Genome-Wide Mining of Chitinase Diversity in the Marine Diatom Thalassiosira weissflogii and Functional Characterization of a Novel GH19 Enzyme
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of Chitinase Family Genes in T. weissflogii
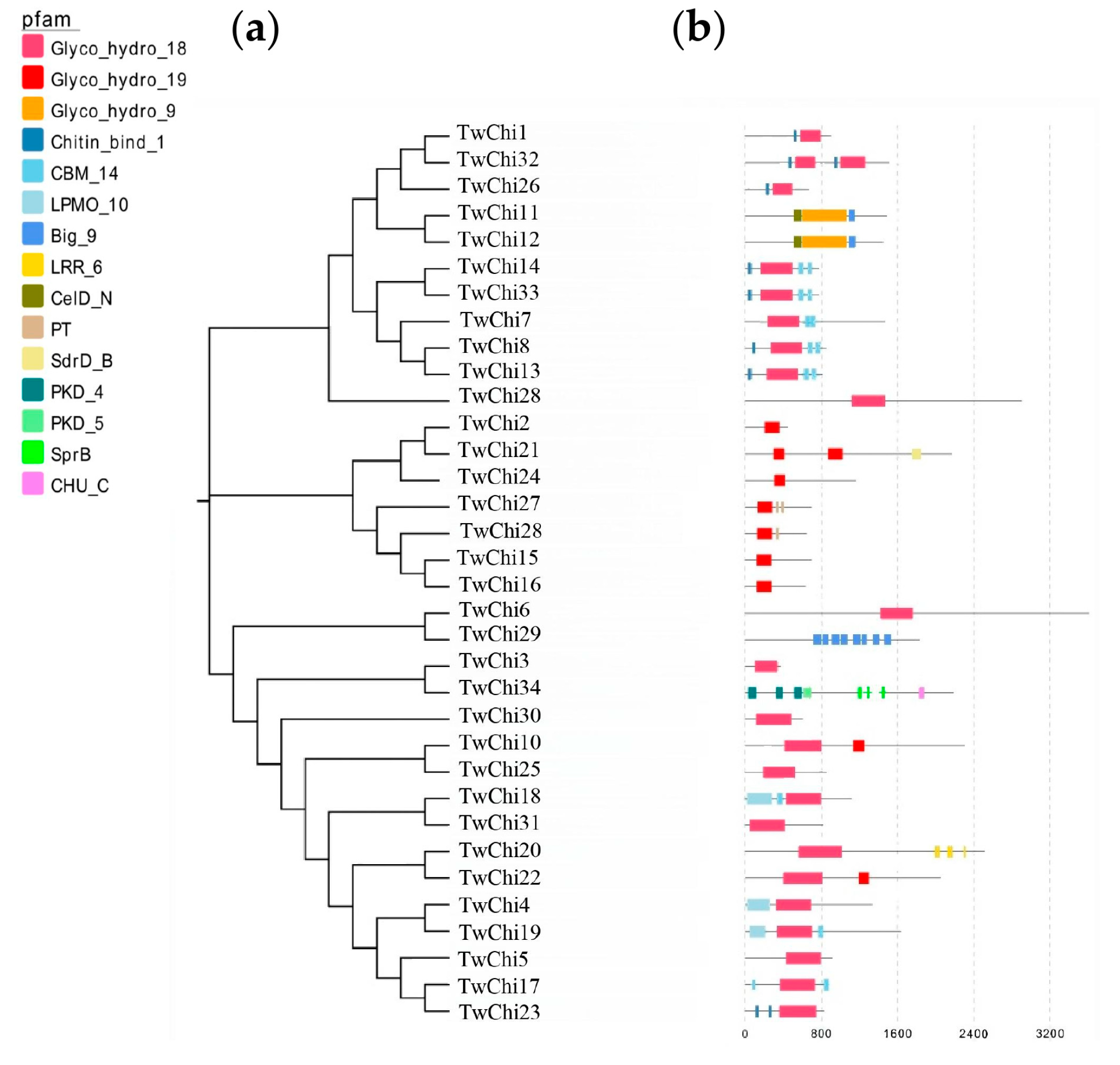
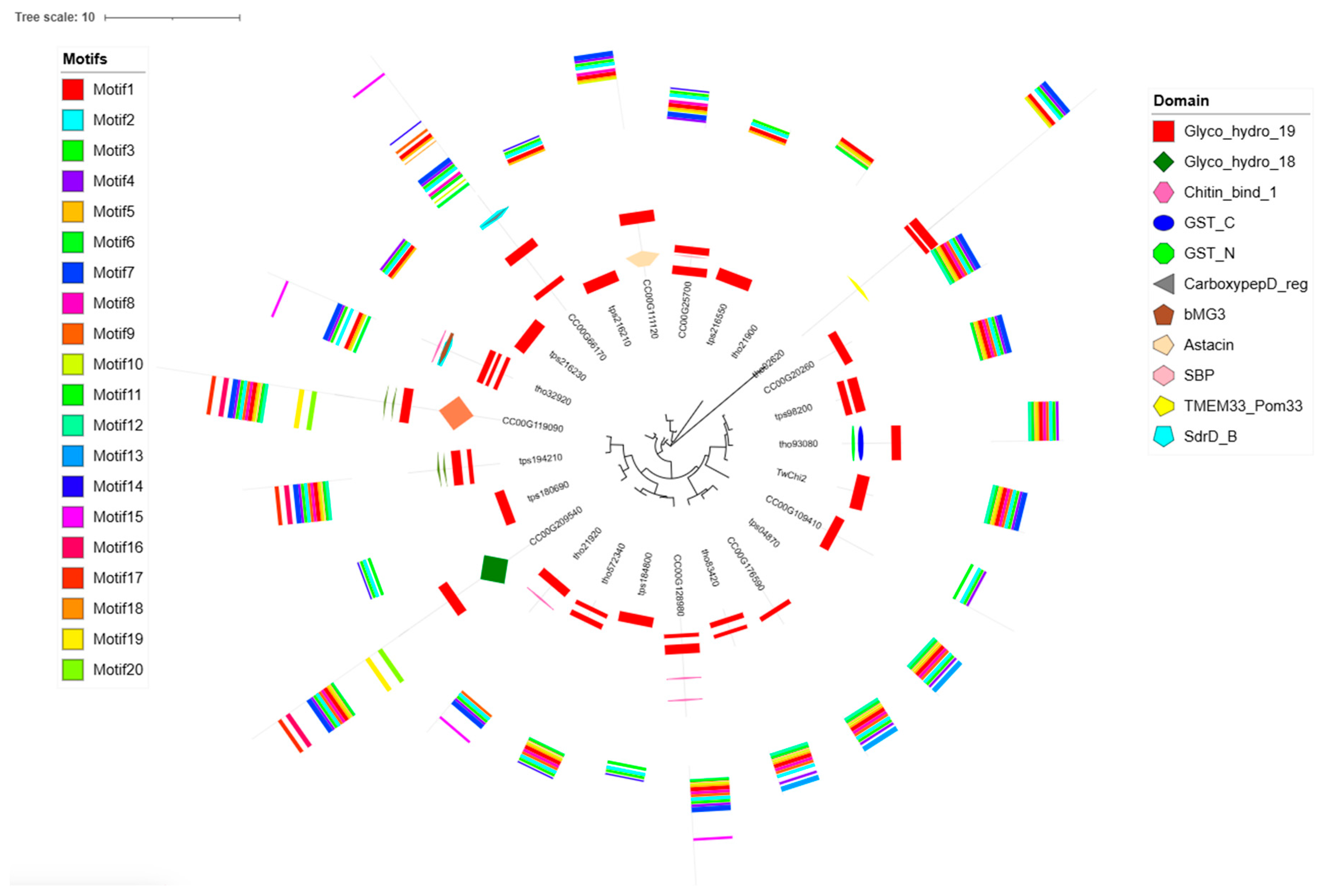
2.2. Phylogenetic Analysis, Gene Structures, Motifs, and Conserved Domains of the TwChis
2.3. Sequence Analysis of TwChi2
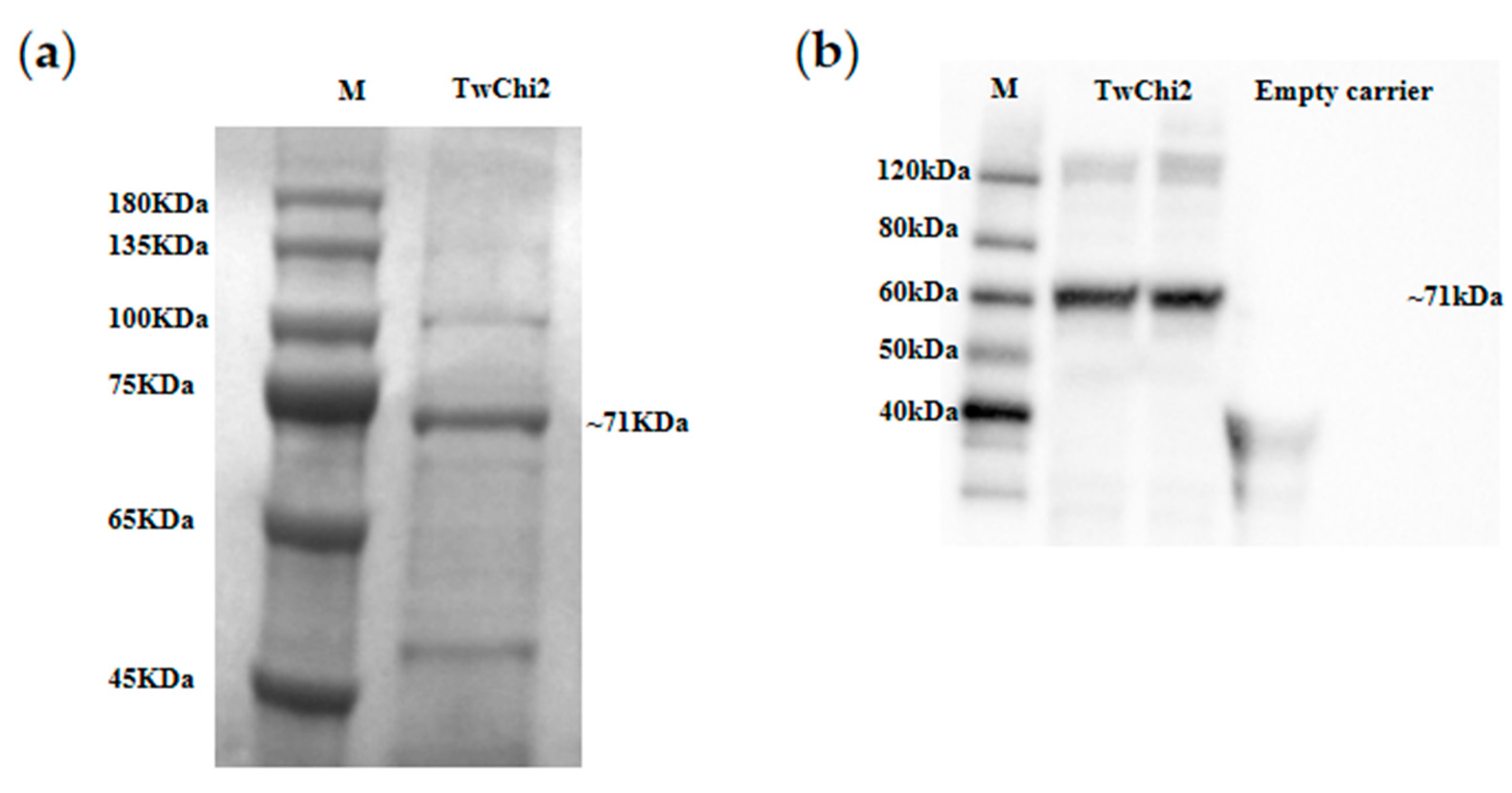
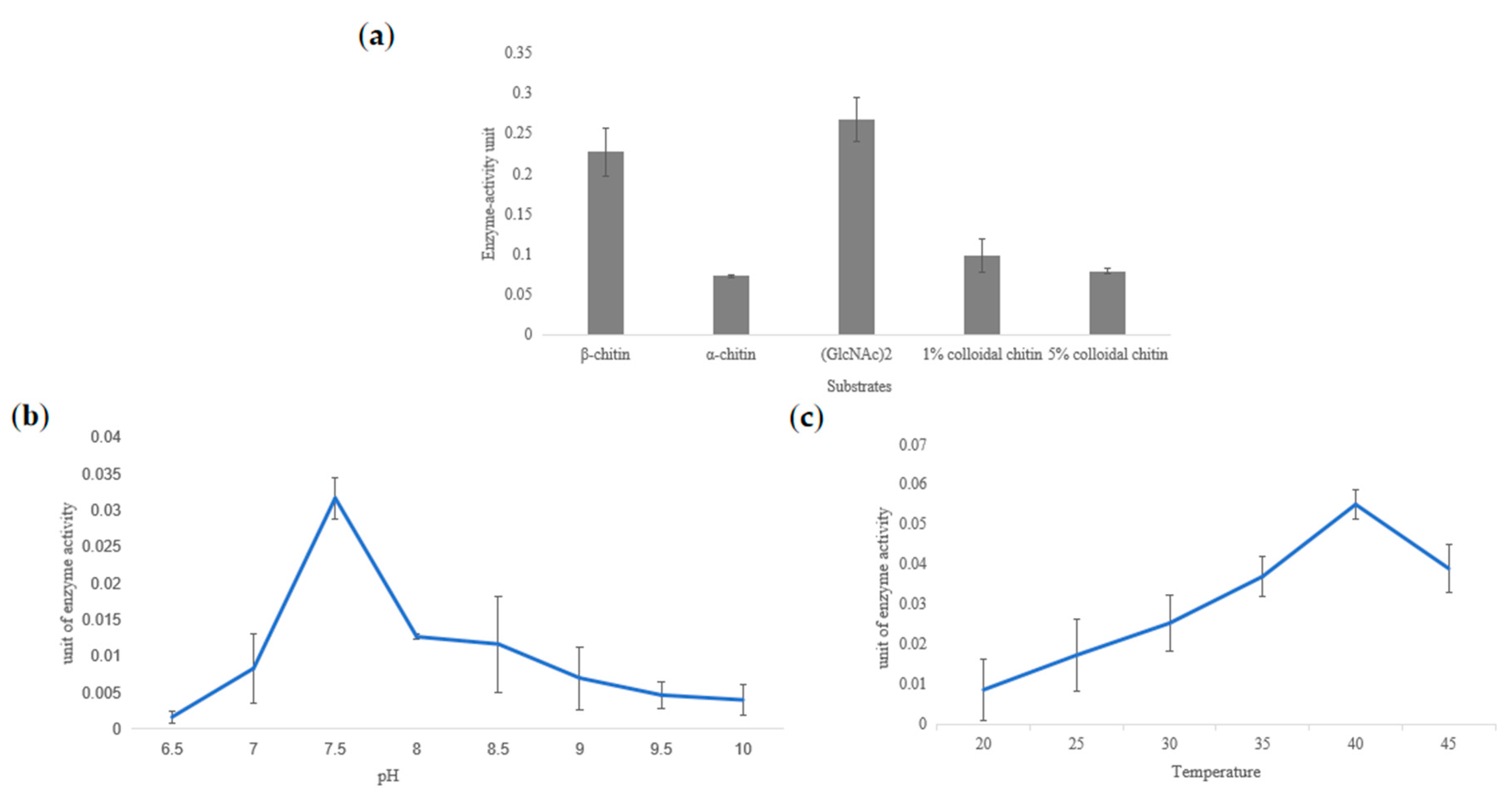
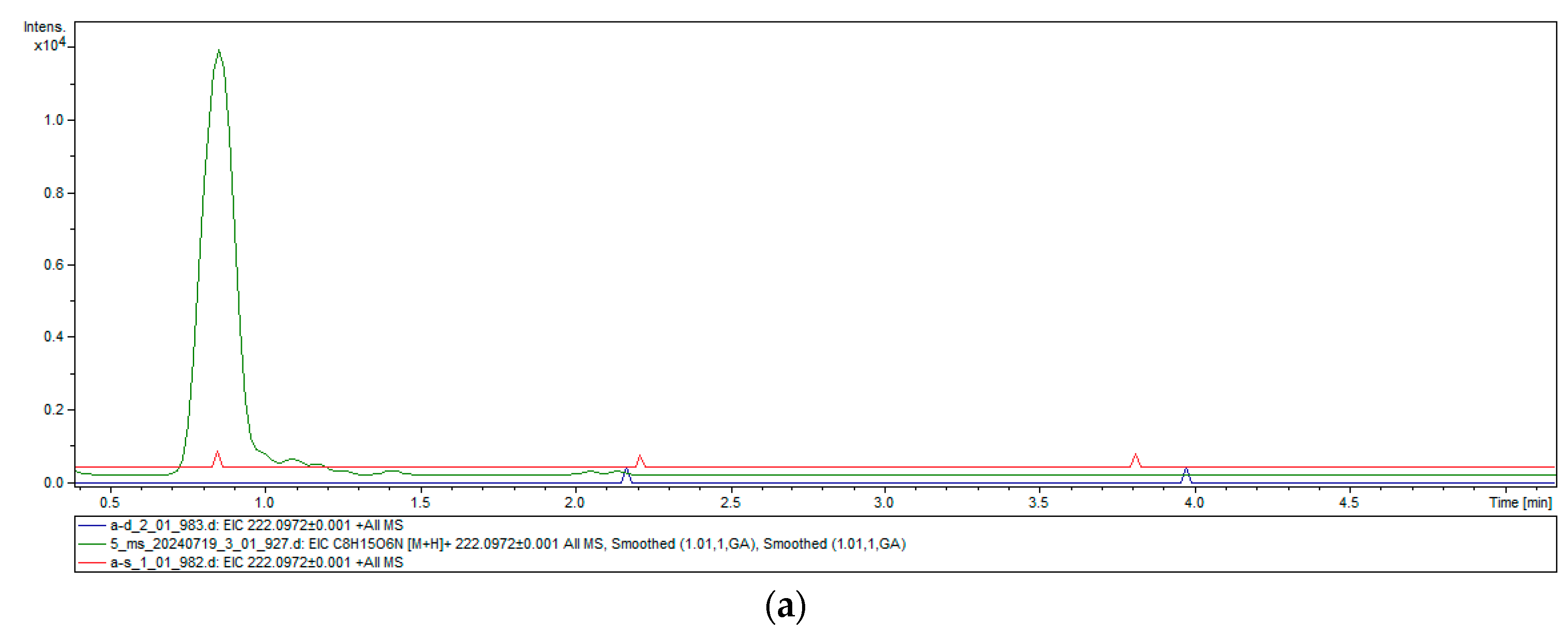
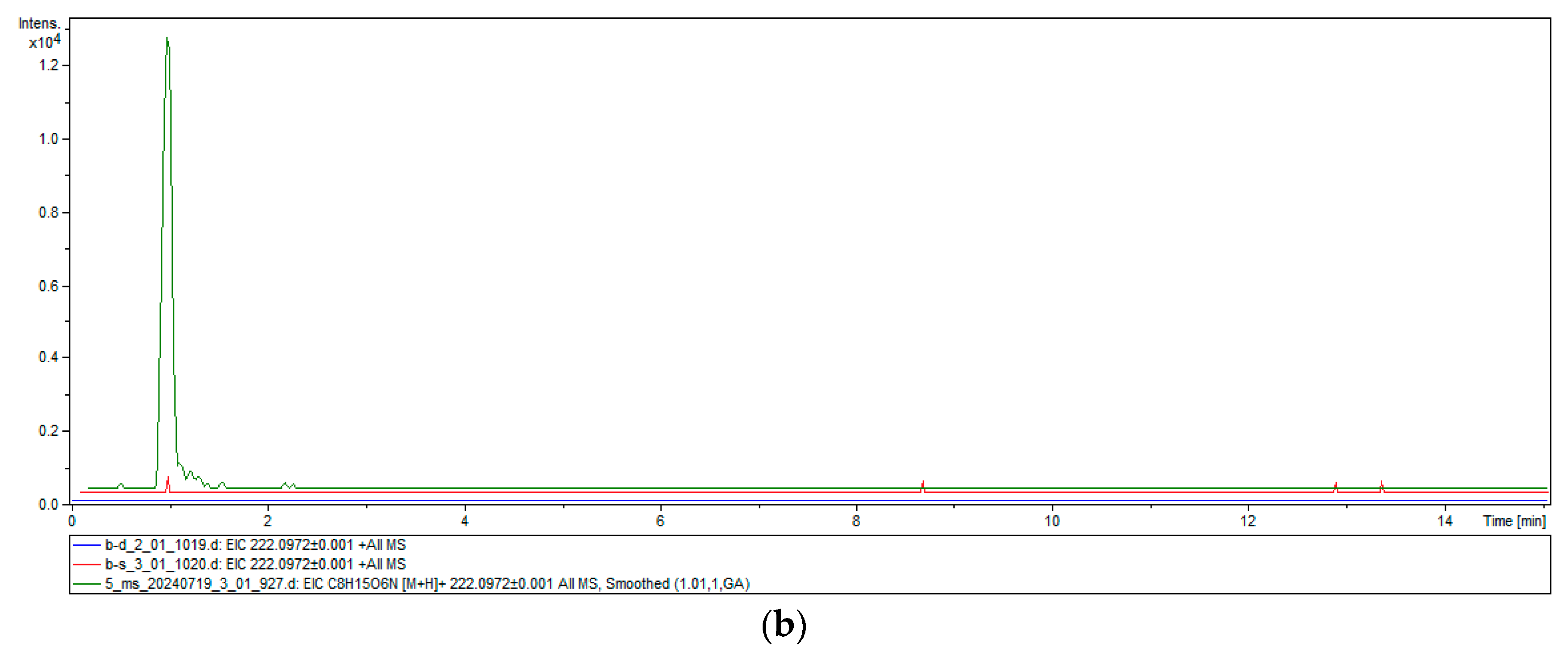
2.4. Heterologous Expression and Chitinase Activity of TwChi2
3. Discussion
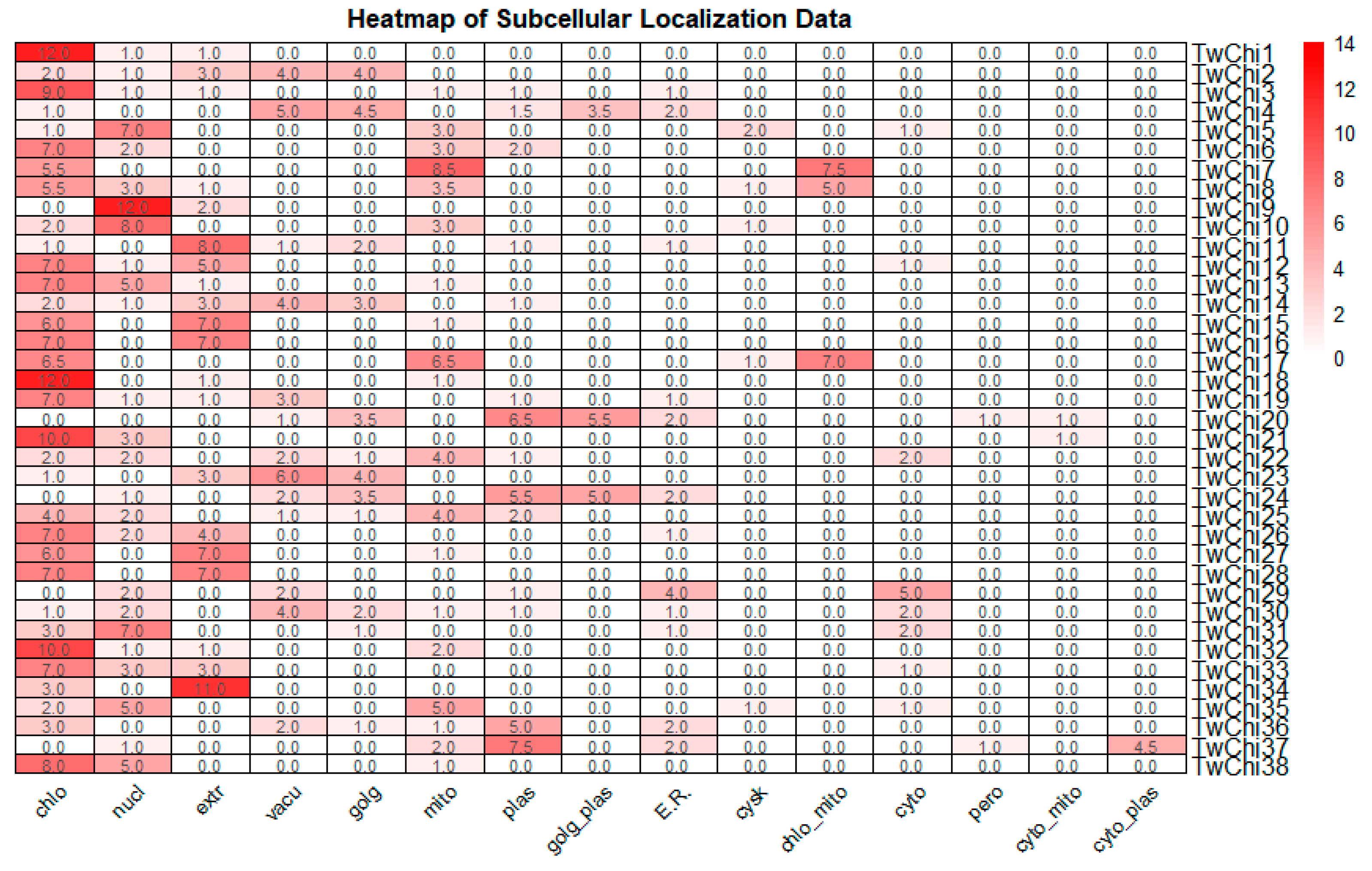
3.1. Diverse Structure and Subcellular Locations of Chitinases in T. weissflogii Showed Their Various Biological Function
3.2. TwChis Principally Belong to the Class II GH19 Family with More CBMs
3.3. TwChi2 Was Proved to Be a Novel Exochitinase Catalyzing Chitin Polymer Degradation
4. Materials and Methods
4.1. Sample Culture and Treatment
4.2. Identification of Chitinase Genes in T. weissflogii
4.3. Sequence Analysis and Structural Characterization
4.4. Cloning and Sequence Analysis of TwChi
4.5. Phylogenetic Analysis of TwChi2 Homologous Sequences in Diatoms
4.6. Expression and Purification of Recombinant TwChi2
4.7. SDS-PAGE and Western Blotting
4.8. Chitinase Activity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minke, R.; Blackwell, J. The structure of α-chitin. J. Mol. Biol. 1978, 120, 167–181. [Google Scholar] [PubMed]
- Aam, B.B.; Heggset, E.B.; Norberg, A.L.; Sørlie, M.; Vårum, K.M.; Eijsink, V.G.H. Production of chitooligosaccharides and their potential applications in medicine. Mar. Drugs 2010, 8, 1482–1517. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, R.A.G.; Tuveng, T.R.; Antonsen, S.G.; Eijsink, V.G.H.; Sorlie, M. Can we make chitosan by enzymatic deacetylation of chitin? Molecules 2019, 24, 3862. [Google Scholar] [CrossRef] [PubMed]
- Rudall, K.M. The chitin/protein complexes of insect cuticles. Adv. Insect Physiol. 1963, 1, 257–313. [Google Scholar]
- Lamarque, G.; Cretenet, M.; Viton, C.; Domard, A. New route of deacetylation of alpha- and beta-chitins by means of freeze-pump out-thaw cycles. Biomacromolecules 2005, 6, 1380–1388. [Google Scholar] [CrossRef]
