Synthesis and Evaluation of Aquatic Antimicrobial Peptides Derived from Marine Metagenomes Using a High-Throughput Screening Approach
Abstract
1. Introduction
2. Results
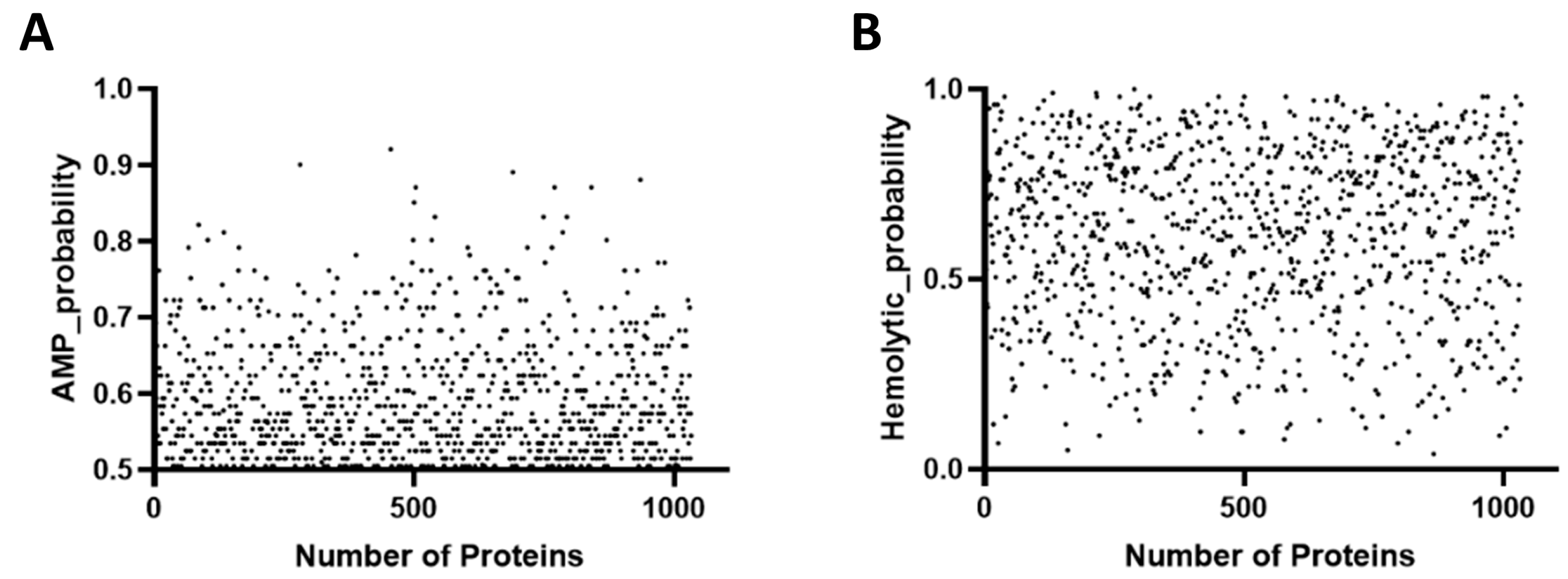
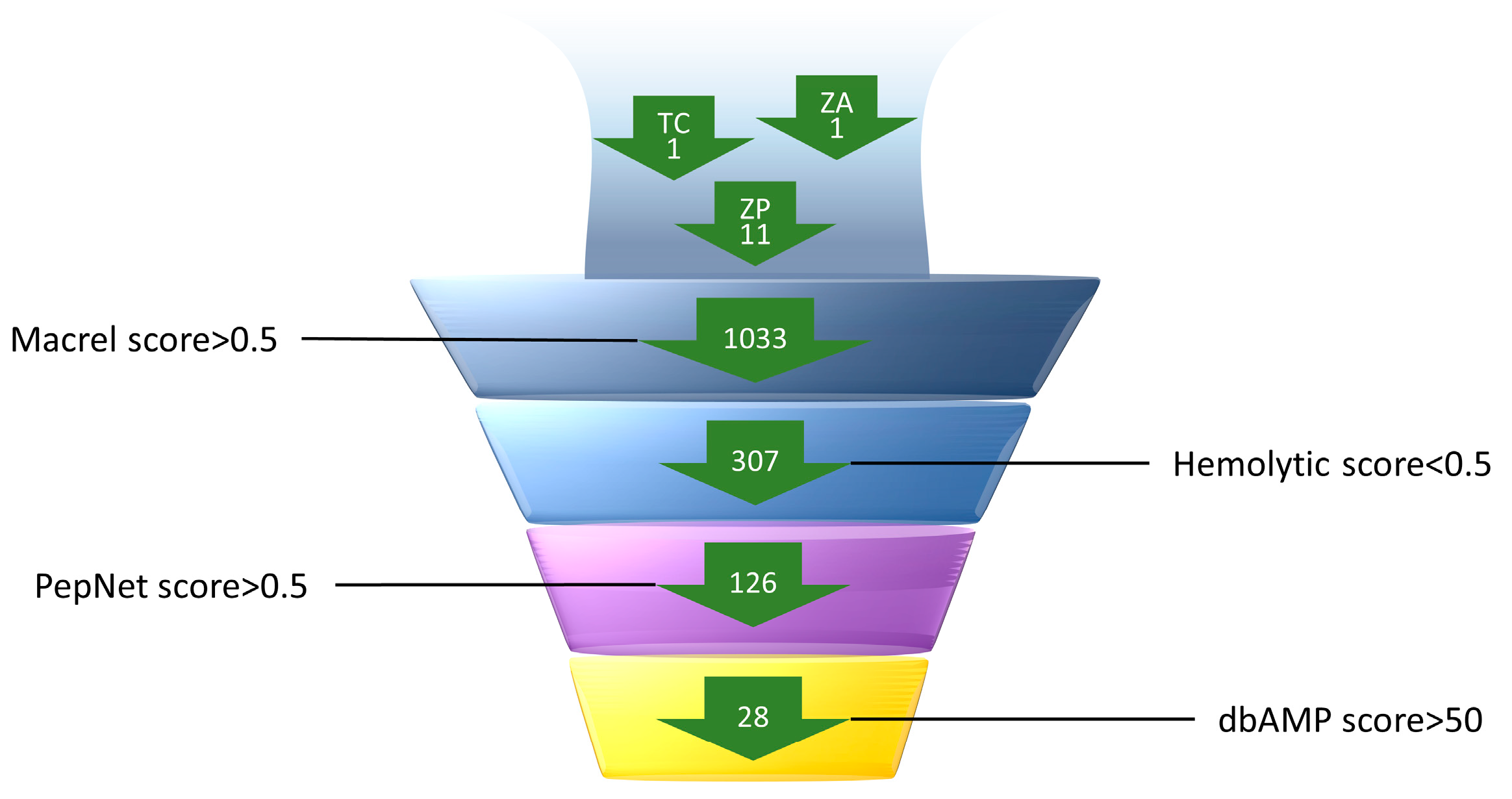
2.1. Prediction and Screening of AMPs in the Macrogenome
2.2. Physicochemical Properties of Candidate AMPs
2.3. The Rapid Synthesis and Screening of AMPs Is Based on Cell-Free Expression
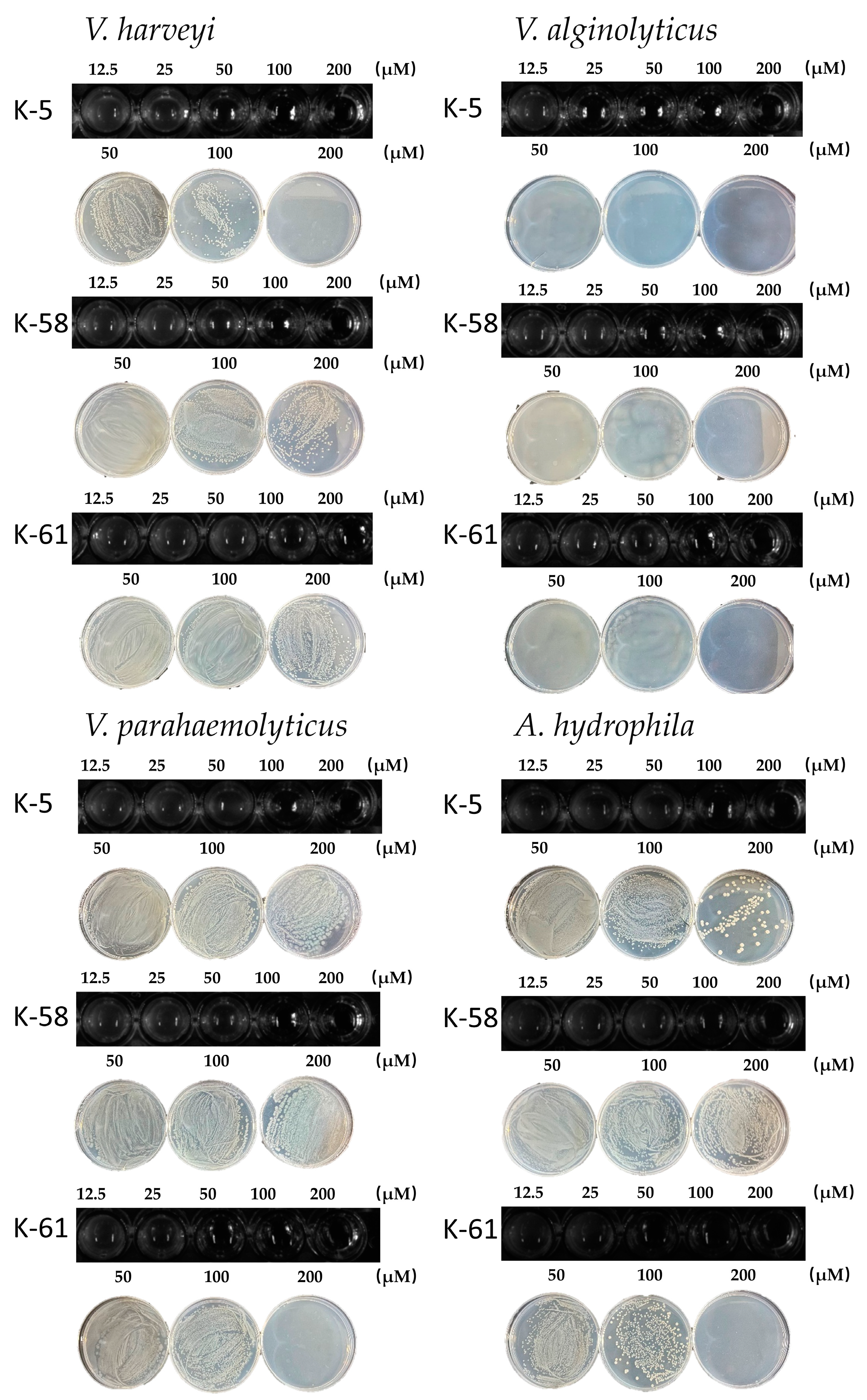
2.4. Inhibition of V. harveyi, V. alginolyticus, V. parahaemolyticus, and A. hydrophila by Three AMPs
2.5. Evaluation of In Vitro Cytotoxicity and Hemolytic Activity
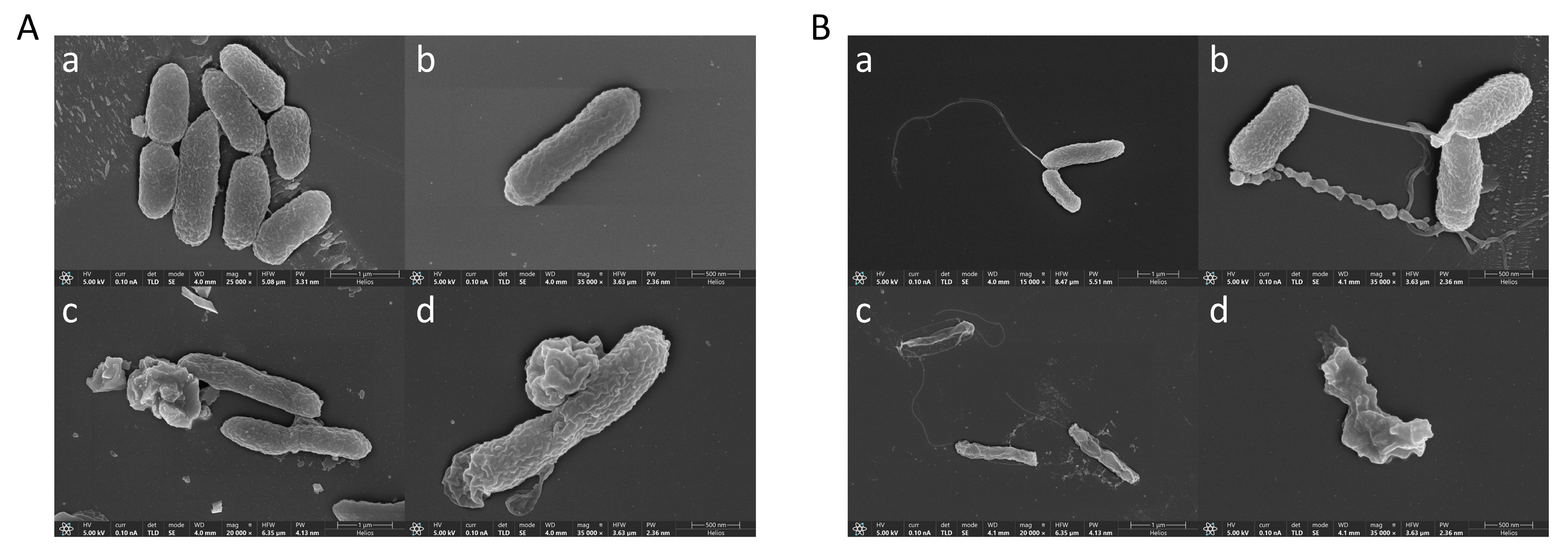
2.6. Effect of Peptide K-5 on V. alginolyticus and V. harvey Ultrastructure via Scanning Electron Microscopy
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Prediction and Screening of Candidate AMPs
4.3. Prediction of Three-Dimensional Structures and Physicochemical Properties of Candidate AMPs
4.4. Construction of a Cell-Free Expression System
4.5. Rapid Synthesis and Screening of AMP Candidates
4.6. Chemical Synthesis of Antimicrobial Peptides K-5, K-58, and K-61
4.7. Minimum Inhibitory Concentration and Minimum Bactericidal Concentration of Synthetic Peptides
4.8. Cytotoxicity Assay
4.9. Hemolysis Assay
4.10. Effects of Antimicrobial Peptide K-5 on Bacterial Morphology and Structure as Observed by Scanning Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Liu, H.; Liu, S.; Mao, J. Impact of Bacteriocins on Multidrug-resistant Bacteria and Their Application in Aquaculture Disease Prevention and Control. Rev. Aquac. 2024, 16, 1286–1307. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, Y.; Song, Z.; Tan, Z.; Cheng, J. Recent Advances in Design of Antimicrobial Peptides and Polypeptides toward Clinical Translation. Adv. Drug Deliv. Rev. 2021, 170, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shang, L.; Lan, J.; Chou, S.; Feng, X.; Shi, B.; Wang, J.; Lyu, Y.; Shan, A. Targeted and Intracellular Antibacterial Activity against S. agalactiae of the Chimeric Peptides Based on Pheromone and Cell-Penetrating Peptides. ACS Appl. Mater. Interfaces 2020, 12, 44459–44474. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jia, Y.; Sun, Y.; Liu, K.; Zhou, C.; Liu, C.; Li, D.; Liu, G.; Zhang, C.; Yang, T.; et al. Global Marine Microbial Diversity and Its Potential in Bioprospecting. Nature 2024, 633, 371–379. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial Peptides (AMPs): A Promising Class of Antimicrobial Compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef]
- Chen, N.; Jiang, C. Antimicrobial Peptides: Structure, Mechanism, and Modification. Eur. J. Med. Chem. 2023, 255, 115377. [Google Scholar] [CrossRef]
- Al-Omari, A.M.; Akkam, Y.H.; Zyout, A.; Younis, S.; Tawalbeh, S.M.; Al-Sawalmeh, K.; Al Fahoum, A.; Arnold, J. Accelerating Antimicrobial Peptide Design: Leveraging Deep Learning for Rapid Discovery. PLoS ONE 2024, 19, e0315477. [Google Scholar] [CrossRef]
- Kim, H.K.; Min, S.; Song, M.; Jung, S.; Choi, J.W.; Kim, Y.; Lee, S.; Yoon, S.; Kim, H. (Henry) Deep Learning Improves Prediction of CRISPR–Cpf1 Guide RNA Activity. Nat. Biotechnol. 2018, 36, 239–241. [Google Scholar] [CrossRef]
- Lopez, R.; Regier, J.; Cole, M.B.; Jordan, M.I.; Yosef, N. Deep Generative Modeling for Single-Cell Transcriptomics. Nat. Methods 2018, 15, 1053–1058. [Google Scholar] [CrossRef]
