A Marine-Derived Steroid from Rhodococcus sp., 3,12-Dioxochola-4,6-dien-24-oic Acid, Enhances Skin Re-Epithelialization and Tissue Repair
Abstract
1. Introduction
2. Results and Discussion
2.1. Structural Determination
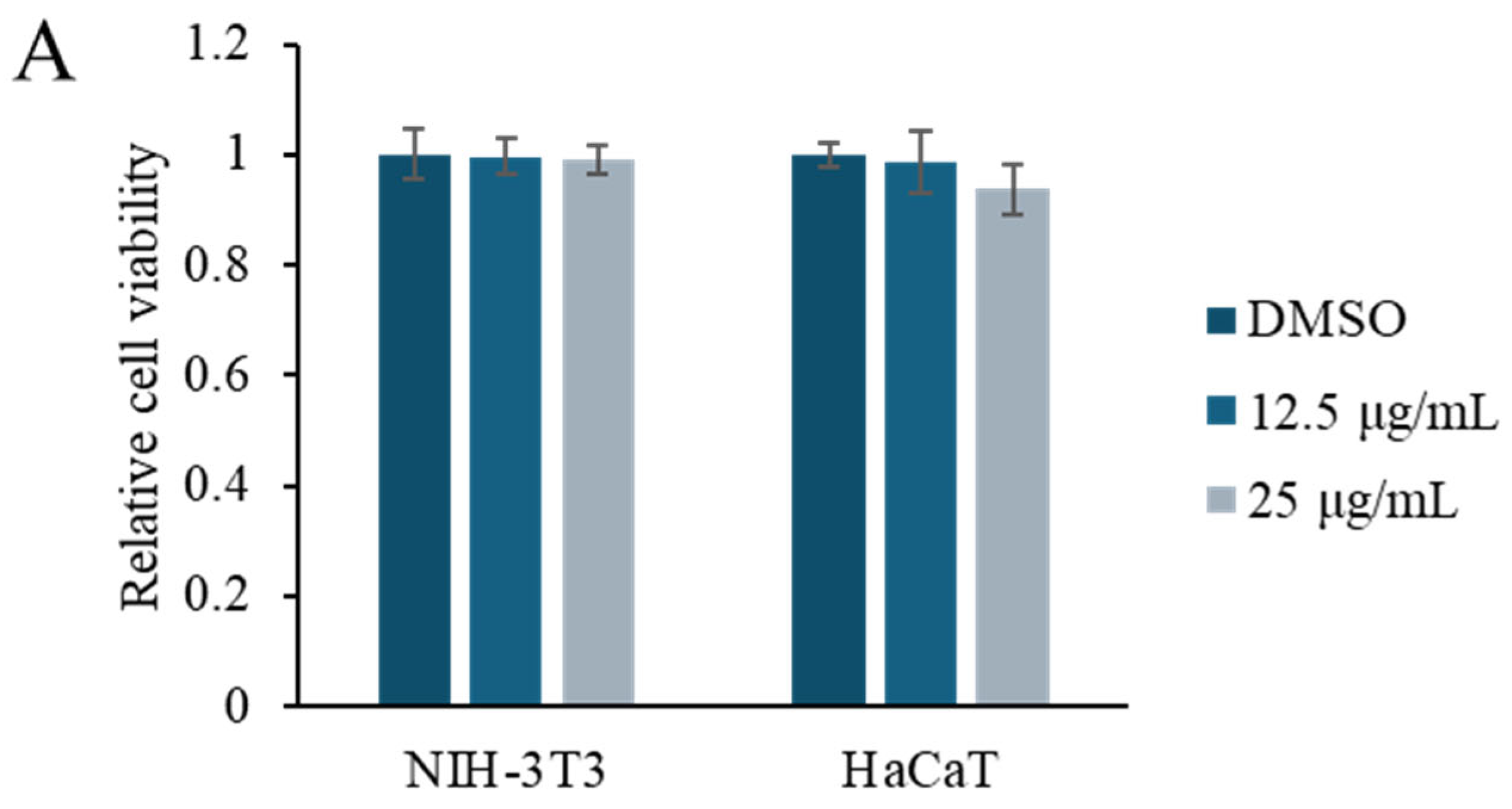
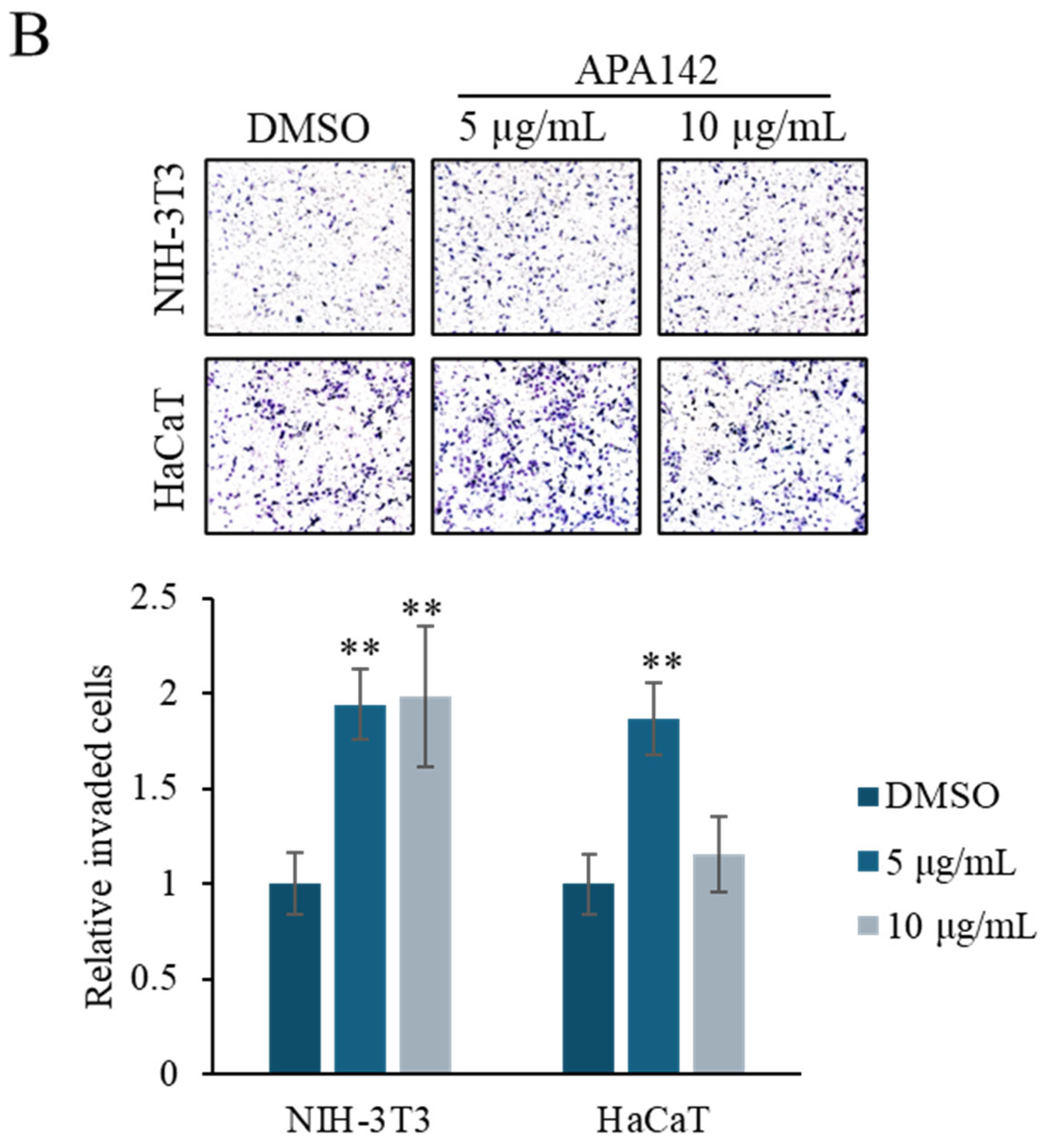
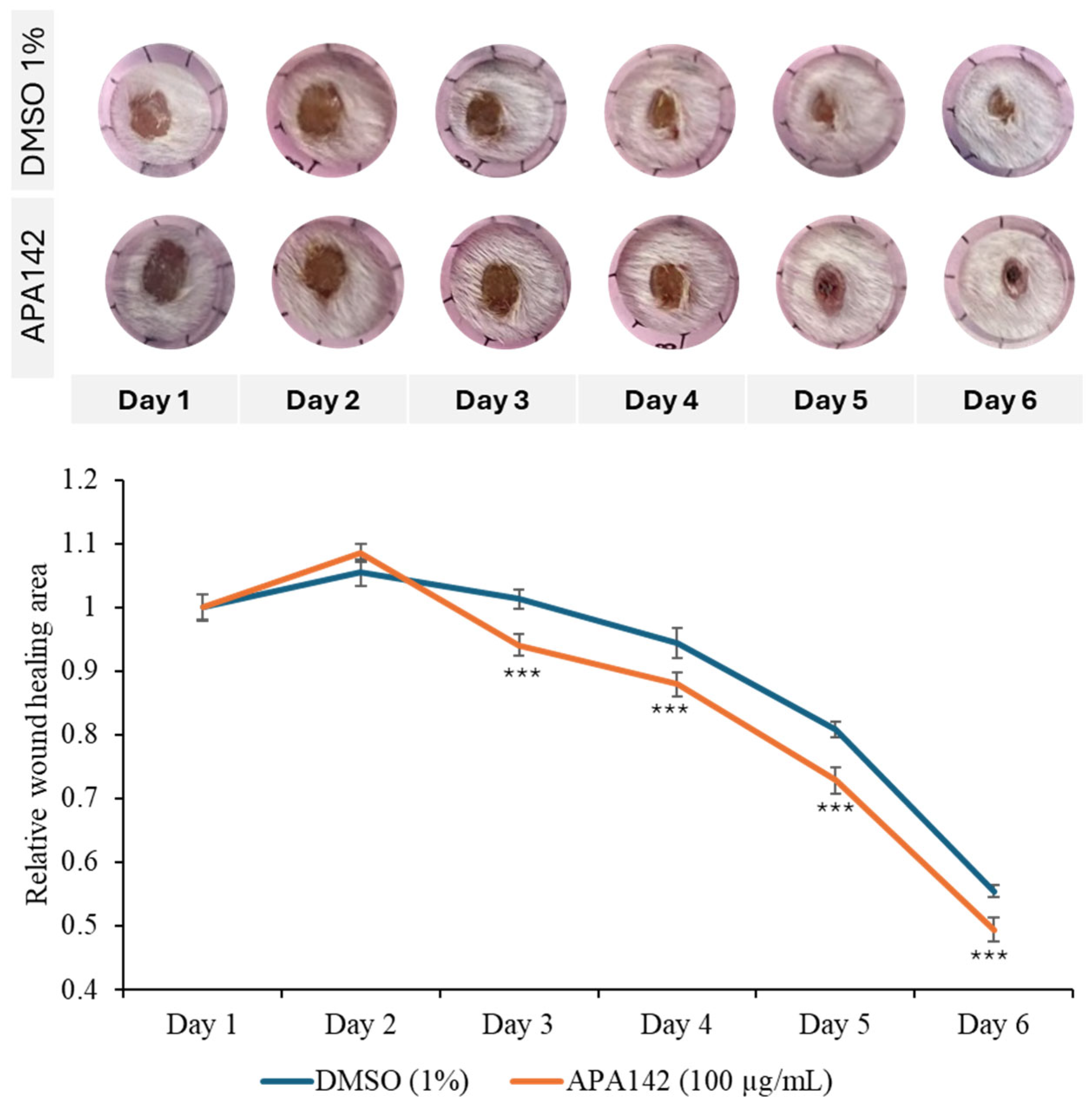
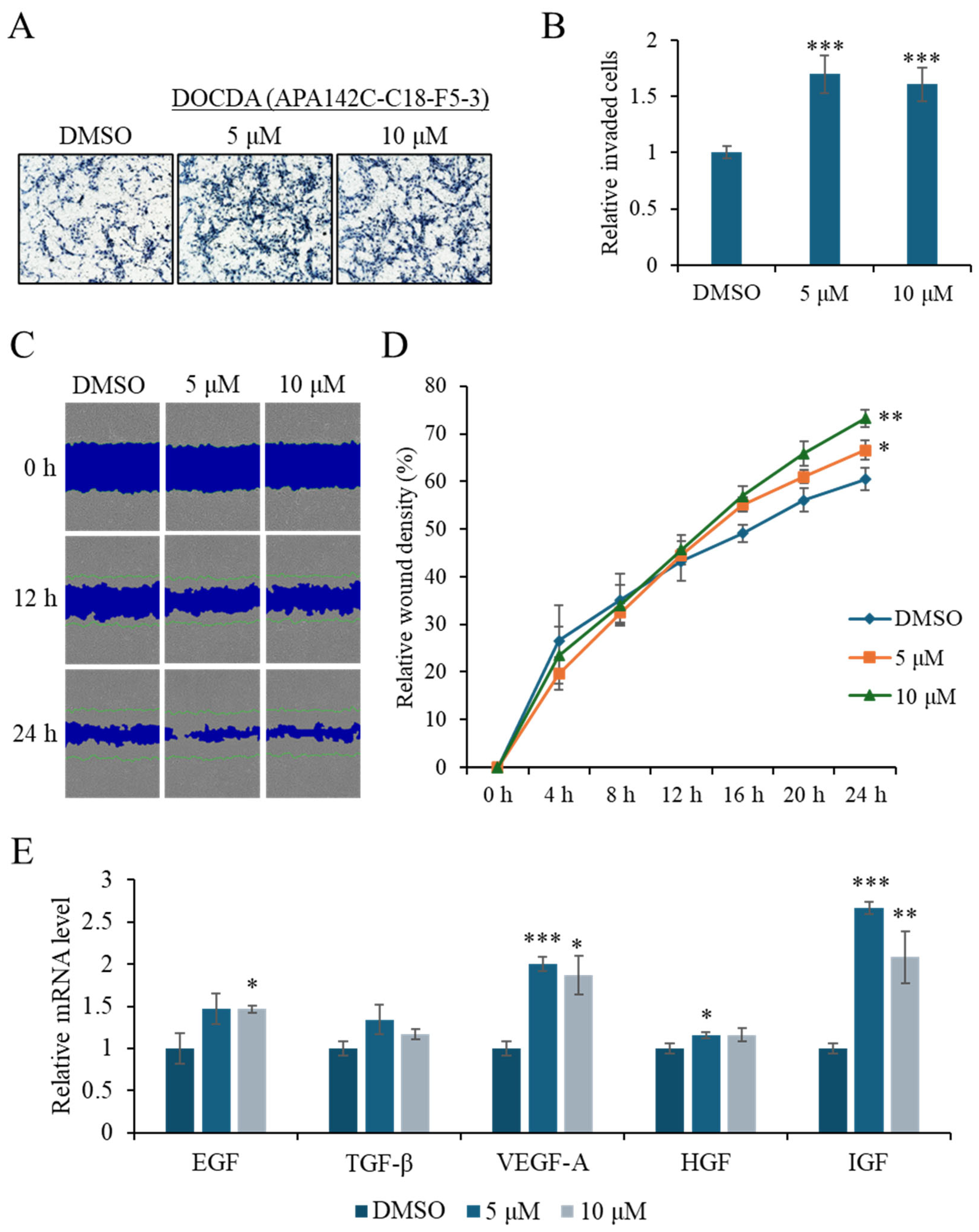
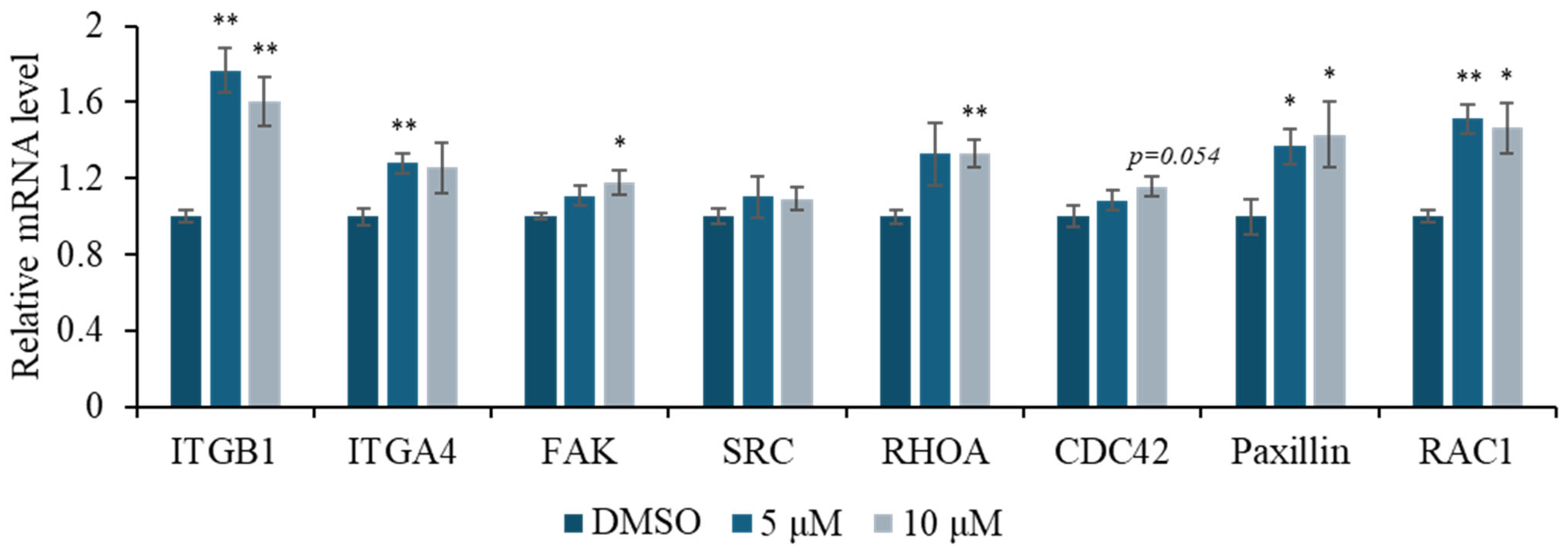
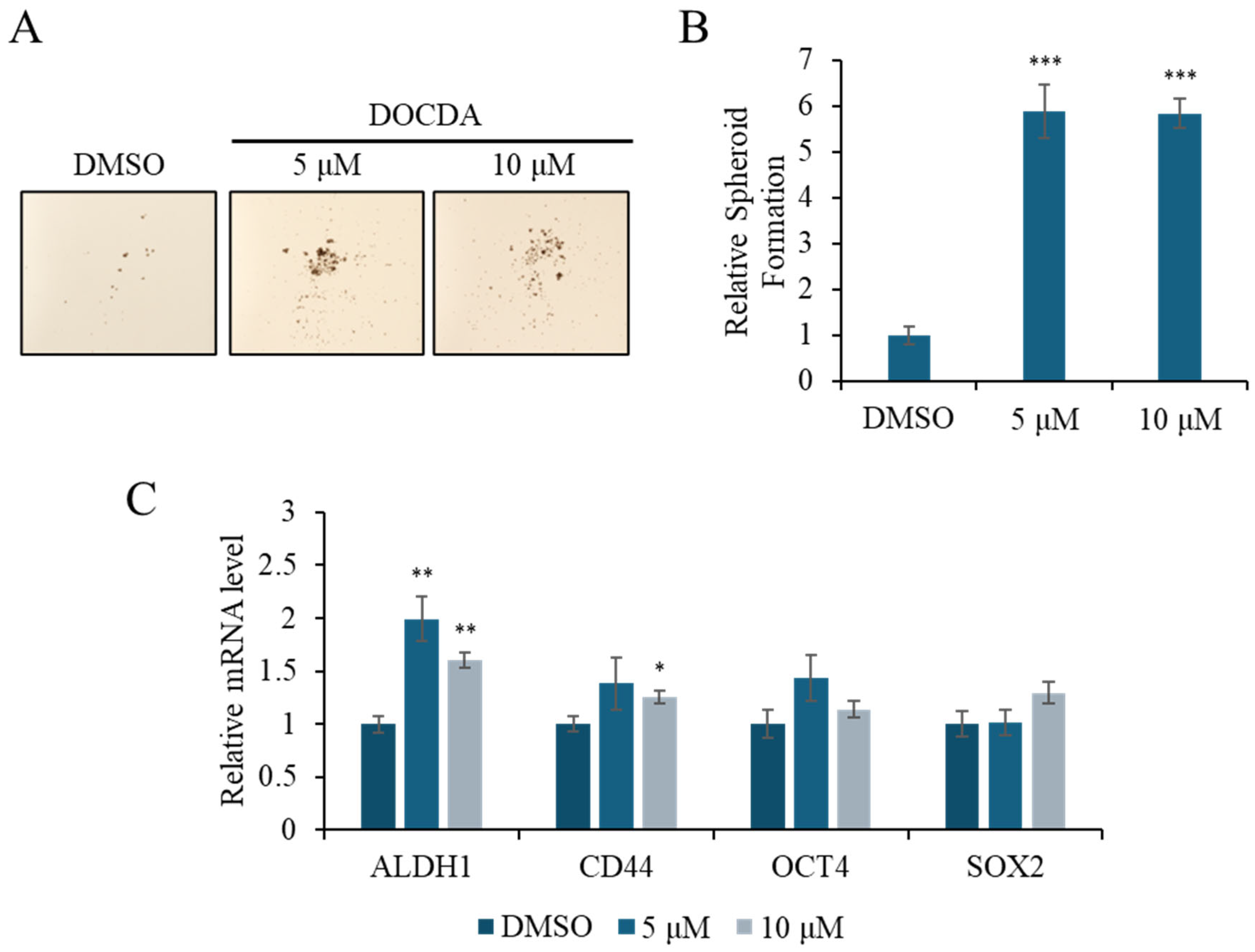
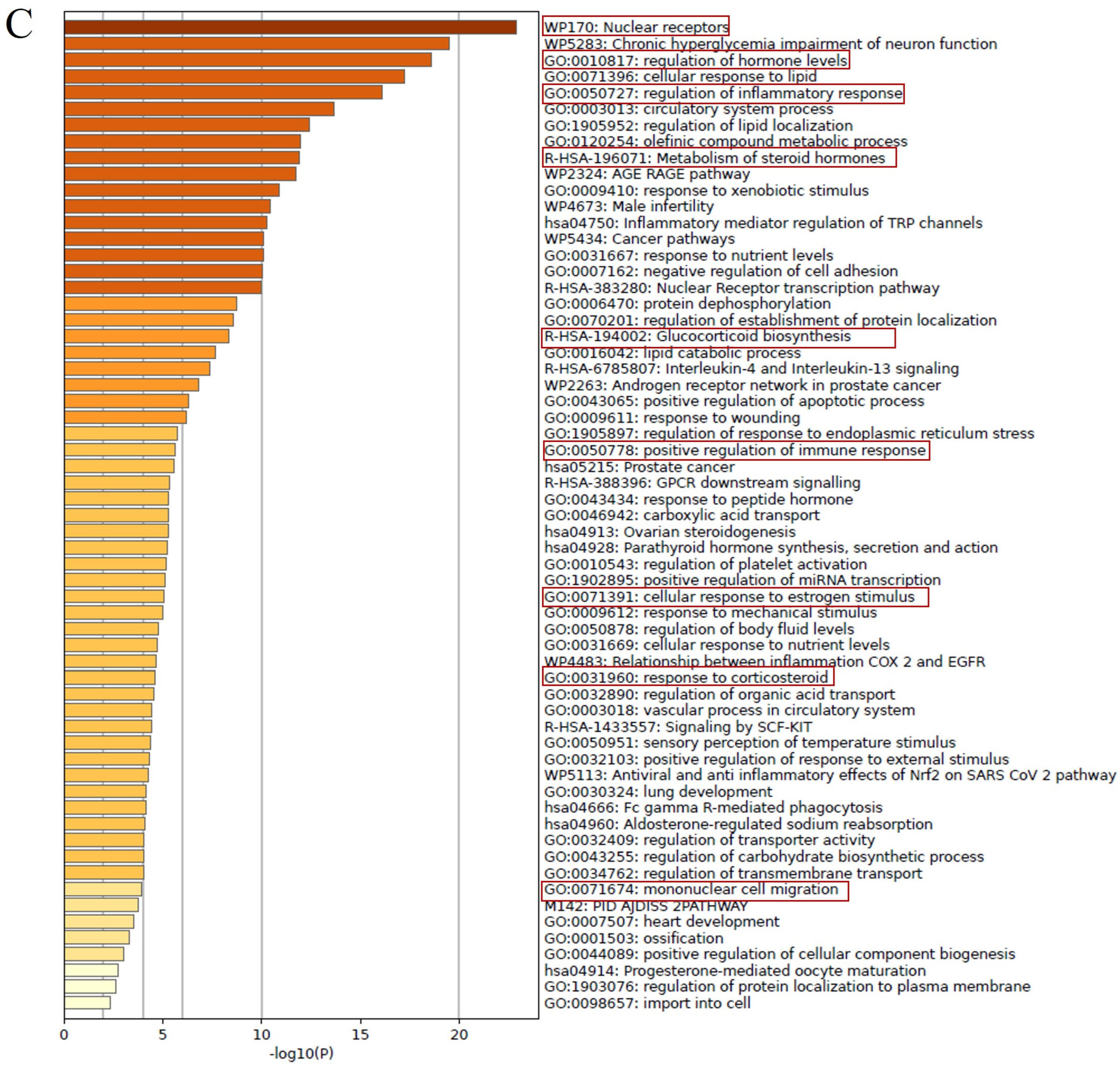
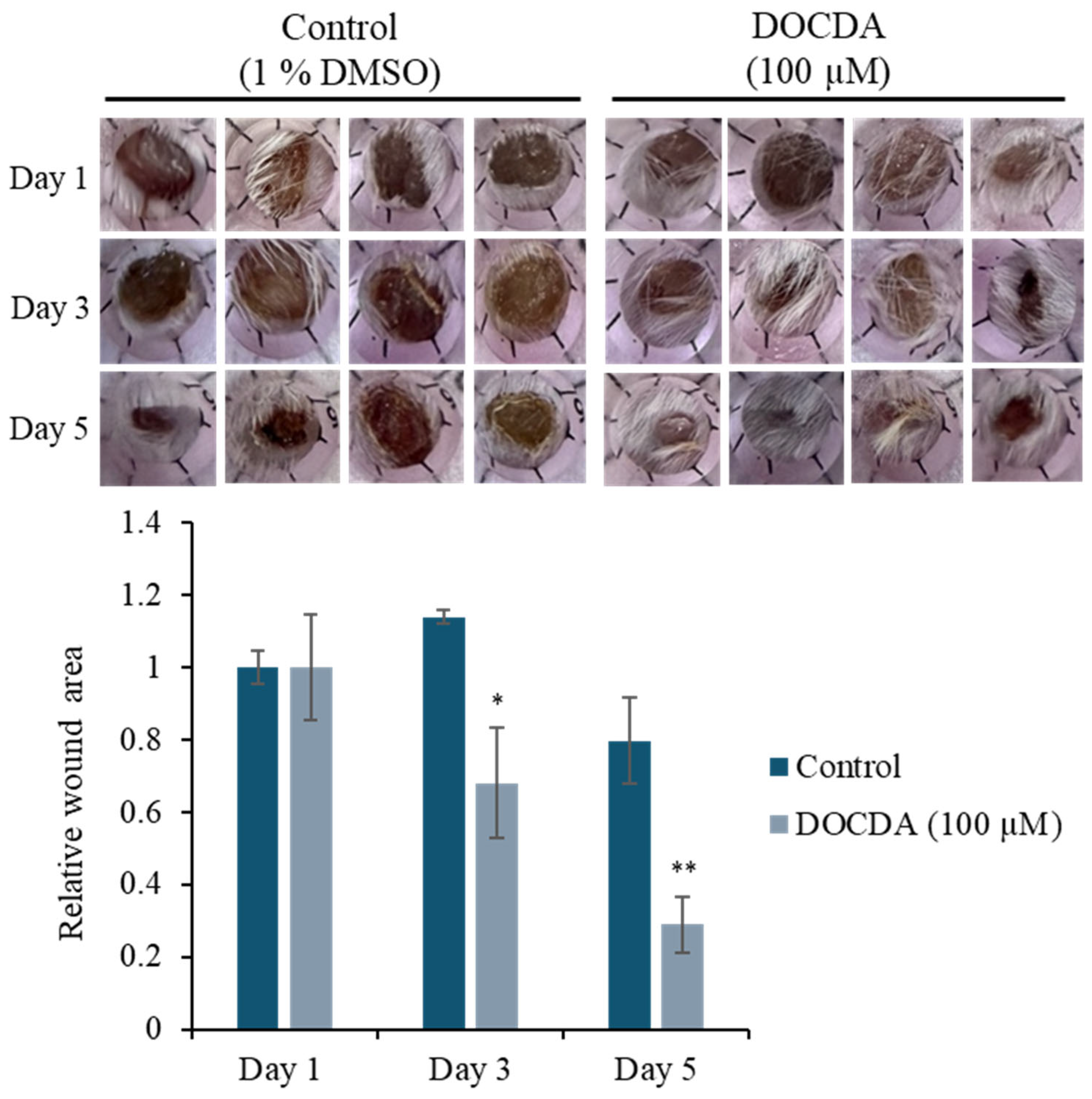
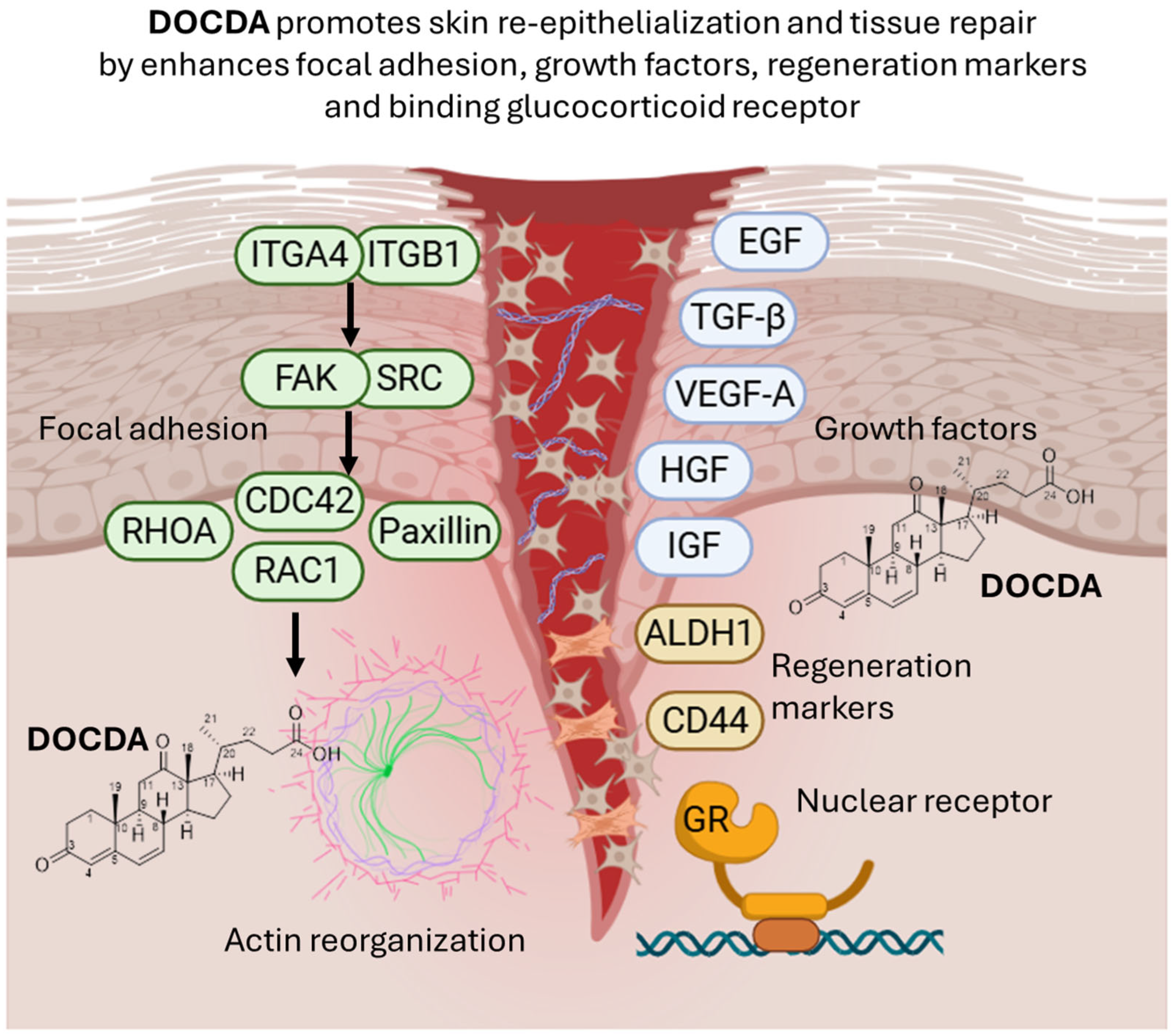
2.2. Bioactivity Assay
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Bacterial Isolation and Identification
3.3. Cultivation, Extraction, and Isolation
3.4. Cell Culture
3.5. Cell Viability
3.6. Invasion
3.7. Scratch Wound Healing Assay
3.8. Spheroid Formation Assay
3.9. qRT-PCR
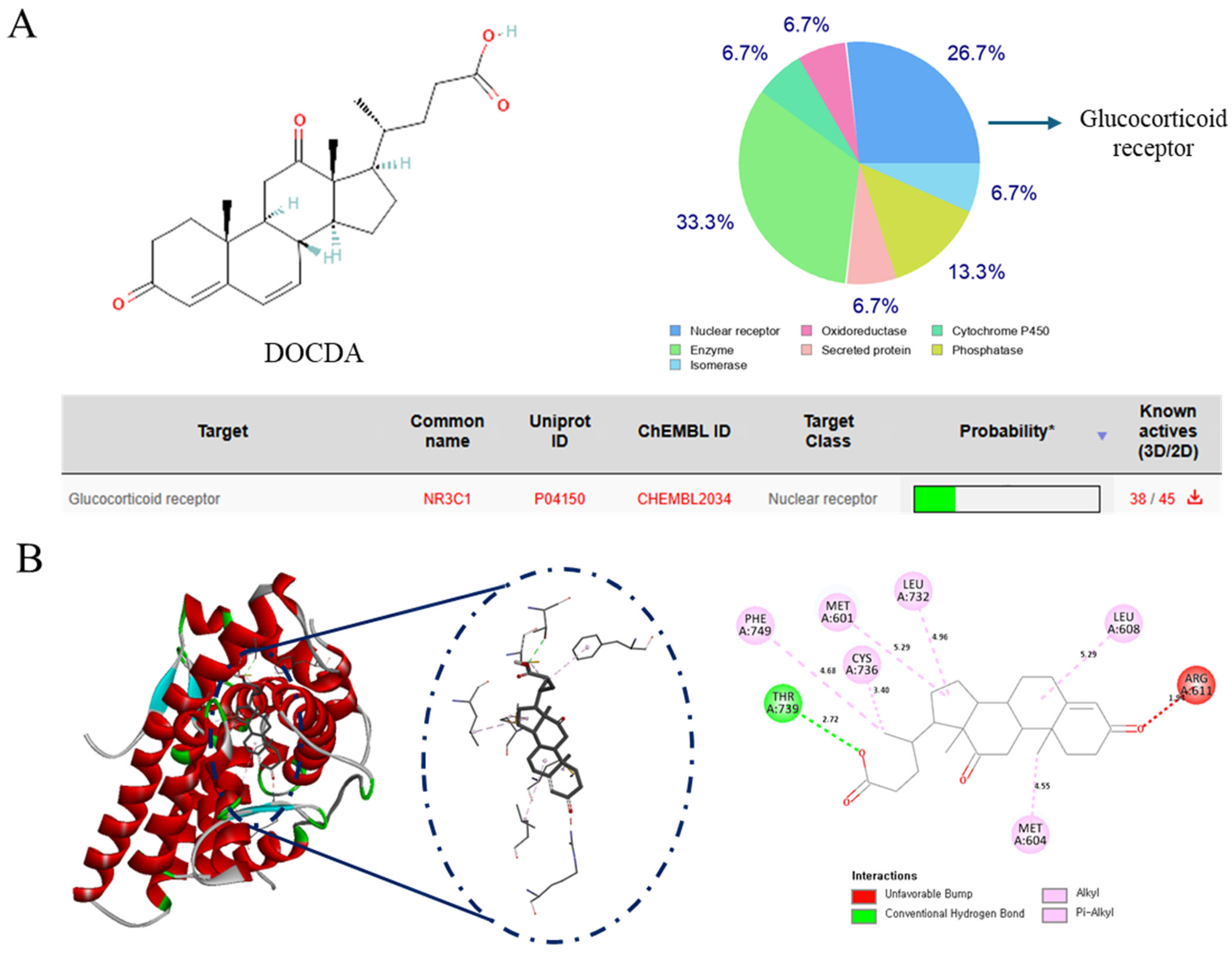
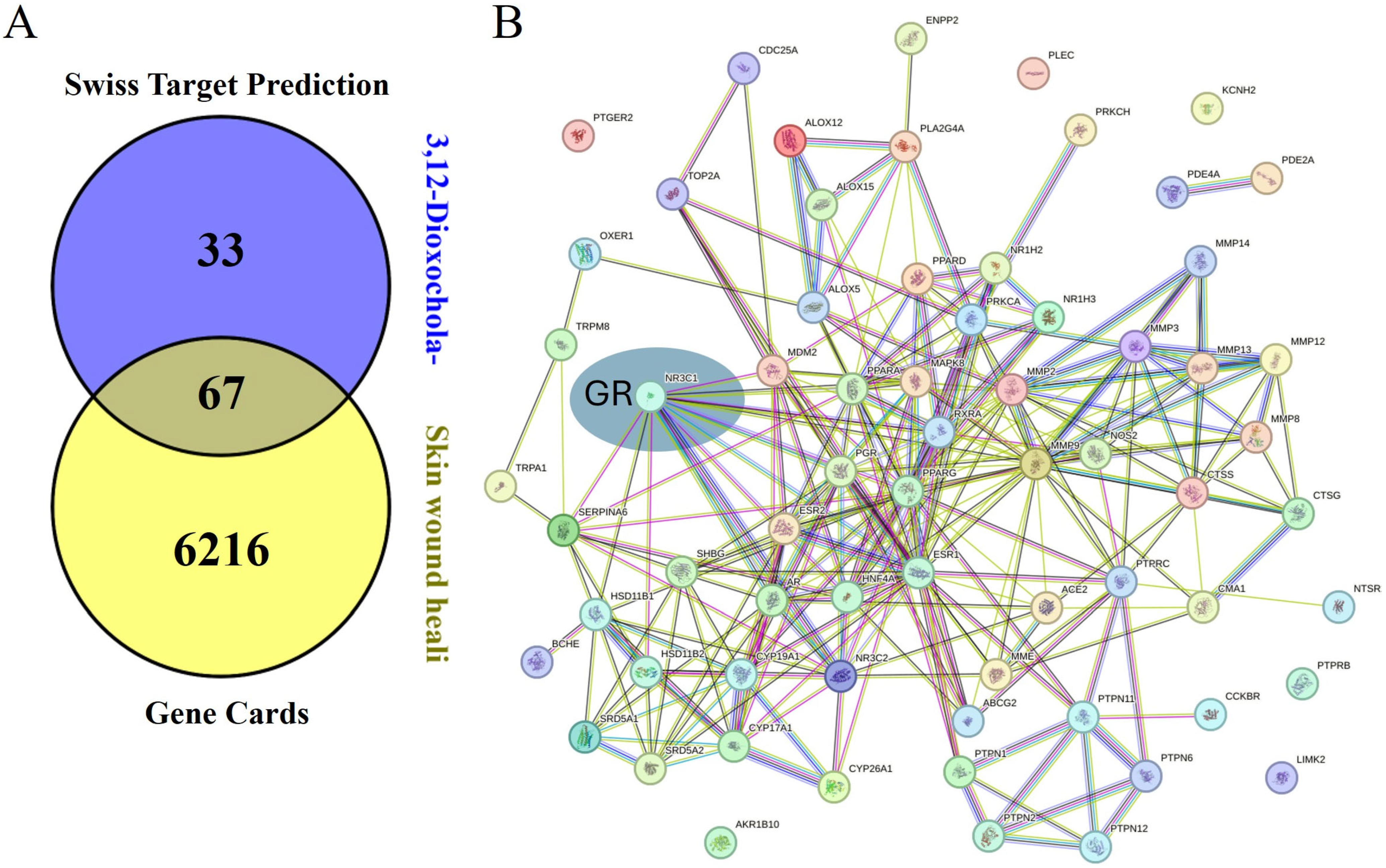
3.10. Target Prediction and Networking Pharmacology
3.11. Molecular Docking
3.12. In Silico Toxicity Assessment
3.13. Animal Model for Topical Wound Healing Evaluation
3.14. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DOCDA | 3,12-dioxochola-4,6-dien-24-oic acid |
| HaCaT | Human Adult Low Calcium Temperature Keratinocytes |
| EGF | Epidermal Growth Factor |
| VEGF-A | Vascular Endothelial Growth Factor A |
| IGF | Insulin-like Growth Factor |
| TGF-β | Transforming Growth Factor Beta |
| HGF | Hepatocyte Growth Factor |
| ITGB1 | Integrin Subunit Beta 1 |
| ITGA4 | Integrin Subunit Alpha 4 |
| FAK | Focal Adhesion Kinase |
| SRC | Proto-oncogene tyrosine-protein kinase Src |
| RHOA | Ras Homolog Family Member A |
| CDC42 | Cell Division Cycle 42 |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 |
| GR | Glucocorticoid Receptor |
| ALDH1 | Aldehyde Dehydrogenase 1 |
| CD44 | Cluster of Differentiation 44 |
References
- Jiang, X.; Zeng, Y.E.; Li, C.; Wang, K.; Yu, D.G. Enhancing diabetic wound healing advances in electrospun scaffolds from pathogenesis to therapeutic applications. Front. Bioeng. Biotechnol. 2024, 12, 1354286. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; DiPietro, L.A. Factors affecting wound healing. J. Dent. Res. 2010, 89, 219. [Google Scholar] [CrossRef]
- Kemin, L.; Rutie, Y. A critical review of the progress in prevention and treatment of radiation-induced skin damage. Front. Oncol. 2024, 14, 1395778. [Google Scholar] [CrossRef]
- Kim, J.; Kim, E.H.; Lee, H.; Sung, J.H.; Bang, O.Y. Clinical-scale mesenchymal stem cell-derived extracellular vesicle therapy for wound healing. Int. J. Mol. Sci. 2023, 24, 4273. [Google Scholar] [CrossRef]
- Hwang, J.H.; Kang, Y.; Park, H.J.; Kim, S.; Lee, S.H.; Kim, H.; Nam, S.J.; Lim, K.M. Skin wound healing effects of (+)-syringaresinol from ginseng berry. J. Ginseng Res. 2023, 47, 654–661. [Google Scholar] [CrossRef]
- Januszyk, M.; Kwon, S.H.; Wong, V.W.; Padmanabhan, J.; Maan, Z.N.; Whittam, A.J.; Major, M.R.; Gurtner, G.C. The role of focal adhesion kinase in keratinocyte fibrogenic gene expression. Int. J. Mol. Sci. 2017, 18, 1915. [Google Scholar] [CrossRef]
