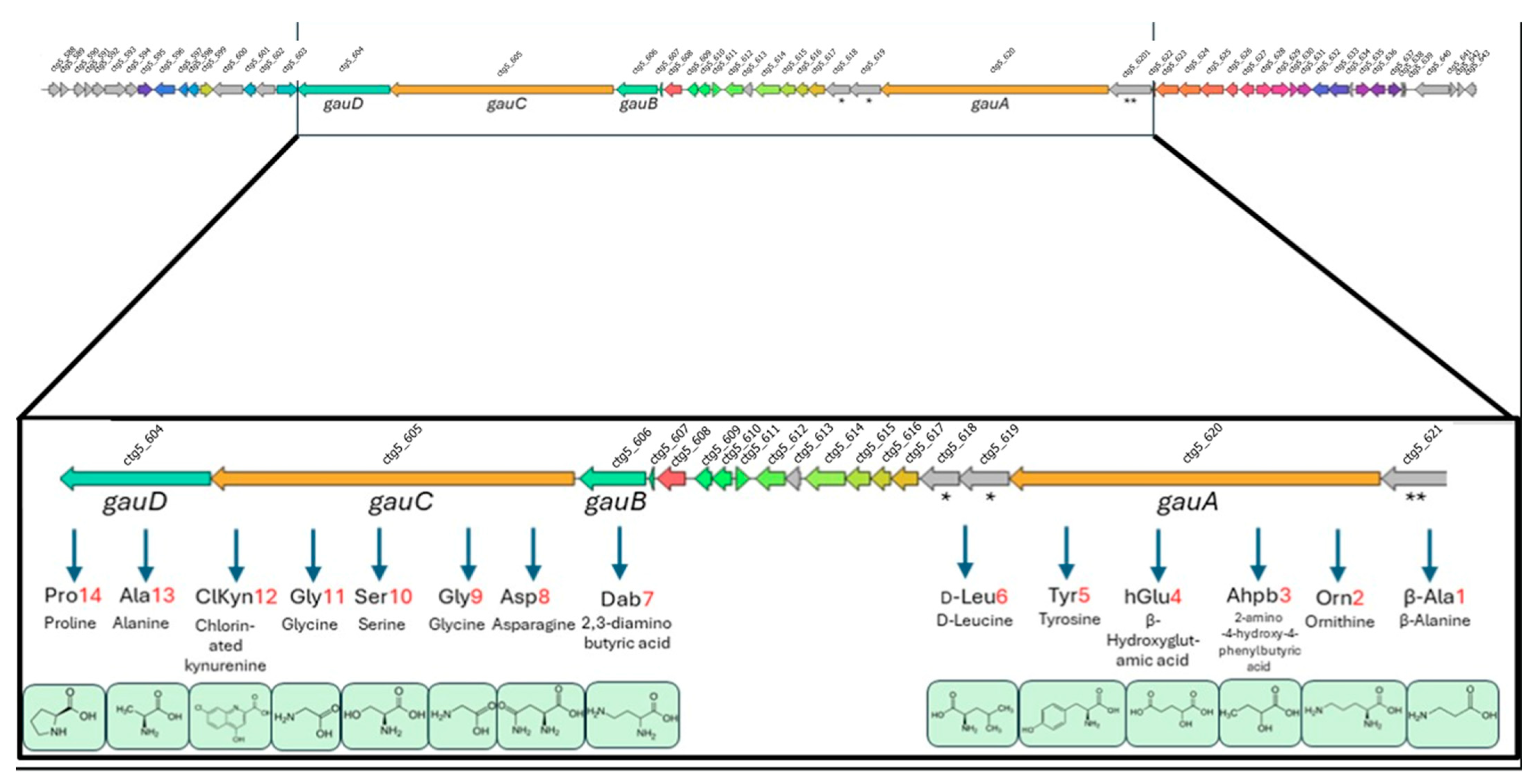
2.5. Genomics of B188M101
2.6. The Proposed Biosynthesis of Ageloline A-D (1–4)
2.7. Biological Evaluation of Compounds 1–5
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Microbial Isolation and Identification
3.3. Small Fermentation of Streptomyces sp. B188M101 Using an OSMAC Approach
3.4. Disc Diffusion Assay of Crude Extracts
3.5. Large-Scale Fermentation of Streptomyces sp. B188M101
3.6. Fractionation and Isolation of Metabolites from Streptomyces sp. B188M101
3.7. Antimicrobial Activity
3.8. MZmine 4.20 Data Pre-Processing, MetaboAnalyst 5.0, and Feature-Based Molecular Networking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FishStatJ-Software for Fishery and Aquaculture Statistical Time Series. Available online: https://sextant.ifremer.fr/geonetwork/srv/api/records/92c9fae9-fb8e-4ce3-80b0-bf7af46353d1 (accessed on 18 July 2025).