- Wu, R.; Ding, F.; Wang, R.; Shen, R.; Zhang, X.; Luo, S.; Su, C.; Wu, Z.; Xie, Q.; Berger, B.; et al. High-Resolution de novo Structure Prediction from Primary Sequence. bioRxiv 2022. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a 3-Track Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Tian, T.; Huang, S.; Wan, F.; Li, J.; Li, S.; Wang, X.; Yang, H.; Hong, L.; Wu, N.; et al. An Integrative Drug Repositioning Framework Discovered a Potential Therapeutic Agent Targeting COVID-19. Signal Transduct. Target. Ther. 2021, 6, 165. [Google Scholar] [CrossRef]
- Rajkomar, A.; Oren, E.; Chen, K.; Dai, A.M.; Hajaj, N.; Hardt, M.; Liu, P.J.; Liu, X.; Marcus, J.; Sun, M.; et al. Scalable and Accurate Deep Learning with Electronic Health Records. npj Digit. Med. 2018, 1, 18. [Google Scholar] [CrossRef]
- Fan, S.; Qin, P.; Lu, J.; Wang, S.; Zhang, J.; Wang, Y.; Cheng, A.; Cao, Y.; Ding, W.; Zhang, W. Bioprospecting of Culturable Marine Biofilm Bacteria for Novel Antimicrobial Peptides. iMeta 2024, 3, e244. [Google Scholar] [CrossRef]
- Ji, S.; An, F.; Zhang, T.; Lou, M.; Guo, J.; Liu, K.; Zhu, Y.; Wu, J.; Wu, R. Antimicrobial Peptides: An Alternative to Traditional Antibiotics. Eur. J. Med. Chem. 2024, 265, 116072. [Google Scholar] [CrossRef]
- Deng, T.; Ge, H.; He, H.; Liu, Y.; Zhai, C.; Feng, L.; Yi, L. The Heterologous Expression Strategies of Antimicrobial Peptides in Microbial Systems. Protein Expr. Purif. 2017, 140, 52–59. [Google Scholar] [CrossRef]
- Nirenberg, M.W.; Matthaei, J.H. The Dependence of Cell-Free Protein Synthesis in E. coli upon Naturally Occurring or Synthetic Polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef]
- Kightlinger, W.; Duncker, K.E.; Ramesh, A.; Thames, A.H.; Natarajan, A.; Stark, J.C.; Yang, A.; Lin, L.; Mrksich, M.; DeLisa, M.P.; et al. A Cell-Free Biosynthesis Platform for Modular Construction of Protein Glycosylation Pathways. Nat. Commun. 2019, 10, 5404. [Google Scholar] [CrossRef]