- Szabo, A.Z.; Fong, S.; Yue, L.; Zhang, K.; Strachan, L.R.; Scalapino, K.; Mancianti, M.L.; Ghadially, R. The CD44+ALDH+ population of human keratinocytes is enriched for epidermal stem cells with long-term repopulating ability. Stem Cells 2013, 31, 786–799. [Google Scholar] [CrossRef]
- Hu, Y.L.; Lu, S.; Szeto, K.W.; Sun, J.; Wang, Y.; Lasheras, J.C.; Chien, S. FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci. Rep. 2014, 4, 6024. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Gao, C.; Chen, R.; Jin, C.; Li, H.; Yan, H.; Wang, X. Focal adhesion kinase and paxillin promote migration and adhesion to fibronectin by swine skeletal muscle satellite cells. Oncotarget 2016, 7, 30845–30854. [Google Scholar] [CrossRef] [PubMed]
- López-Colomé, A.M.; Lee-Rivera, I.; Benavides-Hidalgo, R.; López, E. Paxillin: A crossroad in pathological cell migration. J. Hematol. Oncol. 2017, 10, 50. [Google Scholar] [CrossRef]
- Yoshida, S.; Matsumoto, K.; Tomioka, D.; Bessho, K.; Itami, S.; Yoshikawa, K.; Nakamura, T. Recombinant hepatocyte growth factor accelerates cutaneous wound healing in a diabetic mouse model. Growth Factors 2004, 22, 111–119. [Google Scholar] [CrossRef]
- Gilbert, R.W.D.; Vickaryous, M.K.; Viloria-Petit, A.M. Signalling by transforming growth factor beta isoforms in wound healing and tissue regeneration. J. Dev. Biol. 2016, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Hardwicke, J.; Schmaljohann, D.; Boyce, D.; Thomas, D. Epidermal growth factor therapy and wound healing—past, present and future perspectives. Surgeon 2008, 6, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, L.M.; Pérez, P.; Sevilla, L.M.; Pérez, P. Glucocorticoid receptor signaling in skin barrier function. In Keratin; InTech: London, UK, 2018. [Google Scholar]
- Sevilla, L.M.; Pérez, P. Roles of the Glucocorticoid and Mineralocorticoid Receptors in Skin Pathophysiology. Int. Mol. Sci. 2018, 19, 1906. [Google Scholar] [CrossRef] [PubMed]
- Stojadinovic, O.; Lee, B.; Vouthounis, C.; Vukelic, S.; Pastar, I.; Blumenberg, M.; Brem, H.; Tomic-Canic, M. Novel genomic effects of glucocorticoids in epidermal keratinocytes: Inhibition of apoptosis, interferon-γ pathway, and wound healing along with promotion of terminal differentiation. J. Biol. Chem. 2007, 282, 4021–4034. [Google Scholar] [CrossRef]
- Jozic, I.; Vukelic, S.; Stojadinovic, O.; Liang, L.; Ramirez, H.A.; Pastar, I.; Canic, M.T. Stress signals, mediated by membranous glucocorticoid receptor, activate PLC/PKC/GSK-3β/β-catenin pathway to inhibit wound closure. J. Invest. Dermatol. 2017, 137, 1144–1154. [Google Scholar] [CrossRef]
- Reichardt, S.D.; Amouret, A.; Muzzi, C.; Vettorazzi, S.; Tuckermann, J.P.; Lühder, F.; Reichardt, H.M. The role of glucocorticoids in inflammatory diseases. Cells 2021, 10, 2921. [Google Scholar] [CrossRef]
- Alotiby, A. Immunology of stress: A review article. J. Clin. Med. 2024, 13, 6394. [Google Scholar] [CrossRef]
- Yasir, M.; Goyal, A.; Sonthalia, S. Corticosteroid Adverse Effects; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Wang, A.S.; Armstrong, E.J.; Armstrong, A.W. Corticosteroids and wound healing: Clinical considerations in the perioperative period. Am. J. Surg. 2013, 206, 410–417. [Google Scholar] [CrossRef]