- Garlock, T.; Asche, F.; Anderson, J.; Ceballos-Concha, A.; Love, D.C.; Osmundsen, T.C.; Pincinato, R.B.M. Aquaculture: The Missing Contributor in the Food Security Agenda. Glob. Food Sec. 2022, 32, 100620. [Google Scholar] [CrossRef]
- Sasikumar, R.; Saranya, S.; Lourdu Lincy, L.; Thamanna, L.; Chellapandi, P. Genomic Insights into Fish Pathogenic Bacteria: A Systems Biology Perspective for Sustainable Aquaculture. Fish. Shellfish. Immunol. 2024, 154, 109978. [Google Scholar] [CrossRef]
- Reducing Disease Risk in Aquaculture. Available online: https://documents.worldbank.org/en/publication/documents-reports/documentdetail/110681468054563438/reducing-disease-risk-in-aquaculture (accessed on 18 July 2025).
- Fečkaninová, A.; Koščová, J.; Mudroňová, D.; Popelka, P.; Toropilová, J. The Use of Probiotic Bacteria against Aeromonas Infections in Salmonid Aquaculture. Aquaculture 2017, 469, 1–8. [Google Scholar] [CrossRef]
- Hossain, A.; Habibullah-Al-Mamun, M.; Nagano, I.; Masunaga, S.; Kitazawa, D.; Matsuda, H. Antibiotics, Antibiotic-Resistant Bacteria, and Resistance Genes in Aquaculture: Risks, Current Concern, and Future Thinking. Environ. Sci. Pollut. Res. 2022, 29, 11054–11075. [Google Scholar] [CrossRef]
- Vincent, A.T.; Hosseini, N.; Charette, S.J. The Aeromonas salmonicida Plasmidome: A Model of Modular Evolution and Genetic Diversity. Ann. N. Y. Acad. Sci. 2021, 1488, 16–32. [Google Scholar] [CrossRef]
- Fournier, K.C.; Paquet, V.E.; Attéré, S.A.; Farley, J.; Marquis, H.; Gantelet, H.; Ravaille, C.; Vincent, A.T.; Charette, S.J. Expansion of the PRAS3 Plasmid Family in Aeromonas salmonicida Subsp. Salmonicida and Growing Evidence of Interspecies Connections for These Plasmids. Antibiotics 2022, 11, 1047. [Google Scholar] [CrossRef]
- Liu, L.; Li, J.; Tu, M.; Gao, L.; Zhang, Y.; Rao, Y.; Rao, L.; Gui, M. Complete Genome Sequence Provides Information on Quorum Sensing Related Spoilage and Virulence of Aeromonas salmonicida GMT3 Isolated from Spoiled Sturgeon. Food Res. Int. 2024, 196, 115039. [Google Scholar] [CrossRef]
- Bello-López, J.M.; Sánchez-Garibay, C.; Ibáñez-Cervantes, G.; León-García, G.; Gonzalez-Avila, L.U.; Hernández-Cortez, C.; Barbosa, P.A.; Castro-Escarpulli, G. Genetic and Phenotypic Determinants of Resistance to Antibiotics in Aeromonas Spp., Strains Isolated from Pediatric Patients. J. Infect. Dev. Ctries. 2020, 14, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Liu, Y.; Wiegertjes, G.F.; Raaijmakers, J.M. Exploring Fish Microbial Communities to Mitigate Emerging Diseases in Aquaculture. FEMS Microbiol. Ecol. 2018, 94, fix161. [Google Scholar] [CrossRef] [PubMed]
- Rahayu, S.; Amoah, K.; Huang, Y.; Cai, J.; Wang, B.; Shija, V.M.; Jin, X.; Anokyewaa, M.A.; Jiang, M. Probiotics Application in Aquaculture: Its Potential Effects, Current Status in China and Future Prospects. Front. Mar. Sci. 2024, 11, 1455905. [Google Scholar] [CrossRef]