- Gooday, G.W. The ecology of chitin degradation. Adv. Microb. Ecol. 1990, 11, 387–430. [Google Scholar]
- Brunner, E.; Richthammer, P.; Ehrlich, H.; Paasch, S.; Simon, P.; Ueberlein, S.; van Pee, K.H. Chitin-based organic networks: An integral part of cell wall biosilica in the diatom Thalassiosira pseudonana. Angew. Chem. Int. Ed. 2009, 48, 9724–9727. [Google Scholar] [CrossRef]
- Longnecker, K.; Soule, M.C.K.; Kujawinski, E.B. Dissolved organic matter produced by Thalassiosira pseudonana. Mar. Chem. 2015, 168, 114–123. [Google Scholar]
- Shao, Z.; Ampomah, O.; Vieira, F.R.J.; Dorrell, R.G.; Li, S.; Tirichine, L.; Bulone, V.; Duan, D.; Bowler, C. Characterization of a marine diatom chitin synthase using a combination of meta-omics, genomics, and heterologous expression approaches. Msystems 2023, 8, e0113122. [Google Scholar]
- Cheng, H.; Shao, Z.; Lu, C.; Duan, D. Genome-wide identification of chitinase genes in Thalassiosira pseudonana and analysis of their expression under abiotic stresses. BMC Plant Biol. 2021, 21, 87. [Google Scholar] [CrossRef]
- Cheng, M.; Shao, Z.; Wang, X.; Lu, C.; Li, S.; Duan, D. Novel chitin deacetylase from Thalassiosira weissflogii highlights the potential for chitin derivative production. Metabolites 2023, 13, 429. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.P.; Almeida, B.C.; Colwell, R.R.; Rivera, I.N.G. The importance of chitin in the marine environment. Mar. Biotechnol. 2011, 13, 823–830. [Google Scholar] [CrossRef]
- Akeed, Y.; Atrash, F.; Naffaa, W. Partial purification and characterization of chitinase produced by Bacillus licheniformis B307. Heliyon 2020, 6, e03858. [Google Scholar] [CrossRef]
- Jang, M.K.; Kong, B.G.; Jeong, Y.I.; Lee, C.H.; Nah, J.W. Physicochemical characterization of α-chitin, β-chitin, and γ-chitin separated from natural resources. J. Polym. Sci. Pol. Chem. 2004, 42, 3423–3432. [Google Scholar]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Kaczmarek, M.B.; Struszczyk-Swita, K.; Li, X.K.; Szczesna-Antczak, M.; Daroch, M. Enzymatic modifications of chitin, chitosan, and chitooligosaccharides. Front. Bioeng. Biotechnol. 2019, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Minguet-Lobato, M.; Cervantes, F.V.; Miguez, N.; Plou, F.J.; Fernández-Lobato, M. Chitinous material bioconversion by three new chitinases from the yeast Mestchnikowia pulcherrima. Microb. Cell Factories 2024, 23, 31. [Google Scholar]