- Jung, J.K.; Alam, K.K.; Verosloff, M.S.; Capdevila, D.A.; Desmau, M.; Clauer, P.R.; Lee, J.W.; Nguyen, P.Q.; Pastén, P.A.; Matiasek, S.J.; et al. Cell-Free Biosensors for Rapid Detection of Water Contaminants. Nat. Biotechnol. 2020, 38, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Pandi, A.; Adam, D.; Zare, A.; Trinh, V.T.; Schaefer, S.L.; Burt, M.; Klabunde, B.; Bobkova, E.; Kushwaha, M.; Foroughijabbari, Y.; et al. Cell-Free Biosynthesis Combined with Deep Learning Accelerates de Novo-Development of Antimicrobial Peptides. Nat. Commun. 2023, 14, 7197. [Google Scholar] [CrossRef] [PubMed]
- Suyamud, B.; Lohwacharin, J.; Yang, Y.; Sharma, V.K. Antibiotic Resistant Bacteria and Genes in Shrimp Aquaculture Water: Identification and Removal by Ferrate(VI). J. Hazard. Mater. 2021, 420, 126572. [Google Scholar] [CrossRef]
- Järvå, M.; Lay, F.T.; Phan, T.K.; Humble, C.; Poon, I.K.H.; Bleackley, M.R.; Anderson, M.A.; Hulett, M.D.; Kvansakul, M. X-Ray Structure of a Carpet-like Antimicrobial Defensin–Phospholipid Membrane Disruption Complex. Nat. Commun. 2018, 9, 1962. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial Peptides: Basic Facts and Emerging Concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Wong, G.C.L. Antimicrobial Peptides and Induced Membrane Curvature: Geometry, Coordination Chemistry, and Molecular Engineering. Curr. Opin. Solid State Mater. Sci. 2013, 17, 151–163. [Google Scholar] [CrossRef]
- Houyvet, B.; Zanuttini, B.; Corre, E.; Le Corguillé, G.; Henry, J.; Zatylny-Gaudin, C. Design of Antimicrobial Peptides from a Cuttlefish Database. Amino Acids 2018, 50, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, J.; Tong, Z.; Jia, Y.; Yang, B.; Wang, Z. The Revitalization of Antimicrobial Peptides in the Resistance Era. Pharmacol. Res. 2021, 163, 105276. [Google Scholar] [CrossRef]