- Sam-on, M.F.S.; Mustafa, S.; Mohd Hashim, A.; Wan Mustapha, W.A.; Yusof, M.T.; Mohd Zaini, N.A.; Mohamed Nazir, M.Y. Bibliometric Mapping on the Probiotic Trends in Managing Aquaculture Pathogens. Food Biosci. 2025, 68, 106372. [Google Scholar] [CrossRef]
- Brugman, S.; Ikeda-Ohtsubo, W.; Braber, S.; Folkerts, G.; Pieterse, C.M.J.; Bakker, P.A.H.M. A Comparative Review on Microbiota Manipulation: Lessons From Fish, Plants, Livestock, and Human Research. Front. Nutr. 2018, 5, 80. [Google Scholar] [CrossRef]
- Butt, U.D.; Khan, S.; Liu, X.; Sharma, A.; Zhang, X.; Wu, B. Present Status, Limitations, and Prospects of Using Streptomyces Bacteria as a Potential Probiotic Agent in Aquaculture. Probiotics Antimicrob. Proteins 2024, 16, 426–442. [Google Scholar] [CrossRef]
- James, G.; Prasannan Geetha, P.; Thavarool Puthiyedathu, S.; Vattringal Jayadradhan, R.K. Applications of Actinobacteria in Aquaculture: Prospects and Challenges. 3 Biotech 2023, 13, 42. [Google Scholar] [CrossRef]
- García-Bernal, M.; Campa-Córdova, Á.I.; Saucedo, P.E.; Casanova-González, M.; Medina-Marrero, R.; Mazón-Suástegui, J.M. Isolation and in Vitro Selection of Actinomycetes Strains as Potential Probiotics for Aquaculture. Vet. World 2015, 8, 170–176. [Google Scholar] [CrossRef]
- Caesar, L.K.; Montaser, R.; Keller, N.P.; Kelleher, N.L. Metabolomics and Genomics in Natural Products Research: Complementary Tools for Targeting New Chemical Entities. Nat. Prod. Rep. 2021, 38, 2041–2065. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Bayram, E.; Lukić Bilela, L.; Cueto, M.; Díaz-Marrero, A.R.; Haznedaroglu, B.Z.; Jimenez, C.; Mandalakis, M.; Pereira, F.; Reyes, F.; et al. Advanced Methods for Natural Products Discovery: Bioactivity Screening, Dereplication, Metabolomics Profiling, Genomic Sequencing, Databases and Informatic Tools, and Structure Elucidation. Mar. Drugs 2023, 21, 308. [Google Scholar] [CrossRef]
- Schorn, M.A.; Verhoeven, S.; Ridder, L.; Huber, F.; Acharya, D.D.; Aksenov, A.A.; Aleti, G.; Moghaddam, J.A.; Aron, A.T.; Aziz, S.; et al. A Community Resource for Paired Genomic and Metabolomic Data Mining. Nat. Chem. Biol. 2021, 17, 363–368. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Augustijn, H.E.; Machushynets, N.V.; Bayona, L.M.; Soldatou, S.; de Jonge, N.F.; Casu, S.; Jaspars, M.; van Wezel, G.P.; Medema, M.H.; et al. FERMO: A Dashboard for Automated Prioritization of Molecular Features from Mass Spectral Data. bioRxiv 2022. [Google Scholar] [CrossRef]
