The Impact of Chitinase Binding Domain Truncation on the Properties of CaChi18B from Chitinilyticum aquatile CSC-1
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Analysis

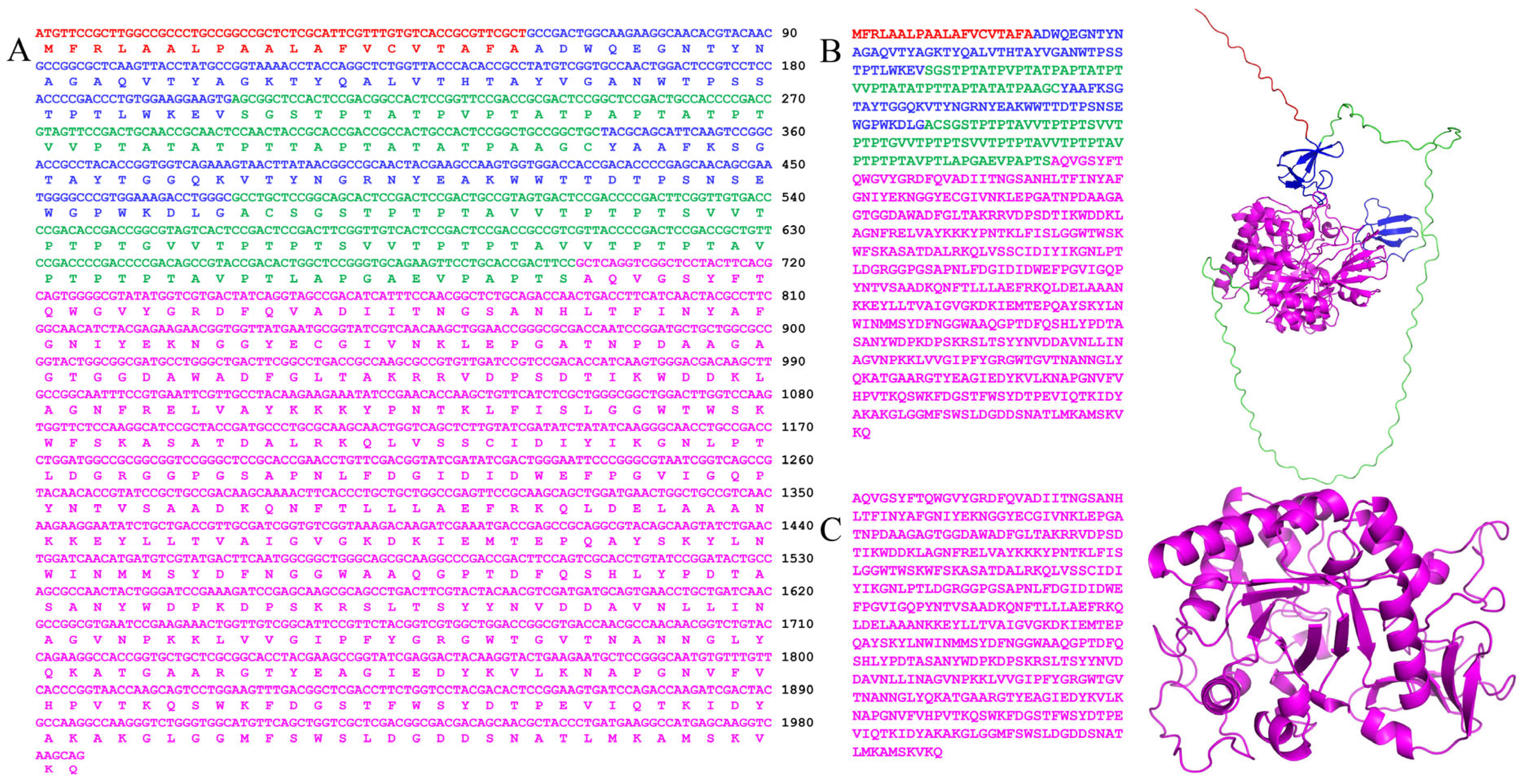
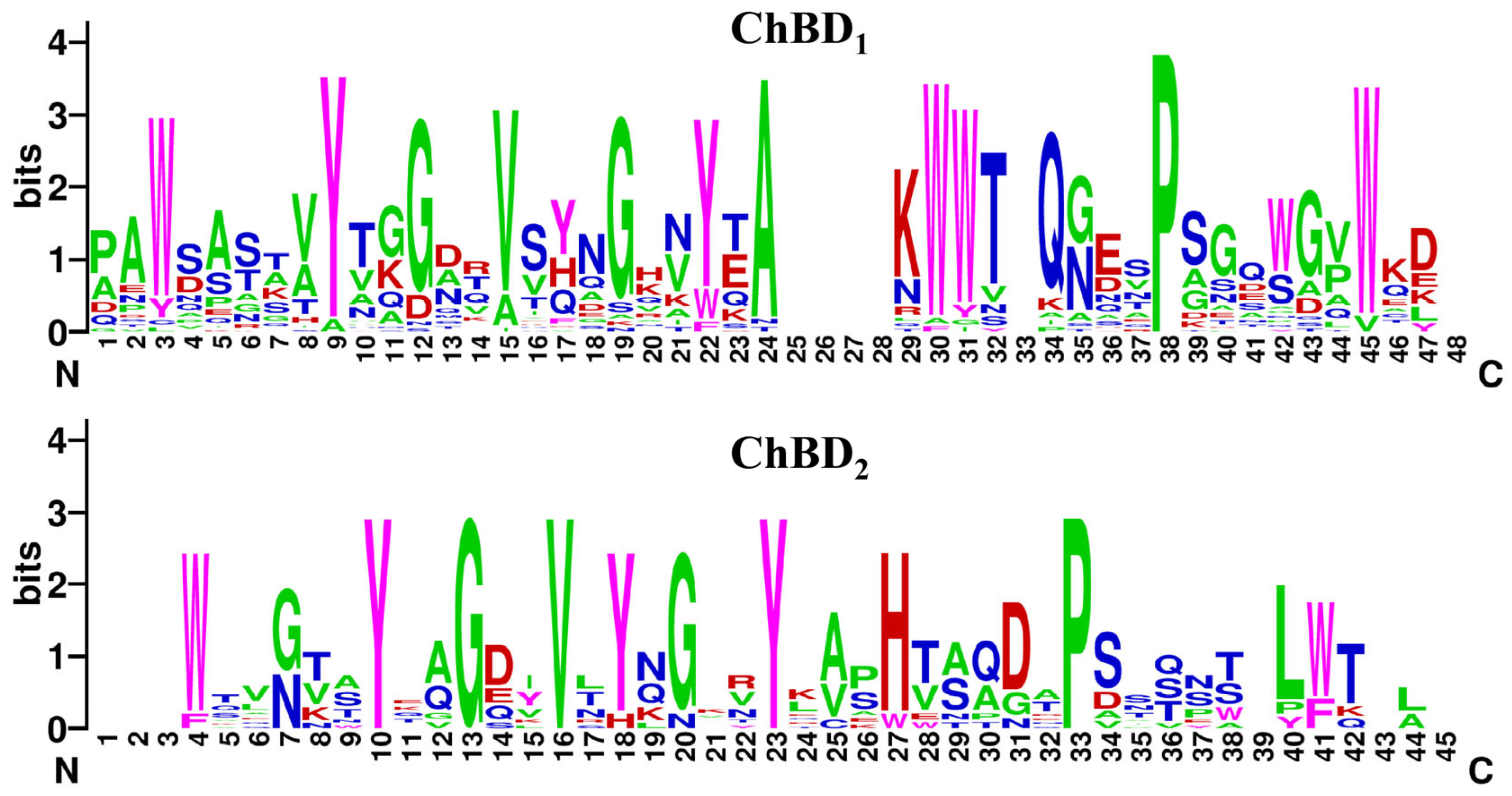
2.2. Induction, Expression, and Purification of Recombinant Chitinase
2.3. Analysis of the Enzymatic Properties of Recombinant Chitinase
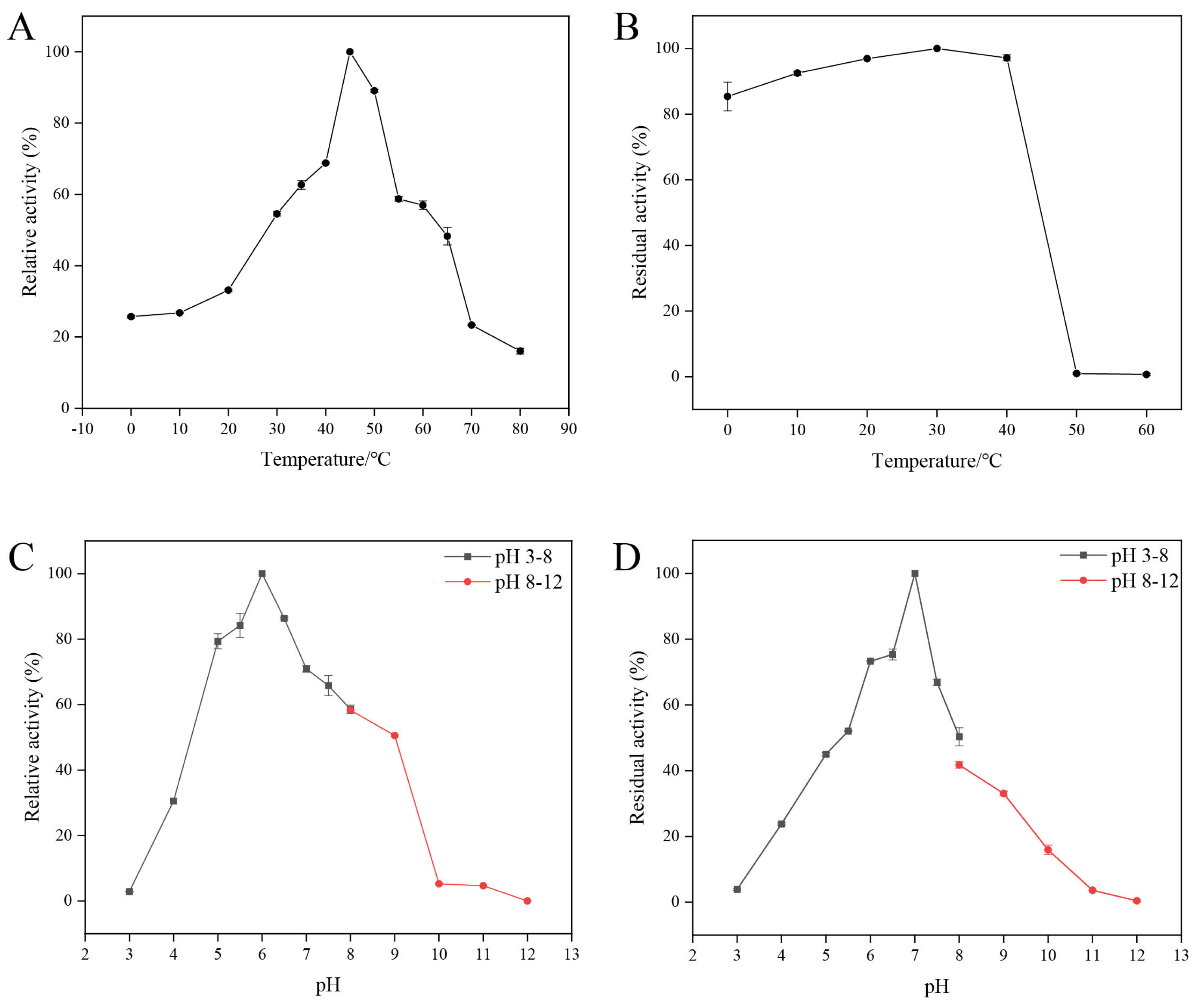
2.3.1. The Effects of Temperature and pH on the Activity of Recombinant Enzyme CaChi18B_ΔChBDs
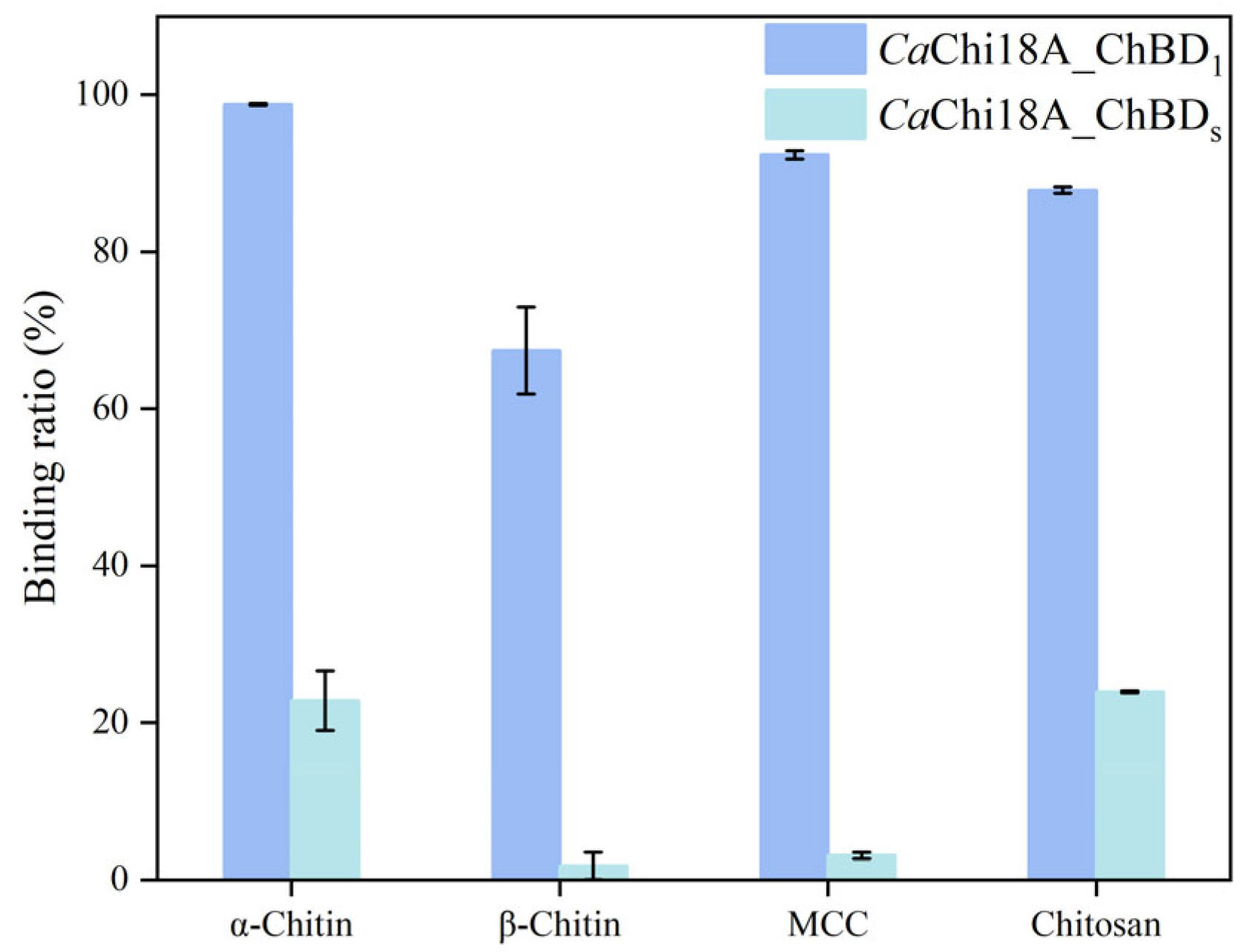
2.3.2. Determination of Enzyme Binding Ability
2.3.3. Substrate Specificity and Kinetic Parameters of Recombinant Chitinase CaChi18B_ΔChBDs
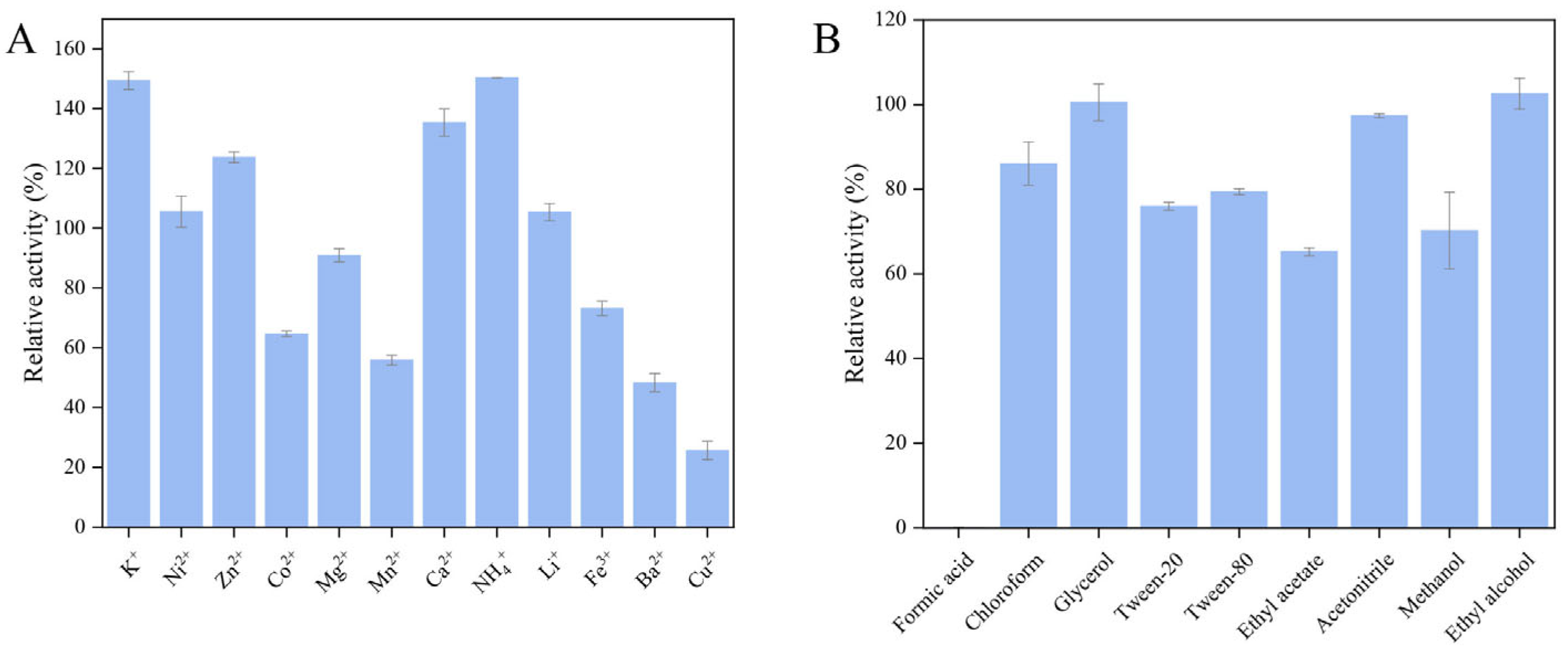
2.3.4. Tolerance of Recombinant Chitinase CaChi18B_ΔChBDs to Metal Ions and Chemical Reagents
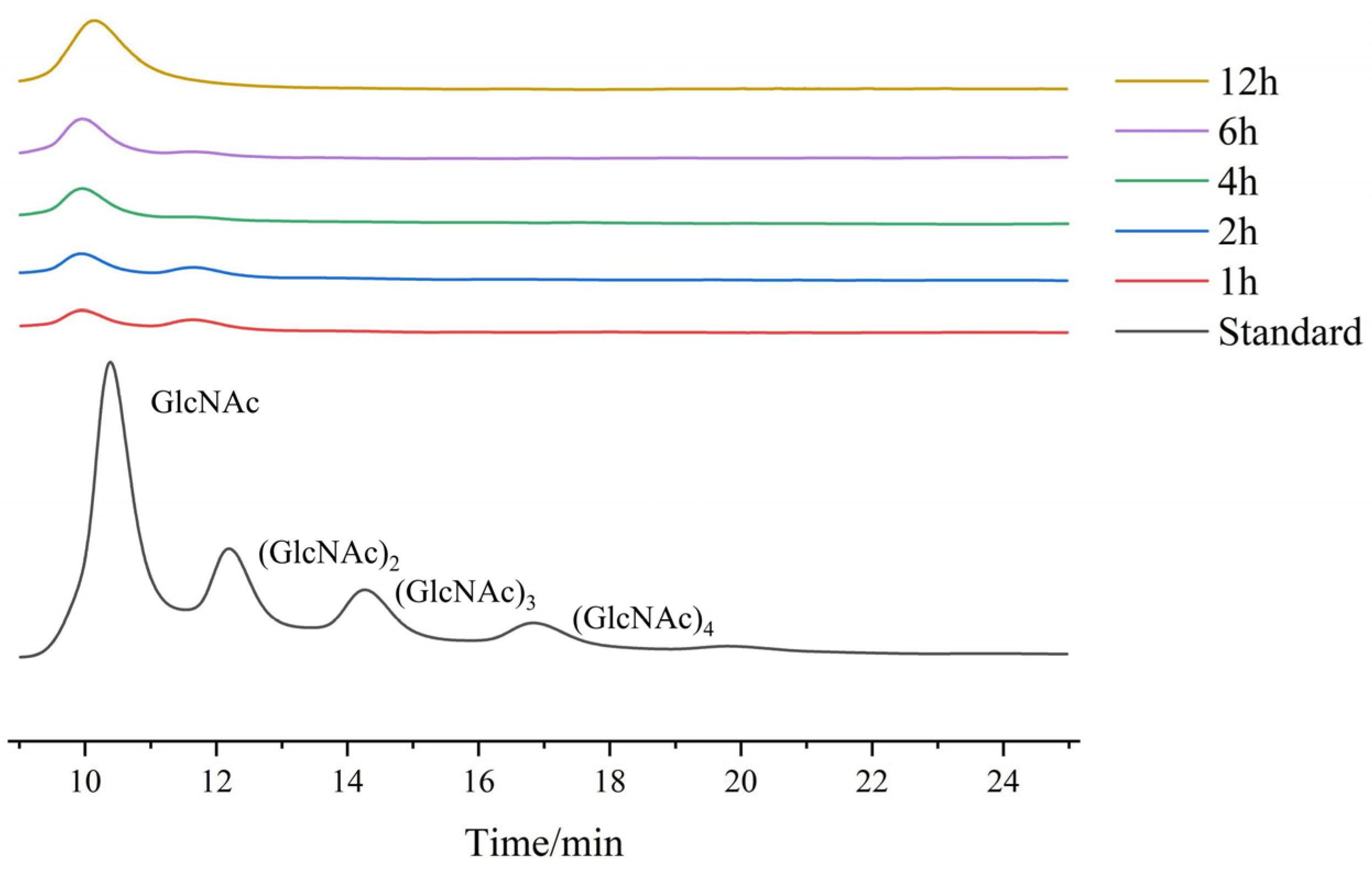
2.3.5. Analysis of Hydrolysis Products
3. Materials and Methods
3.1. Material and Reagents
3.1.1. Strain and Plasmid
3.1.2. Main Reagents and Instruments
3.1.3. Software and Websites
3.2. Experimental Methods
3.2.1. Preparation of Colloidal Chitin
3.2.2. Gene Cloning and Construction of Expression Vectors for CaChi18B_ΔChBDs
3.2.3. Protein Expression and Purification of CaChi18B_ΔChBDs
3.2.4. Enzymatic Property Analysis of CaChi18B_ΔChBDs
Effect of Temperature on Recombinant Enzyme Activity
Effect of pH on Recombinant Enzyme Activity
Determination of Enzyme Binding Capacity
Determination of Substrate Specificity, Km, and Vmax
Effects of Metal Ions and Organic Reagents on Recombinant Enzyme Activity
Analysis of Hydrolysis Products
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jana, S.; Hamre, A.G.; Wildberger, P.; Holen, M.M.; Eijsink, V.G.H.; Beckham, G.T.; Sørlie, M.; Payne, C.M. Aromatic-Mediated Carbohydrate Recognition in Processive Serratia Marcescens Chitinases. J. Phys. Chem. B 2016, 120, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Microbial Chitinases: Properties, Enhancement and Potential Applications. Protoplasma 2021, 258, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Zdarta, J.; Klapiszewski, Ł.; Wysokowski, M.; Norman, M.; Kołodziejczak-Radzimska, A.; Moszyński, D.; Ehrlich, H.; Maciejewski, H.; Stelling, A.; Jesionowski, T. Chitin-Lignin Material as a Novel Matrix for Enzyme Immobilization. Mar. Drugs 2015, 13, 2424–2446. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Goswami, P.; Paritosh, K.; Kumar, M.; Pareek, N.; Vivekanand, V. Seafood Waste: A Source for Preparation of Commercially Employable Chitin/Chitosan Materials. Bioresour. Bioprocess. 2019, 6, 8. [Google Scholar] [CrossRef]
- Ma, Q.; Gao, X. Categories and Biomanufacturing Methods of Glucosamine. Appl. Microbiol. Biotechnol. 2019, 103, 7883–7889. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Liu, Z.; Xie, X. Anti-Tumor Effect of Chitin Oligosaccharide plus Cisplatin In Vitro and In Vivo. OncoTargets Ther. 2019, 12, 7581–7590. [Google Scholar] [CrossRef]
- Kang, K.A.; Lee, K.H.; Chae, S.; Zhang, R.; Jung, M.S.; Lee, Y.; Kim, S.Y.; Kim, H.S.; Joo, H.G.; Park, J.W.; et al. Eckol Isolated from Ecklonia cava Attenuates Oxidative Stress Induced Cell Damage in Lung Fibroblast Cells. FEBS Lett. 2005, 579, 6295–6304. [Google Scholar] [CrossRef]
- Cody, R.M.; Davis, N.D.; Lin, J.; Shaw, D. Screening Microorganisms for Chitin Hydrolysis and Production of Ethanol from Amino Sugars. Biomass 1990, 21, 285–295. [Google Scholar] [CrossRef]
- Maia, A.; Cardona Gloria, Y.; Fuchs, K.; Chang, T.-H.; Engels, P.; Zhou, M.; Hinnenthal, T.; Rusch, E.; Gouttefangeas, C.; Weber, A.N.R. Chitin Oligomers Promote Lymphoid Innate and Adaptive Immune Cell Activation. J. Leukoc. Biol. 2023, 114, 180–186. [Google Scholar] [CrossRef]
- Zheng, J.; Cheng, G.; Li, Q.; Jiao, S.; Feng, C.; Zhao, X.; Yin, H.; Du, Y.; Liu, H. Chitin Oligosaccharide Modulates Gut Microbiota and Attenuates High-Fat-Diet-Induced Metabolic Syndrome in Mice. Mar. Drugs 2018, 16, 66. [Google Scholar] [CrossRef]
- Salvatore, S.; Heuschkel, R.; Tomlin, S.; Davies, S.E.; Edwards, S.; Walker-Smith, J.A.; French, I.; Murch, S.H. A Pilot Study of N-acetyl Glucosamine, a Nutritional Substrate for Glycosaminoglycan Synthesis, in Paediatric Chronic Inflammatory Bowel Disease. Aliment. Pharmacol. Ther. 2000, 14, 1567–1579. [Google Scholar] [CrossRef] [PubMed]
- Shikhman, A.R. Chondroprotective Activity of N-Acetylglucosamine in Rabbits with Experimental Osteoarthritis. Ann. Rheum. Dis. 2005, 64, 89–94. [Google Scholar] [CrossRef]