- Talamantes, D.; Biabini, N.; Dang, H.; Abdoun, K.; Berlemont, R. Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol. Biofuels 2016, 9, 133. [Google Scholar]
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 2018, 38, BSR2018032300. [Google Scholar]
- Konno, N.; Takahashi, H.; Nakajima, M.; Takeda, T.; Sakamoto, Y. Characterization of β-N-acetylhexosaminidase (LeHex20A), a member of glycoside hydrolase family 20, from Lentinula edodes (shiitake mushroom). AMB Express 2012, 2, 29. [Google Scholar] [CrossRef]
- Udaya Prakash, N.A.; Jayanthi, M.; Sabarinathan, R.; Kangueane, P.; Mathew, L.; Sekar, K. Evolution, homology conservation, and identification of unique sequence signatures in GH19 family chitinases. J. Mol. Evol. 2010, 70, 466–478. [Google Scholar] [CrossRef]
- Bai, Y.; Eijsink, V.G.H.; Kielak, A.M.; van Veen, J.A.; de Boer, W. Genomic comparison of chitinolytic enzyme systems from terrestrial and aquatic bacteria. Environ. Microbiol. 2016, 18, 38–49. [Google Scholar]
- Hon, W.C.; Griffith, M.; Mlynarz, A.; Kwok, Y.C.; Yang, D.S. Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol. 1995, 109, 879–889. [Google Scholar] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [PubMed]
- Taira, T.; Gushiken, C.; Sugata, K.; Ohnuma, T.; Fukamizo, T. Unique GH18 chitinase from Euglena gracilis: Full-length cDNA cloning and characterization of its catalytic domain. Biosci. Biotechnol. Biochem. 2018, 82, 1090–1100. [Google Scholar]
- Hossain, M.A.; Roslan, H.A. Heterologous expression, characterisation and 3D-structural insights of GH18 chitinases derived from sago palm (Metroxylon sagu). Int. J. Biol. Macromol. 2024, 279, 135533. [Google Scholar]
- Junges, A.; Boldo, J.T.; Souza, B.K.; Guedes, R.L.M.; Sbaraini, N.; Kmetzsch, L.; Thompson, C.E.; Staats, C.C.; de Almeida, L.G.P.; de Vasconcelos, A.T.R.; et al. Genomic analyses and transcriptional profiles of the glycoside hydrolase family 18 genes of the entomopathogenic fungus Metarhizium anisopliae. PLoS ONE 2014, 9, e107864. [Google Scholar] [CrossRef]