- Lou, M.; Ji, S.; Wu, R.; Zhu, Y.; Wu, J.; Zhang, J. Microbial Production Systems and Optimization Strategies of Antimicrobial Peptides: A Review. World J. Microbiol. Biotechnol. 2025, 41, 66. [Google Scholar] [CrossRef]
- Mao, F.; Bao, Y.; Wong, N.-K.; Huang, M.; Liu, K.; Zhang, X.; Yang, Z.; Yi, W.; Shu, X.; Xiang, Z.; et al. Large-Scale Plasma Peptidomic Profiling Reveals a Novel, Nontoxic, Crassostrea Hongkongensis-Derived Antimicrobial Peptide against Foodborne Pathogens. Mar. Drugs 2021, 19, 420. [Google Scholar] [CrossRef]
- Davagnino, J.; Herrero, M.; Furlong, D.; Moreno, F.; Kolter, R. The DNA Replication Inhibitor Microcin B17 Is a Forty-three-amino-acid Protein Containing Sixty Percent Glycine. Proteins 1986, 1, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Post-Translational Modifications of Natural Antimicrobial Peptides and Strategies for Peptide Engineering. Curr. Biotechnol. 2014, 1, 72–79. [Google Scholar]
- De Medeiros, L.N.; Angeli, R.; Sarzedas, C.G.; Barreto-Bergter, E.; Valente, A.P.; Kurtenbach, E.; Almeida, F.C.L. Backbone Dynamics of the Antifungal Psd1 Pea Defensin and Its Correlation with Membrane Interaction by NMR Spectroscopy. Biochim. Biophys. Acta (BBA)—Biomembr. 2010, 1798, 105–113. [Google Scholar] [CrossRef]
- Schreiber, C.; Müller, H.; Birrenbach, O.; Klein, M.; Heerd, D.; Weidner, T.; Salzig, D.; Czermak, P. A High-Throughput Expression Screening Platform to Optimize the Production of Antimicrobial Peptides. Microb. Cell Fact. 2017, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Pelton, J.M.; Hochuli, J.E.; Sadecki, P.W.; Katoh, T.; Suga, H.; Hicks, L.M.; Muratov, E.N.; Tropsha, A.; Bowers, A.A. Cheminformatics-Guided Cell-Free Exploration of Peptide Natural Products. J. Am. Chem. Soc. 2024, 146, 8016–8030. [Google Scholar] [CrossRef]