- Marchetti, M.; De Berardis, B.; Bigioni, I.; Mariano, A.; Superti, F.; Scotto d’Abusco, A. In Vitro Antiviral and Anti-Inflammatory Activities of N-Acetylglucosamine: Development of an Alternative and Safe Approach to Fight Viral Respiratory Infections. Int. J. Mol. Sci. 2023, 24, 5129. [Google Scholar] [CrossRef] [PubMed]
- Kubomura, D.; Ueno, T.; Yamada, M.; Nagaoka, I. Evaluation of the Chondroprotective Action of N-Acetylglucosamine in a Rat Experimental Osteoarthritis Model. Exp. Ther. Med. 2017, 14, 3137–3144. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jiang, X.; Yang, Q. Glycoside Hydrolase Family 18 Chitinases: The Known and the Unknown. Biotechnol. Adv. 2020, 43, 107553. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Wang, S.; Yang, D.-F.; Yang, L.-Y.; Wang, Q.-Y.; Yu, J.; Li, N.; Pan, L.-X. The Discovery, Enzymatic Characterization and Functional Analysis of a Newly Isolated Chitinase from Marine-Derived Fungus Aspergillus fumigatus Df347. Mar. Drugs 2022, 20, 520. [Google Scholar] [CrossRef]
- Li, J.; Gao, K.; Secundo, F.; Mao, X. Biochemical Characterization of Two β-N-Acetylglucosaminidases from Streptomyces Violascens for Efficient Production of N-Acetyl-d-Glucosamine. Food Chem. 2021, 364, 130393. [Google Scholar] [CrossRef]
- Wang, S.; Fu, G.; Li, J.; Wei, X.; Fang, H.; Huang, D.; Lin, J.; Zhang, D. High-Efficiency Secretion and Directed Evolution of Chitinase BcChiA1 in Bacillus subtilis for the Conversion of Chitinaceous Wastes into Chitooligosaccharides. Front. Bioeng. Biotechnol. 2020, 8, 432. [Google Scholar] [CrossRef]
- Prabavathy, V.R.; Mathivanan, N.; Sagadevan, E.; Murugesan, K.; Lalithakumari, D. Self-Fusion of Protoplasts Enhances Chitinase Production and Biocontrol Activity in Trichoderma harzianum. Bioresour. Technol. 2006, 97, 2330–2334. [Google Scholar] [CrossRef]
- Wang, L.; Xie, Y.; Chang, J.; Wang, J.; Liu, H.; Shi, M.; Zhong, Y. A Novel Sucrose-Inducible Expression System and Its Application for Production of Biomass-Degrading Enzymes in Aspergillus niger. Biotechnol. Biofuels Bioprod. 2023, 16, 23. [Google Scholar] [CrossRef]
- Poshina, D.N.; Raik, S.V.; Poshin, A.N.; Skorik, Y.A. Accessibility of Chitin and Chitosan in Enzymatic Hydrolysis: A Review. Polym. Degrad. Stab. 2018, 156, 269–278. [Google Scholar] [CrossRef]