- Qiu, S.T.; Zhou, S.P.; Tan, Y.; Feng, J.; Bai, Y.; He, J.; Cao, H.; Che, Q.; Guo, J.; Su, Z. Biodegradation and Prospect of Polysaccharide from Crustaceans. Mar. Drugs 2022, 20, 310. [Google Scholar] [CrossRef]
- Hasan, I.; Gai, F.; Cirrincione, S.; Rimoldi, S.; Saroglia, G.; Terova, G. Chitinase and Insect Meal in Aquaculture Nutrition: A Comprehensive Overview of the Latest Achievements. Fishes 2023, 8, 607. [Google Scholar] [CrossRef]
- Traller, J.C.; Cokus, S.J.; Lopez, D.A.; Gaidarenko, O.; Smith, S.R.; McCrow, J.P.; Gallaher, S.D.; Podell, S.; Thompson, M.; Cook, O.; et al. Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol. Biofuels 2016, 9, 258. [Google Scholar]
- Smucker, R.A. Chitin primary production. Biochem. Syst. Ecol. 1991, 19, 357–369. [Google Scholar]
- Passarinho, P.A.; de Vries, S.C. Arabidopsis Chitinases: A genomic survey. Arab. Book 2002, 1, e0023. [Google Scholar]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Sci. 2018, 270, 257–267. [Google Scholar] [PubMed]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant Chitinases. Plant J. 1993, 3, 31–40. [Google Scholar]
- Lacombe-Harvey, M.E.; Brzezinski, R.; Beaulieu, C. Chitinolytic functions in actinobacteria: Ecology, enzymes, and evolution. Appl. Microbiol. Biotechnol. 2018, 102, 7219–7230. [Google Scholar] [CrossRef]
- Huang, G.L.; Zahng, D.W.; Zhao, H.J.; Zhang, H.C.; Wang, P.G. Chemo-enzymatic synthesis of 1,4-oxazepanyl sugar as potent inhibitor of chitinase. Bioorg. Med. Chem. 2006, 14, 2446–2449. [Google Scholar]
- Molinari, L.M.; Pedroso, R.B.; Scoaris, D.D.; Ueda-Nakamura, T.; Nakamura, C.V.; Dias Filho, B.P. Identification and partial characterisation of a chitinase from Nile tilapia, Oreochromis niloticus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 146, 81–87. [Google Scholar]
- Alverson, A.J.; Beszteri, B.; Julius, M.L.; Theriot, E.C. The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol. Biol. 2011, 11, 125. [Google Scholar] [CrossRef]
- Spano, D.; Pospiskova, K.; Safarik, I.; Pisano, M.B.; Pintus, F.; Floris, G.; Medda, R. Chitinase III in Euphorbia characias latex Purification and characterization. Protein Expr. Purif. 2015, 116, 152–158. [Google Scholar]