- Brahma, B.; Patra, M.C.; Karri, S.; Chopra, M.; Mishra, P.; De, B.C.; Kumar, S.; Mahanty, S.; Thakur, K.; Poluri, K.M.; et al. Diversity, Antimicrobial Action and Structure-Activity Relationship of Buffalo Cathelicidins. PLoS ONE 2015, 10, e0144741. [Google Scholar] [CrossRef] [PubMed]
- Santos-Júnior, C.D.; Pan, S.; Zhao, X.-M.; Coelho, L.P. Macrel: Antimicrobial Peptide Screening in Genomes and Metagenomes. PeerJ 2020, 8, e10555. [Google Scholar] [CrossRef]
- Santos-Júnior, C.D.; Torres, M.D.T.; Duan, Y.; Rodríguez Del Río, Á.; Schmidt, T.S.B.; Chong, H.; Fullam, A.; Kuhn, M.; Zhu, C.; Houseman, A.; et al. Discovery of Antimicrobial Peptides in the Global Microbiome with Machine Learning. Cell 2024, 187, 3761–3778.e16. [Google Scholar] [CrossRef]
- Jhong, J.-H.; Yao, L.; Pang, Y.; Li, Z.; Chung, C.-R.; Wang, R.; Li, S.; Li, W.; Luo, M.; Ma, R.; et al. dbAMP 2.0: Updated Resource for Antimicrobial Peptides with an Enhanced Scanning Method for Genomic and Proteomic Data. Nucleic Acids Res. 2022, 50, D460–D470. [Google Scholar] [CrossRef]
- Han, J.; Kong, T.; Liu, J. PepNet: An Interpretable Neural Network for Anti-Inflammatory and Antimicrobial Peptides Prediction Using a Pre-Trained Protein Language Model. Commun. Biol. 2024, 7, 1198. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L.; Scientific, D.; Carlos, S. PyMOL: An Open-Source Molecular Graphics Tool. Available online: https://legacy.ccp4.ac.uk/newsletters/newsletter40/11_pymol.pdf (accessed on 19 March 2025).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Peng, S.; Yang, J.; Zhou, F.; Suo, H. Isolation and Identification of Novel Antibacterial Peptides Produced by Lactobacillus Fermentum SHY10 in Chinese Pickles. Food Chem. 2021, 348, 129097. [Google Scholar] [CrossRef]