- Itoh, T.; Hibi, T.; Fujii, Y.; Sugimoto, I.; Fujiwara, A.; Suzuki, F.; Iwasaki, Y.; Kim, J.-K.; Taketo, A.; Kimoto, H. Cooperative Degradation of Chitin by Extracellular and Cell Surface-Expressed Chitinases from Paenibacillus sp. Strain FPU-7. Appl. Environ. Microbiol. 2013, 79, 7482–7490. [Google Scholar] [CrossRef] [PubMed]
- Hammami, I.; Siala, R.; Jridi, M.; Ktari, N.; Nasri, M.; Triki, M.A. Partial Purification and Characterization of ChiIO8, a Novel Antifungal Chitinase Produced by Bacillus cereus IO8. J. Appl. Microbiol. 2013, 115, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhang, A.; Chen, K.; Hao, Z.; Tong, J.; Ouyang, P. Characterization of Extracellular Chitinase from Chitinibacter sp. GC72 and Its Application in GlcNAc Production from Crayfish Shell Enzymatic Degradation. Biochem. Eng. J. 2015, 97, 59–64. [Google Scholar] [CrossRef]
- Shibasaki, H.; Uchimura, K.; Miura, T.; Kobayashi, T.; Usami, R.; Horikoshi, K. Highly Thermostable and Surfactant-Activated Chitinase from a Subseafloor Bacterium, Laceyella putida. Appl. Microbiol. Biotechnol. 2014, 98, 7845–7853. [Google Scholar] [CrossRef]
- Adrangi, S.; Faramarzi, M.A.; Shahverdi, A.R.; Sepehrizadeh, Z. Purification and Characterization of Two Extracellular Endochitinases from Massilia timonae. Carbohydr. Res. 2010, 345, 402–407. [Google Scholar] [CrossRef]
- Kim, J.; Kim, S.I.; Hong, E.; Ryu, Y. Strategies for Increasing Heterologous Expression of a Thermostable Esterase from Archaeoglobus fulgidus in Escherichia coli. Protein Expr. Purif. 2016, 127, 98–104. [Google Scholar] [CrossRef]
- Harnischfeger, J.; Beutler, M.; Salzig, D.; Rahlfs, S.; Becker, K.; Grevelding, C.G.; Czermak, P. Biochemical Characterization of the Recombinant Schistosome Tegumental Protein SmALDH_312 Produced in E. Coli and Baculovirus Expression Vector System. Electron. J. Biotechnol. 2021, 54, 26–36. [Google Scholar] [CrossRef]
- Cardozo, F.A.; Facchinatto, W.M.; Colnago, L.A.; Campana-Filho, S.P.; Pessoa, A. Bioproduction of N-Acetyl-Glucosamine from Colloidal α-Chitin Using an Enzyme Cocktail Produced by Aeromonas caviae CHZ306. World J. Microbiol. Biotechnol. 2019, 35, 114. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Wang, S.