- Tanaka, T.; Fukui, T.; Imanaka, T. Different cleavage specificities of the dual catalytic domains in chitinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biol. Chem. 2001, 276, 35629–35635. [Google Scholar]
- Park, H.Y.; Pan, C.H.; So, M.Y.; Ah, J.H.; Jo, D.H.; Kim, S.I. Purification, characterization, and cDNA cloning of rice class III chitinase. Mol. Cells. 2002, 13, 69–76. [Google Scholar] [PubMed]
- Chuang, H.H.; Lin, F.P. New role of C-terminal 30 amino acids on the insoluble chitin hydrolysis in actively engineered chitinase from Vibrio parahaemolyticus. Appl. Microbiol. Biotechnol. 2007, 76, 123–133. [Google Scholar] [PubMed]
- Jankiewicz, U.; Brzezinska, M.S. Purification, characterization, and gene cloning of a chitinase from Stenotrophomonas maltophilia N4. J. Basic Microbiol. 2015, 55, 709–717. [Google Scholar] [CrossRef]
- Sekiguchi, J.; Matsumiya, M.; Mochizuki, A. Distribution of Chitinolytic Enzymes in Seaweeds. Fish. Sci. 1995, 61, 876–881. [Google Scholar] [CrossRef]
- Essghaier, B.; Rouaissi, M.; Boudabous, A.; Jijakli, H.; Sadfi-Zouaoui, N. Production and partial characterization of chitinase from a halotolerant Planococcus rifitoensis strain M2-26. World J. Microb. Biotechnol. 2010, 26, 977–984. [Google Scholar]
- Lestari, P.; Prihatiningsih, N.; Djatmiko, H.A. Partial biochemical characterization of crude extract extracellular chitinase enzyme from Bacillus subtilis B 298. In IOP Conference Series: Materials Science and Engineering, Proceedings of the 11th Joint Conference on Chemistry in Conjunction with the 4th Regional Biomaterials Scientific Meeting, Purwokerto, Indonesia, 15–16 September 2016; IOP Publishing: Bristol, UK, 2017; Volume 172, p. 012041. [Google Scholar]
- Zhu, Q.S.; Arakane, Y.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc. Natl. Acad. Sci. USA 2008, 105, 6650–6655. [Google Scholar] [PubMed]
- Scheerer, D.; Levy, D.; Casier, R.; Riven, I.; Mazal, H.; Haran, G. Interplay between conformational dynamics and substrate binding regulates enzymatic activity: A single-molecule FRET study. Chem. Sci. 2025, 16, 7. [Google Scholar]