- Nuñez Santiago, I.; Machushynets, N.V.; Mladic, M.; van Bergeijk, D.A.; Elsayed, S.S.; Hankemeier, T.; van Wezel, G.P. NanoRAPIDS as an Analytical Pipeline for the Discovery of Novel Bioactive Metabolites in Complex Culture Extracts at the Nanoscale. Commun. Chem. 2024, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Bell, B.A.; Yan, J.X.; Chevrette, M.G.; Brittin, N.J.; Zhu, Y.; Chanana, S.; Maity, M.; Braun, D.R.; Wheaton, A.M.; et al. Metabolomics and Genomics Enable the Discovery of a New Class of Nonribosomal Peptidic Metallophores from a Marine Micromonospora. J. Am. Chem. Soc. 2023, 145, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Goering, A.W.; McClure, R.A.; Doroghazi, J.R.; Albright, J.C.; Haverland, N.A.; Zhang, Y.; Ju, K.S.; Thomson, R.J.; Metcalf, W.W.; Kelleher, N.L. Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. ACS Cent. Sci. 2016, 2, 99–108. [Google Scholar] [CrossRef]
- Caesar, L.K.; Butun, F.A.; Robey, M.T.; Ayon, N.J.; Gupta, R.; Dainko, D.; Bok, J.W.; Nickles, G.; Stankey, R.J.; Johnson, D.; et al. Correlative Metabologenomics of 110 Fungi Reveals Metabolite–Gene Cluster Pairs. Nat. Chem. Biol. 2023, 19, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.A.; Crossman, L.; Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A.D.W. Diverse and Abundant Secondary Metabolism Biosynthetic Gene Clusters in the Genomes of Marine Sponge Derived Streptomyces Spp. Isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef]
- Al-Ansari, M.; Kalaiyarasi, M.; Almalki, M.A.; Vijayaraghavan, P. Optimization of Medium Components for the Production of Antimicrobial and Anticancer Secondary Metabolites from Streptomyces Sp. AS11 Isolated from the Marine Environment. J. King Saud. Univ. Sci. 2020, 32, 1993–1998. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering Cryptic Natural Product Biosynthesis in Microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Rateb, M.E.; Ebel, R.; Jaspars, M. Natural Product Diversity of Actinobacteria in the Atacama Desert. Antonie van Leeuwenhoek, Int. J. General. Mol. Microbiol. 2018, 111, 1467–1477. [Google Scholar] [CrossRef]
- Goodfellow, M.; Fiedler, H.P. A Guide to Successful Bioprospecting: Informed by Actinobacterial Systematics. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2010, 98, 119–142. [Google Scholar] [CrossRef] [PubMed]
- Boudemagh, A.; Kitouni, M.; Boughachiche, F.; Hamdiken, H.; Oulmi, L.; Reghioua, S.; Zerizer, H.; Couble, A.; Mouniee, D.; Boulahrouf, A.; et al. Isolation and Molecular Identification of Actinomycete Microflora, of Some Saharian Soils of South East Algeria (Biskra, EL-Oued and Ourgla) Study of Antifungal Activity of Isolated Strains. J. Mycol. Med. 2005, 15, 39–44. [Google Scholar] [CrossRef]
- Chanana, S.; Thomas, C.S.; Zhang, F.; Rajski, S.R.; Bugni, T.S. HCAPCA: Automated Hierarchical Clustering and Principal Component Analysis of Large Metabolomic Datasets in R. Metabolites 2020, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial Strain Prioritization Using Metabolomics Tools for the Discovery of Natural Products. Anal. Chem. 2012, 84, 4277–4283. [Google Scholar] [CrossRef]
- Clark, C.M.; Costa, M.S.; Conley, E.; Li, E.; Sanchez, L.M.; Murphy, B.T. Using the Open-Source MALDI TOF-MS IDBac Pipeline for Analysis of Microbial Protein and Specialized Metabolite Data. J. Vis. Exp. 2019, 2019, e59219. [Google Scholar] [CrossRef]