-L. Production of Thermophilic Chitinase by Paenibacillus sp. TKU052 by Bioprocessing of Chitinous Fishery Wastes and Its Application in N-Acetyl-D-Glucosamine Production. Polymers 2021, 13, 3048. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Madhuprakash, J.; El Gueddari, N.E.; Moerschbacher, B.M.; Podile, A.R. Catalytic Efficiency of Chitinase-D on Insoluble Chitinous Substrates Was Improved by Fusing Auxiliary Domains. PLoS ONE 2015, 10, e0116823. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-Binding Modules: Fine-Tuning Polysaccharide Recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Qiu, M.; Zhang, X.; Chen, J. Expression and Characterization of an Enhanced Recombinant Heparinase I with Chitin Binding Domain. Int. J. Biol. Macromol. 2017, 105, 1250–1258. [Google Scholar] [CrossRef]
- Itoh, Y.; Watanabe, J.; Fukada, H.; Mizuno, R.; Kezuka, Y.; Nonaka, T.; Watanabe, T. Importance of Trp59 and Trp60 in Chitin-Binding, Hydrolytic, and Antifungal Activities of Streptomyces griseus Chitinase C. Appl. Microbiol. Biotechnol. 2006, 72, 1176–1184. [Google Scholar] [CrossRef]
- Nakamura, T.; Mine, S.; Hagihara, Y.; Ishikawa, K.; Uegaki, K. Structure of the Catalytic Domain of the Hyperthermophilic Chitinase from Pyrococcus Furiosus. Acta Cryst. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 7–11. [Google Scholar] [CrossRef]
- Pham, M.-L.; Leister, T.; Nguyen, H.A.; Do, B.-C.; Pham, A.-T.; Haltrich, D.; Yamabhai, M.; Nguyen, T.-H.; Nguyen, T.-T. Immobilization of β-Galactosidases from Lactobacillus on Chitin Using a Chitin-Binding Domain. J. Agric. Food Chem. 2017, 65, 2965–2976. [Google Scholar] [CrossRef]
- Chenxi, G.; Jianrong, C.; Yinghua, T.; Quanfeng, W.; Mingguo, J.; Lixia, P. Heterologous Expression and Enzymatic Properties of Truncated Mutant of Bi-Functional Chitinase from Chitinilyticum Aquatile. Guangxi Sci. 2024, 32, 1–15. [Google Scholar] [CrossRef]
- Zhang, A.; He, Y.; Wei, G.; Zhou, J.; Dong, W.; Chen, K.; Ouyang, P. Molecular Characterization of a Novel Chitinase CmChi1 from Chitinolyticbacter meiyuanensis SYBC-H1 and Its Use in N-Acetyl-d-Glucosamine Production. Biotechnol. Biofuels 2018, 11, 179. [Google Scholar] [CrossRef]