- Zhang, A.; He, Y.M.; Wei, G.G.; Zhou, J.; Dong, W.L.; Chen, K.Q.; Ouyang, P.K. Molecular characterization of a novel chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and its use in N-acetyl-d-glucosamine production. Biotechnol. Biofuels 2018, 11, 179. [Google Scholar] [CrossRef]
- Tzelepis, G.; Dubey, M.; Jensen, D.F.; Karlsson, M. Identifying glycoside hydrolase family 18 genes in the mycoparasitic fungal species Clonostachys rosea. Microbiology 2015, 161, 1407–1419. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar]
- Li, P.; Pei, Y.; Sang, X.; Ling, Y.; Yang, Z.; He, G. Transgenic indica rice expressing a bitter melon (Momordica charantia) class I chitinase gene (McCHIT1) confers enhanced resistance to Magnaporthe grisea and Rhizoctonia solani. Eur. J. Plant Pathol. 2009, 125, 533–543. [Google Scholar] [CrossRef]
- Prihoda, J.; Tanaka, A.; de Paula, W.B.; Allen, J.F.; Tirichine, L.; Bowler, C. Chloroplast-mitochondria cross-talk in diatoms. J. Exp. Bot. 2012, 63, 1543–1557. [Google Scholar] [CrossRef]
- Kashyap, P.; Deswal, R. A novel class I Chitinase from Hippophae rhamnoides: Indications for participating in ICE-CBF cold stress signaling pathway. Plant Sci. 2017, 259, 62–70. [Google Scholar]
- Gupta, R.; Deswal, R. Low temperature stress modulated secretome analysis and purification of antifreeze protein from Hippophae rhamnoides, a Himalayan wonder plant. J. Proteome Res. 2012, 11, 2684–2696. [Google Scholar]
- Mock, T.; Samanta, M.P.; Iverson, V.; Berthiaume, C.; Robison, M.; Holtermann, K.; Colleen, D.; BonDurant, S.S.; Richmond, K.; Rodesch, M.; et al. Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc. Natl. Acad. Sci. USA 2008, 105, 1579–1584. [Google Scholar]
- Schneider, S.; Ullrich, W.R. Differential induction of resistance and enhanced enzyme activities in cucumber and tobacco caused by treatment with various abiotic and biotic inducers. Physiol. Mol. Plant Pathol. 1994, 45, 291–304. [Google Scholar] [CrossRef]
- Sahoo, P.R.; Mishra, S.R.; Mohapatra, S.; Sahu, S.; Sahoo, G.; Behera, P.C. In silico structural and phylogenetic analysis of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in domestic animals. Indian J. Anim. Res. 2019, 53, 1607–1612. [Google Scholar]
- Tzelepis, G.D.; Melin, P.; Jensen, D.F.; Stenlid, J.; Karlsson, M. Functional analysis of glycoside hydrolase family 18 and 20 genes in Neurospora crassa. Fungal Genet. Biol. 2012, 49, 717–730. [Google Scholar]