- McVean, G. A Genealogical Interpretation of Principal Components Analysis. PLoS Genet. 2009, 5, e1000686. [Google Scholar] [CrossRef]
- Cabezas, L.M.C.; Izbicki, R.; Stern, R.B. Hierarchical Clustering: Visualization, Feature Importance and Model Selection. Appl. Soft Comput. 2023, 141, 110303. [Google Scholar] [CrossRef]
- Boyko, N.I.; Tkachyk, O.A. Hierarchical Clustering Algorithm for Dendrogram Construction and Cluster Counting. Inform. Math. Methods Simul. 2023, 13, 5–15. [Google Scholar] [CrossRef]
- Kimes, P.K.; Liu, Y.; Neil Hayes, D.; Marron, J.S. Statistical Significance for Hierarchical Clustering. Biometrics 2017, 73, 811–821. [Google Scholar] [CrossRef]
- Islam, M.S.; Akhtar, M.M.; Parvez, M.S.; Alam, M.J.; Alam, M.F. Antitumor and Antibacterial Activity of a Crude Methanol Leaf Extract of Vitex negundo L. Arch. Biol. Sci. 2013, 65, 229–238. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffmann, N.; Lu, M.; et al. Integrative Analysis of Multimodal Mass Spectrometry Data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef]
- Ono, K.; Fong, D.; Gao, C.; Churas, C.; Pillich, R.; Lenkiewicz, J.; Pratt, D.; Pico, A.R.; Hanspers, K.; Xin, Y.; et al. Cytoscape Web: Bringing Network Biology to the Browser. Nucleic Acids Res. 2025, 53, W203–W212. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Uehara, T.; Yanagida, M. Highly Accurate Chemical Formula Prediction Tool Utilizing High-Resolution Mass Spectra, MS/MS Fragmentation, Heuristic Rules, and Isotope Pattern Matching. Anal. Chem. 2012, 84, 4396–4403. [Google Scholar] [CrossRef]
- Celma, A.; Sancho, J.V.; Schymanski, E.L.; Fabregat-Safont, D.; Ibáñez, M.; Goshawk, J.; Barknowitz, G.; Hernández, F.; Bijlsma, L. Improving Target and Suspect Screening High-Resolution Mass Spectrometry Workflows in Environmental Analysis by Ion Mobility Separation. Environ. Sci. Technol. 2020, 54, 15120–15131. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Metabolomic Database Annotations via Query of Elemental Compositions: Mass Accuracy Is Insufficient Even at Less than 1 Ppm. BMC Bioinform. 2006, 7, 234. [Google Scholar] [CrossRef] [PubMed]
- Tyurin, A.; Alferova, V.; Paramonov, A.; Shuvalov, M.; Kudryakova, G.; Rogozhin, E.; Zherebker, A.; Brylev, V.; Chistov, A.; Baranova, A.; et al. Gausemycins A,B–Cyclic Lipoglycopeptides from Streptomyces sp. Angew. Chem. Int. Ed. Engl. 2021, 60, 18694–18703. [Google Scholar] [CrossRef] [PubMed]
- Kravchenko, T.V.; Paramonov, A.S.; Kudzhaev, A.M.; Efimova, S.S.; Khorev, A.S.; Kudryakova, G.K.; Ivanov, I.A.; Chistov, A.A.; Baranova, A.A.; Krasilnikov, M.S.; et al. Gausemycin Antibiotic Family Acts via Ca2+-Dependent Membrane Targeting. J. Nat. Prod. 2024, 87, 664–674. [Google Scholar] [CrossRef]