- Chen, J.; Yang, D.; Zhang, Y.; Yang, L.; Wang, Q.; Jiang, M.; Pan, L. A Novel Bi-Functional Cold-Adaptive Chitinase from Chitinilyticum aquatile CSC-1 for Efficient Synthesis of N-Acetyl-D-Glucosaminidase. Int. J. Biol. Macromol. 2024, 259, 129063. [Google Scholar] [CrossRef]
- Glavina Del Rio, T.; Abt, B.; Spring, S.; Lapidus, A.; Nolan, M.; Tice, H.; Copeland, A.; Cheng, J.-F.; Chen, F.; Bruce, D.; et al. Complete Genome Sequence of Chitinophaga pinensis Type Strain (UQM 2034T). Stand Genom. Sci. 2010, 2, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, A.; Aslam, M.; Kanai, T.; Atomi, H. A Structurally Novel Chitinase from the Chitin-Degrading Hyperthermophilic Archaeon Thermococcus chitonophagus. Appl. Environ. Microbiol. 2016, 82, 3554–3562. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhao, X.; Xu, B.; Su, E.; Kovalevsky, A.; Shen, Q.; Liu, D.; Wan, Q. Discovering and Designing a Chimeric Hyperthermophilic Chitinase for Crystalline Chitin Degradation. ACS Sustain. Chem. Eng. 2023, 11, 4690–4698. [Google Scholar] [CrossRef]
- Deng, J.-J.; Zhang, M.-S.; Li, Z.-W.; Lu, D.-L.; Mao, H.-H.; Zhu, M.-J.; Li, J.-Z.; Luo, X.-C. One-Step Processing of Shrimp Shell Waste with a Chitinase Fused to a Carbohydrate-Binding Module. Green Chem. 2020, 22, 6862–6873. [Google Scholar] [CrossRef]
- Wang, F.P.; Li, Q.; Zhou, Y.; Li, M.G.; Xiao, X. The C-Terminal Module of Chi1 from Aeromonas Caviae CB101 Has a Function in Substrate Binding and Hydrolysis. Proteins Struct. Funct. Genet. 2003, 53, 908–916. [Google Scholar] [CrossRef]
- Li, J.; Zheng, J.; Liang, Y.; Yan, R.; Xu, X.; Lin, J. Expression and Characterization of a Chitinase from Serratia marcescens. Protein Expr. Purif. 2020, 171, 105613. [Google Scholar] [CrossRef]
- Ueda, M.; Shioyama, T.; Nakadoi, K.; Nakazawa, M.; Sakamoto, T.; Iwamoto, T.; Sakaguchi, M. Cloning and Expression of a Chitinase Gene from Eisenia fetida. Int. J. Biol. Macromol. 2017, 104, 1648–1655. [Google Scholar] [CrossRef]
- Fu, X.; Yan, Q.; Wang, J.; Yang, S.; Jiang, Z. Purification and Biochemical Characterization of Novel Acidic Chitinase from Paenicibacillus barengoltzii. Int. J. Biol. Macromol. 2016, 91, 973–979. [Google Scholar] [CrossRef]
- Novotná, Z.; Fliegerová, K.; Šimůnek, J. Characterization of Chitinases of Polycentric Anaerobic Rumen Fungi. Folia Microbiol 2008, 53, 241–245. [Google Scholar] [CrossRef]