- Cai, D.; Chen, S.; Wu, B.; Chen, J.; Tao, D.; Li, Z.; Dong, Q.; Zou, Y.; Chen, Y.; Bi, C.; et al. Construction of Multifunctional Porcine Acellular Dermal Matrix Hydrogel Blended with Vancomycin for Hemorrhage Control, Antibacterial Action, and Tissue Repair in Infected Trauma Wounds. Mater. Today Bio 2021, 12, 100127. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Su, B.; Dai, J.; Li, P.; Wang, R.; Yang, X. Anti-Biofilm and Anti-Hemolysis Activities of 10-Hydroxy-2-Decenoic Acid against Staphylococcus Aureus. Molecules 2022, 27, 1485. [Google Scholar] [CrossRef]


| Peptide | Sequence | Structure | Molecular Weight |
|---|---|---|---|
| K-5 | QPYGSQGFYGQRKWGNGQGVPLSQSNGLGGRGGGGGQRLVSKCL | Rich in Gly | 4494.980 |
| K-8 | NSYHIYRCTHCAVKQGGQPPSCNTLICPGKAS | Cys | 3434.949 |
| K-17 | AECADLRGRRGGERRGILCGGEKGGSAGSGRVPILGRVG | Rich in Gly | 3881.428 |
| K-34 | VLSLRRVALEDKGGLGPGAGFKKGLKVTAPARGQDKTA | helix | 3863.511 |
| K-35 | VAVVVVGEVQRKKTGVALKQKQRAGSSGGGGGGRGGAEA | Rich in Gly | 3707.203 |
| K-37 | FGIKGLKGEQLPEPKAPKGKYKSIGFGDLKESINDFFTNKK | helix | 4556.257 |
| K-54 | ALVSDIIKNAKLDDSYGKNARGIPQTSDKLNGCSEKRAK | helix | 4205.746 |
| K-58 | AKGKYCPYCKRPMFAQSEKQFPAGTEVIYTCTCGHKEKVFEDK | Cys | 4948.750 |
| K-61 | LGGKKKKVLKAANDYVAKPRDEYEWRIYWRDMGKLLDDAR | helix | 4797.546 |
| K-62 | LGLLKDLKARYPDAIIQGHRDFPNVKKSCPRFNAKEEYNF | helix | 4693.403 |
| No. | Charge | pI | GRAVY | Aliphatic Index | Boman Index | Hydrophobic Ratio | Similar |
|---|---|---|---|---|---|---|---|
| K-5 | +5 | 10.45 | −0.816 | 48.64 | 1.56 kcal/mol | 41.67% | 41.67% |
| K-8 | +3.5 | 8.89 | −0.453 | 51.88 | 1.40 kcal/mol | 37.14% | 37.14% |
| K-17 | +4 | 10.58 | −0.467 | 72.56 | 2.41 kcal/mol | 39.58% | 39.58% |
| K-34 | +5 | 10.55 | −0.311 | 87.37 | 1.46 kcal/mol | 40.48% | 40.48% |
| K-35 | +5 | 11.07 | −0.284 | 74.87 | 1.38 kcal/mol | 34.00% | 34% |
| K-37 | +4 | 9.63 | −0.878 | 59.51 | 1.64 kcal/mol | 37.78% | 37.78% |