- Cheng, C.; Othman, E.M.; Reimer, A.; Grüne, M.; Kozjak-Pavlovic, V.; Stopper, H.; Hentschel, U.; Abdelmohsen, U.R. Ageloline A, New Antioxidant and Antichlamydial Quinolone from the Marine Sponge-Derived Bacterium Streptomyces sp. SBT345. Tetrahedron Lett. 2016, 57, 2786–2789. [Google Scholar] [CrossRef]
- Mordhorst, S.; Ruijne, F.; Vagstad, A.L.; Kuipers, O.P.; Piel, J. Emulating Nonribosomal Peptides with Ribosomal Biosynthetic Strategies. RSC Chem. Biol. 2022, 4, 7–36. [Google Scholar] [CrossRef]
- Xu, F.; Butler, R.; May, K.; Rexhepaj, M.; Yu, D.; Zi, J.; Chen, Y.; Liang, Y.; Zeng, J.; Hevel, J.; et al. Modified Substrate Specificity of a Methyltransferase Domain by Protein Insertion into an Adenylation Domain of the Bassianolide Synthetase. J. Biol. Eng. 2019, 13, 65. [Google Scholar] [CrossRef]
- McErlean, M.; Overbay, J.; Van Lanen, S. Refining and Expanding Nonribosomal Peptide Synthetase Function and Mechanism. J. Ind. Microbiol. Biotechnol. 2019, 46, 493–513. [Google Scholar] [CrossRef]
- Kupchan, S.M.; Stevens, K.L.; Rohlfing, E.A.; Sickles, B.R.; Sneden, A.T.; Miller, R.W.; Bryan, R.F. New Cytotoxic Neolignans from Aniba Megaphylla Mez.1’2. J. Org. Chem. 1978, 43, 586–590. [Google Scholar] [CrossRef]
- Alferova, V.A.; Shuvalov, M.V.; Suchkova, T.A.; Proskurin, G.V.; Aparin, I.O.; Rogozhin, E.A.; Novikov, R.A.; Solyev, P.N.; Chistov, A.A.; Ustinov, A.V.; et al. 4-Chloro-l-Kynurenine as Fluorescent Amino Acid in Natural Peptides. Amino Acids 2018, 50, 1697–1705. [Google Scholar] [CrossRef]
- Wu, C.; Shang, Z.; Lemetre, C.; Ternei, M.A.; Brady, S.F. Cadasides, Calcium-Dependent Acidic Lipopeptides from the Soil Metagenome That Are Active against Multidrug-Resistant Bacteria. J. Am. Chem. Soc. 2019, 141, 3910–3919. [Google Scholar] [CrossRef]
- Hover, B.M.; Kim, S.H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-Independent Discovery of the Malacidins as Calcium-Dependent Antibiotics with Activity against Multidrug-Resistant Gram-Positive Pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef]
- Heinzelmann, E.; Berger, S.; Müller, C.; Härtner, T.; Poralla, K.; Wohlleben, W.; Schwartz, D. An Acyl-CoA Dehydrogenase Is Involved in the Formation of the Δcis3 Double Bond in the Acyl Residue of the Lipopeptide Antibiotic Friulimicin in Actinoplanes Friuliensis. Microbiology 2005, 151, 1963–1974. [Google Scholar] [CrossRef] [PubMed]
- Martinet, L.; Naômé, A.; Deflandre, B.; Maciejewska, M.; Tellatin, D.; Tenconi, E.; Smargiasso, N.; De Pauw, E.; Van Wezel, G.P.; Rigali, S. A Single Biosynthetic Gene Cluster Is Responsible for the Production of Bagremycin Antibiotics and Ferroverdin Iron Chelators. mBio 2019, 10, e01230-19. [Google Scholar] [CrossRef] [PubMed]
- Koper, K.; Han, S.W.; Pastor, D.C.; Yoshikuni, Y.; Maeda, H.A. Evolutionary Origin and Functional Diversification of Aminotransferases. J. Biol. Chem. 2022, 298, 102122. [Google Scholar] [CrossRef] [PubMed]
- RU2762182C1; Gausemycin A and B. G. F. Gause Research Institute for the Discovery of New Antibiotics: Moscow, Russia, 2021; p. 29.