- Xiayun, J.; Chen, D.; Shenle, H.; Wang, W.; Chen, S.; Zou, S. Identification, Characterization and Functional Analysis of a GH-18 Chitinase from Streptomyces roseolus. Carbohydr. Polym. 2012, 87, 2409–2415. [Google Scholar] [CrossRef]
- Karthik, N.; Binod, P.; Pandey, A. RETRACTED: Purification and Characterisation of an Acidic and Antifungal Chitinase Produced by a Streptomyces sp. Bioresour. Technol. 2015, 188, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.-W. Purification and Characterization of Chitinase from a New Species Strain Pseudomonas sp. TKU008. J. Microbiol. Biotechnol. 2010, 20, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Lee, Y.-S.; Choi, Y.-L. Cloning, Purification, and Characterization of an Organic Solvent-Tolerant Chitinase, MtCh509, from Microbulbifer thermotolerans DAU221. Biotechnol. Biofuels 2018, 11, 303. [Google Scholar] [CrossRef]
- Du, J.; Duan, S.; Miao, J.; Zhai, M.; Cao, Y. Purification and Characterization of Chitinase from Paenibacillus sp. Biotechnol. Appl. Biochem. 2021, 68, 30–40. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Yang, L.; Yang, D.; Jiang, M.; Ling, C.; Chen, H.; Ji, F.; Pan, L. Biochemical Purification and Characterization of a Truncated Acidic, Thermostable Chitinase from Marine Fungus for N-Acetylglucosamine Production. Front. Bioeng. Biotechnol. 2022, 10, 1013313. [Google Scholar] [CrossRef]
- Annamalai, N.; Giji, S.; Muthuvel, A.; Balasubramanian, T. Purification and Characterization of Chitinase from Micrococcus sp. AG84 Isolated from Marine Environment. Afr. J. Microbiol. Res. 2010, 4, 2822–2827. [Google Scholar]
- Aktas, C.; Ruzgar, D.; Gurkok, S.; Gormez, A. Purification and Characterization of Stenotrophomonas Maltophilia Chitinase with Antifungal and Insecticidal Properties Prep. Biochem. Biotechnol. 2022, 53, 797–806. [Google Scholar]
- Cheba, B.A.; Zaghloul, T.I.; EL-Massry, M.H.; EL-Mahdy, A.R. Effect of Metal Ions, Chemical Agents, and Organic Solvent on Bacillus Sp.R2 Chitinase Activity. Procedia Technol. 2016, 22, 465–470. [Google Scholar] [CrossRef]
- Daimon, T.; Hamada, K.; Mita, K.; Okano, K.; Suzuki, M.G.; Kobayashi, M.; Shimada, T. A Bombyx Mori Gene, BmChi-h, Encodes a Protein Homologous to Bacterial and Baculovirus Chitinases. Insect Biochem. Mol. Biol. 2003, 33, 749–759. [Google Scholar] [CrossRef]