- Pang, J.P.; Hu, X.P.; Wang, Y.X.; Liao, J.N.; Chai, X.; Wang, X.W.; Shen, C.; Wang, J.J.; Zhang, L.L.; Wang, X.Y.; et al. Discovery of a novel nonsteroidal selective glucocorticoid receptor modulator by virtual screening and bioassays. Acta Pharmacol. Sin. 2022, 43, 2429–2438. [Google Scholar] [CrossRef]
- Jakob, F.; Hennen, S.; Gautrois, M.; Khalil, F.; Lockhart, A. Novel selective glucocorticoid receptor modulator GRM-01 demonstrates dissociation of anti-inflammatory effects from adverse effects on glucose and bone metabolism. Front. Pharmacol. 2025, 16, 1542351. [Google Scholar] [CrossRef]
- Hobson, A. Selective Glucocorticoid Receptor Modulators; Springer: Berlin/Heidelberg, Germany, 2023; pp. 59–97. [Google Scholar]
- Kim, S.; Lee, C.W.; Park, S.Y.; Asolkar, R.N.; Kim, H.; Kim, G.J.; Oh, S.J.; Kim, Y.; Lee, E.Y.; Oh, D.C.; et al. Acremonamide, a cyclic pentadepsipeptide with wound-healing properties isolated from a marine-derived fungus of the genus Acremonium. J. Nat. Prod. 2021, 84, 2249–2255. [Google Scholar] [CrossRef]
- Chen, K.; Li, J.; Chen, Z.; Shen, C.; Li, X.; Li, Y.; Song, D.; Li, X.; Wang, X.; Xia, Y.; et al. Notoginsenoside R1 alleviates blue light-induced corneal injury and wound healing delay by binding to and inhibiting TRIB1. Phytomedicine 2025, 138, 156399. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar, D.; Garg, Y.; Mahmood, S.; Chopra, S.; Bhatia, A. marine-derived polysaccharides and their therapeutic potential in wound healing application—A review. Int. J. Biol. 2023, 253, 127331. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.C.; Philp, J.C.; Aw, D.W.; Christofi, N. The genus Rhodococcus. J. Appl. Microbiol. 1998, 85, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Comparative genomics of the Rhodococcus genus shows wide distribution of biodegradation traits. Microorganism 2020, 8, 774. [Google Scholar] [CrossRef]
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannoni, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594. [Google Scholar] [CrossRef]
- Ceniceros, A.; Dijkhuizen, L.; Petrusma, M.; Medema, M.H. Genome-based exploration of the specialized metabolic capacities of the genus Rhodococcus. BMC Genom. 2017, 18, 593. [Google Scholar] [CrossRef]
- Alvarez, H.M.; Hernández, M.A.; Lanfranconi, M.P.; Silva, R.A.; Villalba, M.S. Rhodococcus as biofactories for microbial oil production. Molecules 2021, 26, 4871. [Google Scholar] [CrossRef]
- Ivshina, I.; Bazhutin, G.; Tyumina, E. Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future. Front. Microbiol. 2022, 13, 967127. [Google Scholar] [CrossRef]
- Maltseva, P.Y.; Plotnitskaya, N.A.; Ivshina, I.B. Transformation of terpenoids and steroids using Actinomycetes of the genus Rhodococcus. Molecules 2024, 29, 3378. [Google Scholar] [CrossRef]
- Williams, D.R.; Trudgill, P.W.; Taylor, D.G. Metabolism of 1,8-cineole by a Rhodococcus species: Ring cleavage reactions. J. Gen. Microbiol. 1989, 135, 1957–1967. [Google Scholar] [CrossRef][Green Version]