| K-54 | +3 | 9.36 | −0.848 | 77.69 | 2.60 kcal/mol | 38.10% | 38.1% |
| K-58 | +3.25 | 8.78 | −0.783 | 29.53 | 1.84 kcal/mol | 36.36% | 36.36% |
| K-61 | +4 | 9.63 | −1.115 | 73.25 | 2.82 kcal/mol | 33.33% | 33.33% |
| K-62 | +3.25 | 9.36 | −0.800 | 73.25 | 2.37 kcal/mol | 36.36% | 36.36% |
| Salt | 1 × Buffer | Nucleotide Solutions and Other | |||
|---|---|---|---|---|---|
| HEPES | 50 mM | tRNA | 0.2 mg/mL | GTP | 3 mM |
| Potassium glutamate | 90 mM | CoA | 0.26 mM | ATP | 1 mM |
| Magnesium glutamate | 15 mM | cAMP | 0.75 mM | UTP | 1 mM |
| folinic acid | 0.068 mM | CTP | 1 mM | ||
| NAD | 0.33 mM | PEG8000 | 2% | ||
| spermidine | 0.66 mM | T7 RNA polymerase | 200 U | ||
| 3-PGA | 30 mM | 20AA | 1.5 mM each | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, K.; Xu, G.; Tian, Y.; Li, G.; Yi, Z.; Tang, X. Synthesis and Evaluation of Aquatic Antimicrobial Peptides Derived from Marine Metagenomes Using a High-Throughput Screening Approach. Mar. Drugs 2025, 23, 178. https://doi.org/10.3390/md23040178
Wu K, Xu G, Tian Y, Li G, Yi Z, Tang X. Synthesis and Evaluation of Aquatic Antimicrobial Peptides Derived from Marine Metagenomes Using a High-Throughput Screening Approach. Marine Drugs. 2025; 23(4):178. https://doi.org/10.3390/md23040178
Chicago/Turabian StyleWu, Kaiyue, Guangxin Xu, Yin Tian, Guizhen Li, Zhiwei Yi, and Xixiang Tang. 2025. "Synthesis and Evaluation of Aquatic Antimicrobial Peptides Derived from Marine Metagenomes Using a High-Throughput Screening Approach" Marine Drugs 23, no. 4: 178. https://doi.org/10.3390/md23040178
APA StyleWu, K., Xu, G., Tian, Y., Li, G., Yi, Z., & Tang, X. (2025). Synthesis and Evaluation of Aquatic Antimicrobial Peptides Derived from Marine Metagenomes Using a High-Throughput Screening Approach. Marine Drugs, 23(4), 178. https://doi.org/10.3390/md23040178