- Fan, Y.; Guo, S.; Pei, X.; Zhang, Y.; Luo, Z.; Pei, Y. Effects of Chitin Binding Domain on Enzymatic Properties and Insecticidal Activity of Bombyx Mori Chitinase. World J. Microbiol. Biotechnol. 2011, 27, 1551–1558. [Google Scholar] [CrossRef]
- Fu, X.; Yan, Q.; Yang, S.; Yang, X.; Guo, Y.; Jiang, Z. An Acidic, Thermostable Exochitinase with β-N-Acetylglucosaminidase Activity from Paenibacillus barengoltzii Converting Chitin to N-Acetyl Glucosamine. Biotechnol. Biofuels 2014, 7, 174. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization by One Table (TvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator: Figure 1. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]

| Substrate | Relative Activity (%) |
|---|---|
| Colloidal Chitin | 100 |
| α-Chitin | 1.71155 ± 0.09682 |
| β-Chitin | 10.37198 ± 1.24252 |
| CMC-Na | 0.3668 ± 0.03463 |
| Chitosan | 3.64104 ± 0.05201 |
| MCC | ND |
| Primers | Sequence |
|---|---|
| CaChi18B_ΔChBDs-F | 5′-cccagccggcgatggccatggCAGGTCGGCTCCTACTTCACG-3′ |
| CaChi18B_ΔChBDs-R | 5′-ctcgagtgcggccgcaagcttCTGCTTGACCTTGCTCATGGC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, C.; Chen, J.; Huang, X.; Jiang, Y.; Ou, N.; Yang, D.; Jiang, M.; Pan, L. The Impact of Chitinase Binding Domain Truncation on the Properties of CaChi18B from Chitinilyticum aquatile CSC-1. Mar. Drugs 2025, 23, 93. https://doi.org/10.3390/md23030093
Gu C, Chen J, Huang X, Jiang Y, Ou N, Yang D, Jiang M, Pan L. The Impact of Chitinase Binding Domain Truncation on the Properties of CaChi18B from Chitinilyticum aquatile CSC-1. Marine Drugs. 2025; 23(3):93. https://doi.org/10.3390/md23030093
Chicago/Turabian StyleGu, Chenxi, Jianrong Chen, Xinyue Huang, Yongqiang Jiang, Na Ou, Dengfeng Yang, Mingguo Jiang, and Lixia Pan. 2025. "The Impact of Chitinase Binding Domain Truncation on the Properties of CaChi18B from Chitinilyticum aquatile CSC-1" Marine Drugs 23, no. 3: 93. https://doi.org/10.3390/md23030093
APA StyleGu, C., Chen, J., Huang, X., Jiang, Y., Ou, N., Yang, D., Jiang, M., & Pan, L. (2025). The Impact of Chitinase Binding Domain Truncation on the Properties of CaChi18B from Chitinilyticum aquatile CSC-1. Marine Drugs, 23(3), 93. https://doi.org/10.3390/md23030093