- Jin, J.; Mazon, H.; Van Den Heuvel, R.H.H.; Janssen, D.B.; Fraaije, M.W. Discovery of a eugenol oxidase from Rhodococcus sp. strain RHA1. FEBS J. 2007, 274, 2311–2321. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.C.C.R.; Da Fonseca, M.M.R. The remarkable Rhodococcus erythropolis. Appl. Microbiol. Biotechnol. 2005, 67, 715–726. [Google Scholar] [CrossRef]
- Busch, H.; Hagedoorn, P.L.; Hanefeld, U. Rhodococcus as a versatile biocatalyst in organic synthesis. Int. J. Mol. Sci. 2019, 20, 4787. [Google Scholar] [CrossRef]
- Bai, L.; Wang, L.; Zhao, M.; Toki, M.; Hasegawa, T.; Ogura, H.; Kataoka, T.; Hirose, H.; Sakai, J.; Bai, J.; et al. Bioactive pregnanes from Nerium oleander. J. Nat. Prod. 2007, 70, 14–18. [Google Scholar] [CrossRef]
- Owen, R.W.; Bilton, R.F. The degradation of cholic acid by Pseudomonas sp. N.C.I.B. 10590 under anaerobic conditions. Biochem. J. 1983, 216, 641–654. [Google Scholar] [CrossRef]
- Dong, J.; Liu, H.; Wang, H.; Lou, H.; Pan, W.; Li, J. Bioactivities of Steroids and Sesquiterpenes from the Branches and Leaves of Aglaia lawii. Molecules 2024, 29, 39. [Google Scholar] [CrossRef]
- Woo, J.K.; Ha, T.K.; Oh, D.C.; Oh, W.K.; Oh, K.B. Polyoxygenated Steroids from the Sponge Clathria gombawuiensis. J. Nat. Prod. 2017, 80, 3224–3233. [Google Scholar] [CrossRef]
- Jin, P.J.; Deng, Z.; Pei, Y.; Fu, H.; Li, J.; van Ofwegen, L.; Proksch, P.; Lin, W. Polyhydroxylated steroids from the soft coral Sinularia dissecta. Steroids 2005, 70, 487–493. [Google Scholar] [CrossRef]
- Tousif, M.I.; Nazir, M.; Riaz, N.; Saleem, M.; Mahmood, M.H.U.R.; Ahsan, M.; Tauseef, S.; Shafiq, N.; Moveed, A.; Zengin, G.; et al. Unrivalled insight into potential biopharmaceutical application of Allardia tridactylites (Kar. & Kir.) Sch. Bip.: Chemodiversity, in vitro bioactivities and computational analysis. Proc. Biochem. 2023, 129, 185–199. [Google Scholar]
- Zappaterra, F.; Costa, S.; Summa, D.; Bertolasi, V.; Semeraro, B.; Pedrini, P.; Buzzi, R.; Vertuani, S. Biotransformation of cortisone with Rhodococcus rhodnii: Synthesis of new steroids. Molecules 2021, 26, 1352. [Google Scholar] [CrossRef]
- Smith, J.; Rai, V. Novel factors regulating proliferation, migration, and differentiation of fibroblasts, keratinocytes, and vascular smooth muscle cells during wound healing. Biomedicines 2024, 12, 1939. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Han, S.H.; Hong, J.P.; Han, S.K.; Lee, D.H.; Kim, B.S.; Ahn, J.H.; Lee, J.W. Topical epidermal growth factor spray for the treatment of chronic diabetic foot ulcers: A phase III multicenter, double-blind, randomized, placebo-controlled trial. Diabetes Res. Clin. Pract. 2018, 142, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Rahim, F.; Yan, X.; Shah, J.A.; Bibi, N.; Khan, Z.U.; Nawaz, S.; Ming, Y. Epidermal growth factor outperforms placebo in the treatment of diabetic foot ulcer: A meta-analysis. F1000Res 2023, 11, 773. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, M.K.; Otero-Vinas, M.; Falanga, V. Transforming growth factor beta (TGF-β) isoforms in wound healing and fibrosis. Wound Repair Regen. 2016, 24, 215–222. [Google Scholar] [CrossRef]