- Ebbensgaard, A.; Mordhorst, H.; Aarestrup, F.M.; Hansen, E.B. The Role of Outer Membrane Proteins and Lipopolysaccharides for the Sensitivity of Escherichia Coli to Antimicrobial Peptides. Front. Microbiol. 2018, 9, 2153. [Google Scholar] [CrossRef]
- Yoon, Y.; Song, S. Structural Insights into the Lipopolysaccharide Transport (Lpt) System as a Novel Antibiotic Target. J. Microbiol. 2024, 62, 261–275. [Google Scholar] [CrossRef]
- Schneider, T.; Sahl, H.G. An Oldie but a Goodie-Cell Wall Biosynthesis as Antibiotic Target Pathway. Int. J. Med. Microbiol. 2010, 300, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ebanks, R.; Goguen, M.; McKinnon, S.; Pinto, D.; Ross, N.W. Identification of the major outer membrane proteins of Aeromonas salmonicida. Dis Aquat. Org. 2005, 68, 29–38. [Google Scholar] [CrossRef]
- Rey-Varela, D.; Cisneros-Sureda, J.; Balado, M.; Rodríguez, J.; Lemos, M.L.; Jiménez, C. The Outer Membrane Protein Fstc of Aeromonas salmonicida Subsp. Salmonicida Acts as Receptor for Amonabactin Siderophores and Displays a Wide Ligand Plasticity. Structure-Activity Relationships of Synthetic Amonabactin Analogues. ACS Infect. Dis. 2019, 5, 1936–1951. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. 2020, 22, 3380–3387. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory Friendly Classification with the Genome Taxonomy Database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Welcome to KBase Predictive Biology|Kbase. Available online: https://www.kbase.us/ (accessed on 18 July 2025).
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Booth, T.J.; Van Wersch, B.; Van Grieken, L.; Medema, M.H.; Chooi, Y.H. Cblaster: A Remote Search Tool for Rapid Identification and Visualization of Homologous Gene Clusters. Bioinform. Adv. 2021, 1, vbab016. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.H. Clinker & Clustermap.Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; De Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of Microbial Metabolites through Database Search of Mass Spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef] [PubMed]
- NMR Guidelines for ACS Journals. Available online: https://pubsapp.acs.org/paragonplus/submission/acs_nmr_guidelines.pdf (accessed on 16 September 2025).