- Mohun, A.F.; Cook, R.J.Y. Improved Dinitrosalicylic Acid Method for Determining Blood and Cerebrospinal Fluid Sugar Levels. Am. J. Clin. Pathol. 1962, 15, 169–180. [Google Scholar]
| Name | Gene ID | Length of CDS (bp) | Number of Amino Acids | Molecular Weight (KDa) | Isoelectric Point (pI) |
|---|---|---|---|---|---|
| TwChi1 | mikado.scaffold_31G129.1 | 2703 | 900 | 95.71 | 4.76 |
| TwChi2 | mikado.scaffold_10G336.1 | 1362 | 453 | 50.39 | 5.03 |
| TwChi3 | mikado.scaffold_10G382.1 | 1116 | 371 | 40.02 | 6.00 |
| TwChi4 | mikado.scaffold_10G599.1 | 4011 | 1336 | 146.91 | 4.46 |
| TwChi5 | mikado.scaffold_11G224.1 | 2748 | 915 | 99.54 | 4.74 |
| TwChi6 | mikado.scaffold_12G339.1 | 10,833 | 3610 | 390.98 | 4.10 |
| TwChi7 | mikado.scaffold_14G114.1 | 4422 | 1473 | 160.22 | 4.55 |
| TwChi8 | mikado.scaffold_17G236.1 | 2565 | 854 | 92.34 | 4.67 |
| TwChi9 | mikado.scaffold_1G646.1 | 8724 | 2907 | 319.50 | 4.25 |
| TwChi10 | mikado.scaffold_21G148.1 | 6921 | 2306 | 249.20 | 4.32 |
| TwChi11 | mikado.scaffold_21G350.2 | 4473 | 1490 | 160.06 | 4.79 |
| TwChi12 | mikado.scaffold_21G351.1 | 4353 | 1450 | 155.53 | 4.63 |
| TwChi13 | mikado.scaffold_26G169.1 | 2442 | 813 | 87.57 | 4.51 |
| TwChi14 | mikado.scaffold_27G208.3 | 2337 | 778 | 83.62 | 4.40 |
| TwChi15 | mikado.scaffold_2G1055.1 | 2100 | 699 | 73.78 | 4.29 |
| TwChi16 | mikado.scaffold_2G1056.1 | 1905 | 634 | 67.60 | 4.41 |
| TwChi17 | mikado.scaffold_2G568.1 | 2655 | 884 | 94.08 | 4.33 |
| TwChi18 | mikado.scaffold_2G719.1 | 3369 | 1122 | 123.54 | 4.28 |
| TwChi19 | mikado.scaffold_30G74.1 | 4911 | 1636 | 179.35 | 4.49 |
| TwChi20 | mikado.scaffold_35G110.1 | 7548 | 2515 | 27.68 | 4.58 |
| TwChi21 | mikado.scaffold_38G186.2 | 6513 | 2170 | 233.58 | 4.52 |
| TwChi22 | mikado.scaffold_3G605.1 | 6159 | 2052 | 222.97 | 4.32 |
| TwChi23 | mikado.scaffold_4G320.1 | 2514 | 837 | 88.54 | 4.74 |
| TwChi24 | mikado.scaffold_5G227.1 | 3507 | 1168 | 125.86 | 4.37 |
| TwChi25 | mikado.scaffold_5G537.1 | 2586 | 861 | 94.28 | 4.60 |
| TwChi26 | mikado.scaffold_5G910.1 | 2025 | 674 | 71.74 | 4.75 |
| TwChi27 | mikado.scaffold_62G1.1 | 2100 | 699 | 73.90 | 4.14 |