- Tejedor, S.; Wågberg, M.; Correia, C.; Åvall, K.; Hölttä, M.; Hultin, L.; Lerche, M.; Davies, N.; Bergenhem, N.; Snijder, A.; et al. The combination of vascular endothelial growth factor a (VEGF-A) and fibroblast growth factor 1 (FGF1) modified mRNA improves wound healing in diabetic mice: An ex vivo and in vivo investigation. Cells 2024, 13, 414. [Google Scholar] [CrossRef]
- Li, J.F.; Duan, H.F.; Wu, C.T.; Zhang, D.J.; Deng, Y.; Yin, H.L.; Han, B.; Gong, H.C.; Wang, H.W.; Wang, Y.L. HGF accelerates wound healing by promoting the dedifferentiation of epidermal cells through β 1 -Integrin/ILK pathway. Biomed. Res. Int. 2013, 2103, 470418. [Google Scholar]
- Zhang, X.; Hu, F.; Li, J.; Chen, L.; Mao, Y.F.; Li, Q.B.; Nie, C.Y.; Lin, C.; Xiao, J. IGF-1 inhibits inflammation and accelerates angiogenesis via Ras/PI3K/IKK/NF-κB signaling pathways to promote wound healing. European. Eur. J. Pharm. Sci. 2024, 200, 106847. [Google Scholar] [CrossRef]
- Liu, F.; Wu, Q.; Dong, Z.; Liu, K. Integrins in cancer: Emerging mechanisms and therapeutic opportunities. Pharmacol. Ther. 2023, 247, 108458. [Google Scholar] [CrossRef]
- Desiderio, U.V.; Zhu, X.; Evans, J.P. ADAM2 interactions with mouse eggs and cell lines expressing α4/α9 (ITGA4/ITGA9) integrins: Implications for integrin-based adhesion and fertilization. PLoS ONE 2010, 5, 13744. [Google Scholar] [CrossRef] [PubMed]
- Aslan, J.E.; Mccarty, O.J.T. Rho GTPases in platelet function. J. Thromb. Haemost. 2013, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Pulat, S.; Yang, I.; Lee, J.; Hwang, S.; Zhou, R.; Gamage, C.D.B.; Varlı, M.; Taş, İ.; Yang, Y.; Park, S.Y.; et al. Anithiactin D, a phenylthiazole natural product from mudflat-derived Streptomyces sp., suppresses motility ofcCancer cells. Mar. Drugs. 2024, 22, 88. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Blanco, M.T.; Verboon, J.M.; Parkhurst, S.M. Coordination of Rho family GTPase activities to orchestrate cytoskeleton responses during cell wound repair. Curr. Biol. 2014, 24, 144–155. [Google Scholar] [CrossRef]
- Zhao, X.; Guan, J.L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011, 63, 610–615. [Google Scholar] [CrossRef]
- Tan, X.; Yan, Y.; Song, B.; Zhu, S.; Mei, Q.; Wu, K. Focal adhesion kinase: From biological functions to therapeutic strategies. Exp. Hematol. Oncol. 2023, 12, 1–19. [Google Scholar] [CrossRef]
- Turner, C.E. Paxillin and focal adhesion signalling. Nat. Cell. Biol. 2000, 2, E231–E236. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, V.; Mohan, A.; Gupta, G.; Kashyap, V. Translational research in the generation of therapeutic medicine for wound healing: A review. Discov. Med. 2024, 1, 158. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Ying, J.; Wei, C.; Wang, L.; Yang, Z.; Qi, F. Alda-1, an aldehyde dehydrogenase 2 agonist, improves cutaneous wound healing by activating epidermal keratinocytes via Akt/GSK-3β/β-catenin pxathway. Aesthetic. Plast. Surg. 2020, 44, 993–1005. [Google Scholar] [CrossRef]
- Pei, Y.; Reins, R.Y.; McDermott, A.M. Aldehyde dehydrogenase (ALDH) 3A1 expression by the human keratocyte and its repair phenotypes. Exp. Eye Res. 2006, 83, 1063–1073. [Google Scholar] [CrossRef]
