Abstract
Three new cyclic lipopeptides, olenamidonins A-C (1–3), in addition to two previously reported metabolites (4 and 5), were accumulated in the ΔdtxRso deletion mutant of deepsea-derived Streptomyces olivaceus SCSIO 1071. The structures of these cyclic lipopeptides were determined by a combination of spectroscopic methods and circular dichroism (CD) measurement. The antibacterial assay results showed that compounds 1–5 displayed different degrees of growth inhibition against multidrug-resistant (MDR) bacterial strains Enterococcus faecalis CCARM 5172 and Enterococcus faecium CCARM 5203 with minimum inhibitory concentrations (MICs) of 1.56−6.25 μg/mL.
1. Introduction
Microbial genome sequencing has unearthed uncountable cryptic secondary metabolite biosynthetic gene clusters (BGCs) in Streptomyces. However, most of them are silent under normal laboratory conditions [1]. Given that secondary metabolism is precisely controlled by a complex regulatory cascade network [2], the inactivation/overexpression of regulatory genes has been proven to be an effective way to unlock silent gene clusters [3].
DtxR (diphtheria toxin repressor) is a metal-dependent transcriptional regulator. DtxR family regulators, which were first identified in Corynebacterium diphtheriae [4], widely exist in high-GC content Gram-positive bacteria. They were found to be involved in the regulation of iron homeostasis, secondary metabolism, and morphological differentiation [5,6,7]. For example, the deletion of a DtxR family regulator gene dmdR1 in Streptomyces coelicolor A3(2) led to a slow rate of spore formation and the loss of pigmented antibiotics undecylprodigiosin and actinorhodin production [8,9]. The DtxR family regulator IdeR from Streptomyces avermitilis could regulate the production of avermectin and oligomycin in a positive and negative way, respectively; additionally, its deletion resulted in a bald phenotype and delayed morphological differentiation in S. avermitilis [5].
In our efforts to discover novel natural products from deepsea-derived Streptomyces strains, pleiotropic regulators were set as target genes for genetic manipulation to activate silent BGCs. A DtxR family regulatory gene dtxRso was identified from the genome of deepsea-derived Streptomyces olivaceus SCSIO 1071. The inactivation of dtxRso led to the activation of cyclic lipopeptide compound olenamidonins, including three new (1–3) and two known (4 and 5) compounds (Figure 1A). The related structural analogs of olenamidonins, enamidonins, were reported to have a variety of bioactivities, including macrophage foam cell formation inhibition [10], acyl-CoA/cholesterol acyltransferase (ACAT) inhibition [11], and anti-Gram-positive bacteria activities [12]. Herein, we describe the isolation, structural identification, and antibacterial activity evaluation of these lipopeptides.
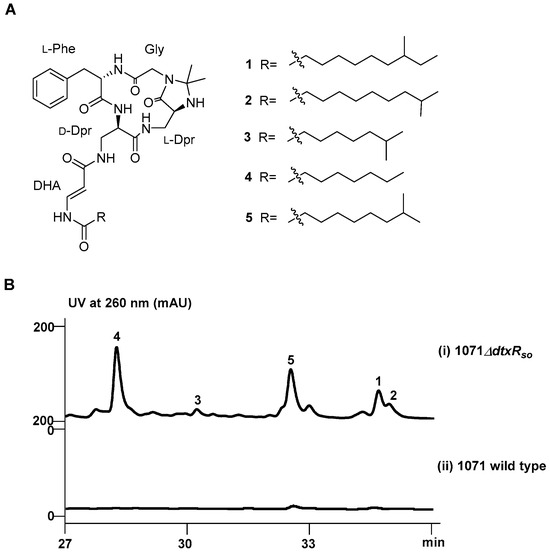
Figure 1.
(A) Chemical structures of 1–5 and (B) HPLC analysis of ΔdtxRso (i) and wild-type (ii) strains.
2. Results
The deepsea-derived S. olivaceus SCSIO 1071 was taxonomically classified through the phylogenetic analysis of its 16S rRNA gene sequence (Table S1, Figure S1) against the EzTaxon-e server Database. DtxRso (Table S2) from S. olivaceus SCSIO 1071 showed 58.3% identity to DtxR (WP_010935052.1) from C. diphtheriae and is highly homologous to IdeR (BAC71567.1) from S. avermitilis (96.0% identity) and DmdR1 (CAC28070.1) from S. coelicolor (98.0% identity). While the deletion of dtxRso had no obvious impact on the growth and morphological differentiation, a series of new peaks were accumulated in the fermentation products of the ΔdtxRso mutant (Figure 1B, panel i) compared to those of the wild-type strain (Figure 1B, panel ii).