| TwChi28 | mikado.scaffold_62G2.1 | 1941 | 646 | 68.94 | 4.38 |
| TwChi29 | mikado.scaffold_6G457.2 | 5508 | 1835 | 199.91 | 4.26 |
| TwChi30 | mikado.scaffold_6G726.1 | 1836 | 611 | 66.72 | 4.71 |
| TwChi31 | mikado.scaffold_7G365.1 | 2478 | 825 | 90.28 | 4.30 |
| TwChi32 | mikado.scaffold_7G450.2 | 4545 | 1514 | 160.16 | 4.77 |
| TwChi33 | mikado.scaffold_23G5.2 | 2337 | 778 | 83.72 | 4.33 |
| TwChi34 | mikado.scaffold_24G266.1 | 6570 | 2189 | 233.94 | 3.61 |
| TwChi35 | mikado.scaffold_2G143.2 | 1332 | 443 | 48.45 | 6.19 |
| TwChi36 | mikado.scaffold_6G219.1 | 1137 | 378 | 40.80 | 9.19 |
| TwChi37 | mikado.scaffold_6G220.1 | 1557 | 518 | 56.98 | 5.00 |
| TwChi38 | mikado.scaffold_48G26.1 | 657 | 218 | 24.34 | 4.77 |
| Name | TwChi2 |
|---|---|
| Molecular weight (KDa) | 50.38808 |
| Number of amino acids | 453 |
| Length of CDS (bp) | 1362 |
| Isoelectric point (pI) | 5.03 |
| Instability index | 34.43 |
| α-helix (%) | 17.44 |
| Extended strand (%) | 6.84 |
| β-turn (%) | 0 |
| Number of predicted TMHs | 0 |
| Conserved domain | Glyco_hydro_19 |
| Subcellular location | Vacuole |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, M.; Li, S.; Wang, J.; Yang, X.; Duan, D.; Shao, Z. Genome-Wide Mining of Chitinase Diversity in the Marine Diatom Thalassiosira weissflogii and Functional Characterization of a Novel GH19 Enzyme. Mar. Drugs 2025, 23, 144. https://doi.org/10.3390/md23040144
Cheng M, Li S, Wang J, Yang X, Duan D, Shao Z. Genome-Wide Mining of Chitinase Diversity in the Marine Diatom Thalassiosira weissflogii and Functional Characterization of a Novel GH19 Enzyme. Marine Drugs. 2025; 23(4):144. https://doi.org/10.3390/md23040144
Chicago/Turabian StyleCheng, Mengzhen, Shuang Li, Jiahui Wang, Xiaoqi Yang, Delin Duan, and Zhanru Shao. 2025. "Genome-Wide Mining of Chitinase Diversity in the Marine Diatom Thalassiosira weissflogii and Functional Characterization of a Novel GH19 Enzyme" Marine Drugs 23, no. 4: 144. https://doi.org/10.3390/md23040144
APA StyleCheng, M., Li, S., Wang, J., Yang, X., Duan, D., & Shao, Z. (2025). Genome-Wide Mining of Chitinase Diversity in the Marine Diatom Thalassiosira weissflogii and Functional Characterization of a Novel GH19 Enzyme. Marine Drugs, 23(4), 144. https://doi.org/10.3390/md23040144