- Michopoulou, A.; Montmasson, M.; Garnier, C.; Lambert, E.; Dayan, G.; Rousselle, P. A novel mechanism in wound healing: Laminin 332 drives MMP9/14 activity by recruiting syndecan-1 and CD44. Matrix Biol. 2020, 94, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, P.; Todd, L.; Shetye, S.; Monslow, J.; Puré, E. CD44-dependent inflammation, fibrogenesis, and collagenolysis regulates extracellular matrix remodeling and tensile strength during cutaneous wound healing. Matrix Biol. 2018, 314, 75–76. [Google Scholar] [CrossRef] [PubMed]
- Klaas, M.; Mäemets-Allas, K.; Heinmäe, E.; Lagus, H.; Arak, T.; Eller, M.; Kingo, K.; Kankuri, E.; Jaks, V. Olfactomedin-4 improves cutaneous wound healing by promoting skin cell proliferation and migration through POU5F1/OCT4 and ESR1 signalling cascades. Cell. Mol. Life Sci. 2022, 79, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, A.; Nayak, S.; Graf, R.; Cross, M.; Hasneen, K.; Gutkind, J.S.; Brooks, S.R.; Morasso, M.I. SOX2 epidermal overexpression promotes cutaneous wound healing via activation of EGFR/MEK/ERK signaling mediated by EGFR ligands. J. Invest. Dermatol. 2019, 139, 1809–1820.e8. [Google Scholar] [CrossRef]
- Rodriguez, G.; Kaya, I.; Jorcano, J.L.; Vassalli, P.; Stamenkovic, I. Selective suppression of CD44 in keratinocytes of mice bearing an antisense CD44 transgene driven by a tissue-specific promoter disrupts hyaluronate metabolism in the skin and impairs keratinocyte proliferation. Genes. Dev. 1997, 11, 996–1007. [Google Scholar]
- Bourguignon, L.Y.W. Matrix hyaluronan-activated CD44 signaling promotes keratinocyte activities and improves abnormal epidermal functions. Am. J. Pathol. 2014, 184, 1912–1919. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Ramez, M.; Gilad, E.; Singleton, P.A.; Man, M.Q.; Crumrine, D.A.; Elias, P.M.; Feingold, K.R. Hyaluronan-CD44 interaction stimulates keratinocyte differentiation, lamellar body formation/secretion, and permeability barrier homeostasis. J. Invest. Dermatol. 2006, 126, 1356–1365. [Google Scholar] [CrossRef]
- Pósa, S.P.; Saskői, É.; Bársony, L.; Pongor, L.; Fekete, F.; Papp, J.; Bozsik, A.; Patócs, A.; Butz, H. The impact of glucocorticoid receptor transactivation on context-dependent cell migration dynamics. Sci. Rep. 2025, 15, 4163. [Google Scholar] [CrossRef]
- Ko, R.F.; Davidson, O.Q.C.; Ahmed, M.A.; Clark, R.M.; Brandenburg, J.S.; Pankratz, V.S.; Sharma, G.; Hathaway, H.J.; Prossnitz, E.R.; Howdieshell, T.R. GPER deficiency impedes murine myocutaneous revascularization and wound healing. Sci. Rep. 2024, 14, 18400. [Google Scholar] [CrossRef]
- Zomer, H.D.; Cooke, P.S. Targeting estrogen signaling and biosynthesis for aged skin repair. Front. Physiol. 2023, 14, 1281071. [Google Scholar] [CrossRef]
- Sriphan, P.; Chruewkamlow, N.; Sathan-ard, C.; Phutthakunphithak, P.; Sampattavanich, S.; Anekpuritanang, T.; Sakamula, R.; Likhityungyuen, T.; Wongwanit, C.; Ruangsetakit, C.; et al. Effectiveness of quality and quantity mononuclear cells for enhancing wound healing in diabetic ischemic limb animal model. Int. Wound J. 2025, 22, e70106. [Google Scholar] [CrossRef] [PubMed]