To elucidate the chemical structures of the activated compounds, a total volume of 35 L fermentation cultures of S. olivaceus SCSIO 1071ΔdtxRso was obtained, from which five compounds (1–5) were isolated. Their structures were identified via high-resolution electrospray ionization mass spectrometry (HRESIMS) and nuclear magnetic resonance (NMR) assignments. Compound 1 was isolated as a colorless oil. The molecular formula of 1 was determined to be C34H51N7O6 according to the HRESIMS data ([M + H]+ at m/z 654.3994, calcd 654.3979, Figure S6). The 1D and 2D NMR (HSQC, COSY, and HMBC) data showed the presence of amino acids phenylalanine (Phe), glycine (Gly), 2,3-diaminopropionic acid (Dpr), and a fatty acid moiety (Figure 2 and Figures S7–S12) [8]. HMBC correlations from H2-16 (ΔH 3.07, 2.78) to C6 (ΔC 171.1), C-17 (ΔC 137.3), and C-18/22 (ΔC 128.8), from H-20 (ΔH 7.17) to C-18/22 (ΔC 128.8), and from H-21 (ΔH 7.24) to C-17 (ΔC 137.3) and C-19 (ΔC 128.3) confirmed the presence of a Phe residue. The HMBC correlations of the methylene signals H2-2 (ΔH 4.17, 3.31) to C-3 (ΔC 168.8) and C-5 (ΔC 53.9) indicated the existence of the Gly residue linked at C-5 of Phe. Two Dpr units were established by COSY signals of H-10 (ΔH 6.98)/H2-11 (ΔH 3.50, 3.24)/H-12 (ΔH 3.77)/H-13 (ΔH 2.99) and H-7 (ΔH 8.7)/H-8(ΔH 4.36)/H2-25(ΔH 3.31)/H-26 (ΔH 7.77). The COSY correlations of H-28 (ΔH 5.55)/H-29 (ΔH 7.59) and the HMBC correlations from H-28 (ΔH 5.55) and H-29 (ΔH 7.59) to C-27 (ΔC 166.6) confirmed the dehydro-β-alanine (DHA) unit. The 1H-1H coupling constant (3JH-28, H-29 =14.0 Hz) and the NOESY correlations for H-28 (ΔH 5.55)/H-26 (ΔH 7.77) defined the double-bond geometry as 28E. The N, N-acetonide group was confirmed by the HMBC correlations from H2-2 (ΔH 4.17, 3.31), H-13 (ΔH 2.99), H3-23 (ΔH 1.26), and H3-24 (ΔH 1.18) to C-14 (ΔC 76.3). Ten carbons of the C11 fatty acyl chain were identified by the COSY correlations, and the HMBC correlations from H2-7′ (ΔH 1.27, 1.07) and H2-9′ (ΔH 1.29, 1.08) to C-11′ (ΔC 19.1) showed that the methyl group was attached to C-8′ (ΔC 33.7). The advanced Marfey’s method [12] was used to determine the absolute configurations of the Phe and Dpr residues, and the result suggested the peptide backbone of compound 1 to be L-Phe-D-Dpr-L-Dpr-Gly (Figures S2 and S3). While the backbone was the same as those reported for autucedines and enamidonins [12,13], compound 1 contained a different 8′-methyldecanoyl side-chain. Therefore, compound 1 was identified as a new cyclic lipopeptide, named olenamidonin A. The 1H and 13C chemical shifts of 1 are listed in Table 1.
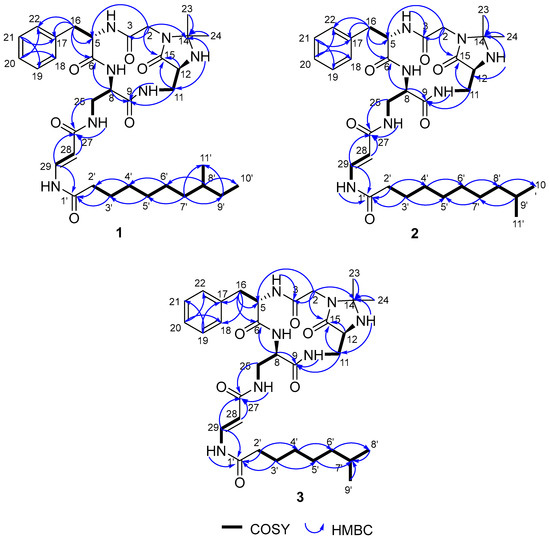
Figure 2.
COSY and key HMBC correlations of 1–3.

Table 1.
1H (600 MHz) and 13C (150 MHz) NMR data of 1–3 in DMSO-d6.