| 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|
| Position | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) |
| 1 | 116.7, C | 116.7, C | 110.3, C | |||
| 2 | 153.3, C | 153.3, C | 153.6, C | |||
| 3 | 116.9, CH | 6.77 (d, 2.2) | 116.9, CH | 6.77 (d, 2.2) | 116.3, CH | 6.74 (d, 2.2) |
| 4 | 141.0, C | 141.0, C | 140.5, C | |||
| 5 | 115.9, CH | 6.56 (dd, 8.7, 2.2) | 115.9, CH | 6.56 (dd, 8.7, 2.2) | 116.0, CH | 6.50 (dd, 8.7, 2.2) |
| 6 | 133.6, CH | 7.72 (d, 8.7) | 133.6, CH | 7.71 (d, 8.7) | 133.9, CH | 7.77 (d, 8.7) |
| 7 | 198.1, C | 198.1, C | 170.8, C | |||
| 8 | 41.2, CH2 | 3.54 (dd, 17.3, 6.3), 3.45 (dd, 16.9, 6.3) | 41.2, CH2 | 3.53 (dd, 17.3, 6.3), 3.46 (dd, 16.9, 6.3) | ||
| 9 | 49.3, CH | 4.89 (m) | 49.3, CH | 4.91 (m) | ||
| 10 | 173.1, C | 173.1, C | ||||
| 11 | 172.9, C | 172.9, C | ||||
| 12 | 21.8, CH3 | 1.97 (s) | 21.8, CH3 | 1.96 (s) | ||
| 13 | 51.5, CH3 | 3.72 (s) | ||||
| Compound MIC (µg/mL) | |||||
|---|---|---|---|---|---|
| Pathogenic Species | 1 | 2 | 3 | 4 | 5 |
| Staphylococcus aureus ATCC6538P | 128 | >128 | 6 | 128 | 32 |
| S. aureus L3797 | >128 | >128 | >128 | >128 | >64 |
| S. pneumoniae L44 | >128 | >128 | 64 | 128 | 128 |
| S. maltophilia L2125 | 128 | 64 | 32 | 64 | nt |
| Aeromonas salmonicida | >128 | >128 | >128 | >128 | 32 |
| Pseudomonas aeruginosa | >128 | >128 | >128 | >128 | >128 |
| E. coli ΔtolC L4242 | >128 | >128 | >128 | >128 | >128 |
| Pathogenic Species | 0 mM CaCl2 | 0.45 mM CaCl2 | 20 mM CaCl2 |
|---|---|---|---|
| Staphylococcus aureus ATCC6538P | 32 | 4 | ≤4 |
| S. aureus GISA L3797 | >64 | 16 | 32 |
| S. pneumoniae L44 | 128 | 128 | nt |
| Aeromonas salmonicida | 32 | 16 | 16 |
| Pseudomonas aeruginosa | >128 | >128 | >128 |
| E. coli ΔtolC L4242 | >128 | >128 | >64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oluwabusola, E.T.; Jackson, S.A.; Brunati, C.; Gackstatter, S.; Vedder, H.; Iorio, M.; Chawande, G.; Margassery, L.M.; Nguyen, G.-S.; Clarke, D.J.; et al. Integrated Omics-Based Discovery of Bioactive Halogenated Metabolites from the Deep-Sea Streptomyces sp. B188M101. Mar. Drugs 2025, 23, 362. https://doi.org/10.3390/md23090362
Oluwabusola ET, Jackson SA, Brunati C, Gackstatter S, Vedder H, Iorio M, Chawande G, Margassery LM, Nguyen G-S, Clarke DJ, et al. Integrated Omics-Based Discovery of Bioactive Halogenated Metabolites from the Deep-Sea Streptomyces sp. B188M101. Marine Drugs. 2025; 23(9):362. https://doi.org/10.3390/md23090362
Chicago/Turabian StyleOluwabusola, Emmanuel Tope, Stephen A. Jackson, Cristina Brunati, Stefanie Gackstatter, Hannah Vedder, Marianna Iorio, Gargee Chawande, Lekha Menon Margassery, Giang-Son Nguyen, David J. Clarke, and et al. 2025. "Integrated Omics-Based Discovery of Bioactive Halogenated Metabolites from the Deep-Sea Streptomyces sp. B188M101" Marine Drugs 23, no. 9: 362. https://doi.org/10.3390/md23090362
APA StyleOluwabusola, E. T., Jackson, S. A., Brunati, C., Gackstatter, S., Vedder, H., Iorio, M., Chawande, G., Margassery, L. M., Nguyen, G.-S., Clarke, D. J., Ebel, R., Jaspars, M., & Dobson, A. D. W. (2025). Integrated Omics-Based Discovery of Bioactive Halogenated Metabolites from the Deep-Sea Streptomyces sp. B188M101. Marine Drugs, 23(9), 362. https://doi.org/10.3390/md23090362