- Varlı, M.; Kim, E.; Oh, S.; Pulat, S.; Zhou, R.; Gamage, C.D.B.; Gökalsın, B.; Sesal, N.C.; Kim, K.K.; Paik, M.J.; et al. Chrysophanol inhibits of colorectal cancer cell motility and energy metabolism by targeting the KITENIN/ErbB4 oncogenic complex. Cancer. Cell Int. 2024, 24, 253. [Google Scholar] [CrossRef]
- Varlı, M.; Lee, K.; Kang, K.B.; Kim, H. Unveiling the antimetastatic activity of monoterpene indole alkaloids targeting MMP9 in cancer cells, with a focus on pharmacokinetic and cellular insights. Mol. Cells 2024, 47, 100143. [Google Scholar] [CrossRef]
- Varlı, M.; Bhosle, S.R.; Jo, E.; Yu, Y.H.; Yang, Y.; Ha, H.H.; Kim, H. Development and synthesis of diffractaic acid analogs as potent inhibitors of colorectal cancer stem cell traits. Sci. Rep. 2025, 15, 6695. [Google Scholar] [CrossRef]
- Varlı, M.; Ji, M.; Kim, E.; Kim, S.J.; Choi, B.; Ha, H.H.; Kim, K.K.; Paik, M.J.; Kim, H. Emodin disrupts the KITENIN oncogenic complex by binding ErbB4 and suppresses colorectal cancer progression in dual blockade with KSRP-binding compound. Phytomedicine 2025, 136, 156247. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic. Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Seaton, W.B.; Burke, S.J.; Fisch, A.R.; Schilletter, W.A.; Beck, M.G.A.; Cassagne, G.A.; Harvey, I.; Fontenot, M.S.; Collier, J.J.; Campagna, S.R. Channel expansion in the ligand-binding domain of the glucocorticoid receptor contributes to the activity of highly potent glucocorticoid analogues. Molecules 2024, 29, 1546. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Grimm, M.; Dai, W.T.; Hou, M.C.; Xiao, Z.X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef] [PubMed]

| PK and Toxicity Category | Property Name | Predicted Value | Property Unit | |
|---|---|---|---|---|
| DOCDA | Hydrocortisone | |||
| Absorption | Caco-2 | −4.94 | −4.55 | logPaap |
| Human Oral Bioavailability 20% | Bioavailable | Bioavailable | Bioavailable/Non-Bioavailable | |
| Human Intestinal Absorption | Absorbed | Absorbed | Absorbed/Non-Absorbed | |
| Madin–Darby Canine Kidney | −4.74 | −4.6 | cm/s | |
| Human Oral Bioavailability 50% | Bioavailable | Bioavailable | Bioavailable/Non-Bioavailable | |
| P-Glycoprotein Inhibitor | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| P-Glycoprotein Substrate | Non-Substrate | Non-Substrate | Substrate/Non-Substrate | |
| Skin Permeability | −1.66 | −2.12 | log Kp | |
| Distribution | Blood–Brain Barrier (Central Nervous System) | −2.48 | −3.05 | log PS |
| Blood-Brain Barrier | Penetrable | Penetrable | Penetrating/Non-Penetrating | |
| Fraction Unbound (Human) | 1.37 | 0.78 | free proportion | |
| Plasma Protein Binding | 84.06 | 67.43 | therapeutic index | |
| Steady-State Volume of Distribution | 0.76 | 0.47 | log VDss | |
| Metabolism | Breast Cancer Resistance Protein | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor |
| CYP 1A2 Inhibitor | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| CYP 1A2_substrate | Non-Substrate | Non-Substrate | Substrate/Non-Substrate | |
| CYP 2C19 Inhibitor | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| CYP 2C19_substrate | Non-Substrate | Substrate | cyp2c19_substrate | |
| CYP 2C9 Inhibitor | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| CYP 2C9 Substrate | Non-Substrate | Non-Substrate | Substrate/Non-Substrate | |
| CYP 2D6 Inhibitor | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| CYP 2D6 Substrate | Non-Substrate | Non-Substrate | Substrate/Non-Substrate | |
| CYP 3A4 Inhibitor | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| CYP 3A4 Substrate | Substrate | Substrate | Substrate/Non-Substrate | |
| OATP1B1 | Inhibitor | Inhibitor | Inhibitor/Non-Inhibitor | |
| OATP1B3 | Non-Inhibitor | Non-Inhibitor | Inhibitor/Non-Inhibitor | |
| Excretion | Clearance | 4.73 | 6.16 | Log (ml/min/kg) |
| Organic Cation Transporter 2 | Non-Inhibitor | Inhibitor | Inhibitor/Non-Inhibitor | |
| Half-Life of Drug | Half-Life < 3 h | Half-Life < 3 h | Half-life ≥ 3 h/Half-life < 3 h | |
| Toxicity | AMES Mutagenesis | Safe | Toxic | Toxic/Safe |
| Bioconcentration Factor | −0.61 | −1.59 | log10(L/kg) | |
| Biodegradation | Safe | Safe | Toxic/Safe | |
| Eye Corrosion | Safe | Safe | Toxic/Safe | |
| Eye irritation | Safe | Safe | Toxic/Safe | |
| Maximum Tolerated Dose | 0.84 | −0.15 | log mg/kg/day | |
| NR-AhR | Safe | Safe | Toxic/Safe | |
| NR-AR | Toxic | Toxic | Toxic/Safe | |
| NR-AR-LBD | Safe | Toxic | Toxic/Safe | |
| NR-Aromatase | Safe | Safe | Toxic/Safe | |
| NR-ER | Safe | Toxic | Toxic/Safe | |
| NR-ER-LBD | Safe | Safe | Toxic/Safe | |
| NR-GR | Safe | Toxic | Toxic/Safe | |
| NR-PPAR-gamma | Safe | Safe | Toxic/Safe | |
| NR-TR | Safe | Safe | Toxic/Safe | |
| Rat (Acute) | 2.2 | 1.95 | log[1/(mol/kg)] | |
| Rat (Chronic Oral) | 2.08 | 2.11 | log(mg/kg_bw/day) | |
| Fathead Minnow | 3.92 | 3.92 | −log10 [(mg/L)/(1000 × MW)] | |
| Skin Sensitization | Safe | Safe | Toxic/Safe | |
| SR-ARE | Toxic | Safe | Toxic/Safe | |
| SR-ATAD5 | Safe | Safe | Toxic/Safe | |
| SR-HSE | Safe | Safe | Toxic/Safe | |
| SR-MMP | Safe | Safe | Toxic/Safe | |
| SR-p53 | Safe | Safe | Toxic/Safe | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varlı, M.; Tan, H.; Lee, C.; Lee, J.; Lee, J.Y.; Kim, J.-H.; Lee, S.; Kim, H.; Nam, S.-J. A Marine-Derived Steroid from Rhodococcus sp., 3,12-Dioxochola-4,6-dien-24-oic Acid, Enhances Skin Re-Epithelialization and Tissue Repair. Mar. Drugs 2025, 23, 292. https://doi.org/10.3390/md23070292
Varlı M, Tan H, Lee C, Lee J, Lee JY, Kim J-H, Lee S, Kim H, Nam S-J. A Marine-Derived Steroid from Rhodococcus sp., 3,12-Dioxochola-4,6-dien-24-oic Acid, Enhances Skin Re-Epithelialization and Tissue Repair. Marine Drugs. 2025; 23(7):292. https://doi.org/10.3390/md23070292
Chicago/Turabian StyleVarlı, Mücahit, Hui Tan, Chaeyoung Lee, Jeongyun Lee, Ji Young Lee, Jeong-Hyeon Kim, Songyi Lee, Hangun Kim, and Sang-Jip Nam. 2025. "A Marine-Derived Steroid from Rhodococcus sp., 3,12-Dioxochola-4,6-dien-24-oic Acid, Enhances Skin Re-Epithelialization and Tissue Repair" Marine Drugs 23, no. 7: 292. https://doi.org/10.3390/md23070292
APA StyleVarlı, M., Tan, H., Lee, C., Lee, J., Lee, J. Y., Kim, J.-H., Lee, S., Kim, H., & Nam, S.-J. (2025). A Marine-Derived Steroid from Rhodococcus sp., 3,12-Dioxochola-4,6-dien-24-oic Acid, Enhances Skin Re-Epithelialization and Tissue Repair. Marine Drugs, 23(7), 292. https://doi.org/10.3390/md23070292