Compound 2 was isolated as a colorless oil. The molecular formula of 2 was determined to be C34H51N7O6 according to the HRESIMS data ([M + H]+ at m/z 654.3991, calcd 654.3979, Figure S13), which was the same as 1. The NMR data showed that the backbone of 2 was the same as that of 1 (Figure 2 and Figures S14–S17). While 2 also contained a C11 fatty acyl chain, the COSY correlations of H2-8′ (ΔH 1.49)/H-9′ (ΔH 1.49)/H3-10′/11′ (ΔH 0.84) combined with the HMBC correlations from H2-8′ (ΔH 1.13) to C-10′/11′ (ΔC 22.5) showed that the methyl group was attached to C-9′ (ΔC 27.3) instead of C-8′ (ΔC 33.7) compared with 1. The CD spectra of 2 and 1 in MeOH display similar Cotton effects, supporting that 2 shares the same absolute configurations as 1 (Figure S4). Thus, compound 2 was identified as a new olenamidonin analog, named olenamidonin B. The 1H and 13C chemical shifts of 2 are listed in Table 1.
Compound 3 was isolated as a colorless oil. The molecular formula of 3 was determined to be C32H47N7O6 according to the HRESIMS data ([M + H]+ at m/z 626.3675, calcd 626.3666, Figure S18), which had two less methyl groups than 1. The NMR data showed that the backbone of 3 was the same as 1 (Figure 2 and Figures S19–S23). Eight carbons of the C9 fatty acyl chain were identified by the COSY correlations, and two double-peak methyl signals with their HMBC correlations to C-7′ (ΔC 27.3) confirmed that the methyl group was attached to C-7′ (ΔC 27.3). The CD spectra of 3 and 1 in MeOH display similar Cotton effects, supporting that 3 shares the same absolute configurations as 1 (Figure S4). Compound 3 contained a different 7′-methyloctanoyl side-chain. Thus, compound 3 was identified as a new olenamidonin analog, named olenamidonin C. The 1H and 13C chemical shifts of 3 are listed in Table 1.
Compounds 4 and 5 were isolated as a colorless oil. The molecular formulas of 4 and 5 were determined to be C31H45N7O6 ([M + H]+ at m/z 612.3522, calcd 612.3510, Figure S24) and C33H49N7O6 ([M + H]+ at m/z 640.3840, calcd 640.3823, Figure S31), respectively. By comparing the NMR data (Figure 2 and Figures S25–S30 and S32–S37) with those reported in the literature, 4 and 5 were respectively confirmed to be known compounds autucedine A and autucedine D previously isolated from Streptomyces olivaceus SCSIO T05 [13], which is the same species as that of 1071 but is a different strain. The 1H and 13C NMR data for 4 and 5 are listed in Table S3.
We further analyzed the olenamidonins’ BGC (Table S4) from 1071 and found that it is identical to the aut BGC from T05 (GenBank ID: WP_194276129.1-WP_194276149.1) [13] except a couple of nucleotides in between open reading frames. Based on the previously proposed biosynthetic pathway of autucedines [13], the 3-oxoacyl-ACP synthase OleH, the homolog of AutS, may display flexible substrate specificity, ligating different acyl-CoAs to DHA-ACP to finally generate lipopeptides with different lipid chains. While a molecular network analysis of autucedine analogs showed that AutS was able to use saturated/unsaturated lipochains with a length of C7 to C13, these compounds were not structurally characterized except autucedines A-E [13].
The antibacterial activities of compounds 1–5 were evaluated against three Gram-positive (Micrococcus luteus ML01, Enterococcus faecalis CCARM 5172, and Enterococcus faecium CCARM 5203) and two Gram-negative (Escherichia coli CCARM 1009 and Pseudomonas aeruginosa 15690) multidrug-resistant (MDR) bacterial strains. As shown in Table 2, while no antibacterial activities toward M. luteus ML01, E. coli CCARM 1009, or P. aeruginosa 15690 were detected for all the compounds, considerable growth inhibitions against E. faecalis CCARM 5172 and E. faecium CCARM 5203 were observed. Noticeably, compounds 1–2 and 5 exhibited stronger anti-E. faecalis CCARM 5172 activities (MICs = 1.56−3.12 μg/mL) than the positive control ciprofloxacin (MIC = 6.25 μg/mL), while 3–4 displayed comparable inhibition activities to that of ciprofloxacin. Compounds 1–2 and 5 also showed similar anti-E. faecium CCARM 5203 activities to that of the positive control tetracycline (MIC = 1.56 μg/mL), which were stronger than those of 3–4 (MIC = 3.12–6.25 μg/mL). The comparison of the structures and antibacterial activities of these compounds showed lipopeptides with C11- and C10-fatty acyl chains (1–2, 5) have better inhibition potentials against E. faecalis CCARM 5172 and E. faecium CCARM 5203 than C9- and C8-fatty acyl chain analogs (3–4), suggesting that the length of the fatty acyl chain probably plays an important role in antibacterial activities.

Table 2.
Antibacterial activities of compounds 1–5 against MDR bacterial strains (MIC, μg/mL).
We further analyzed the distribution of the DtxRso homologs across diverse bacteria. A sequence similarity network (SSN) analysis of 2375 DtxRso homologs from the UniProt database was performed. As shown in Figure 3, these homologs mostly exist in Actinomycetes (in red), including families of Microbacteriaceae, Streptosporangiaceae, Nocardiaceae, Micrococcaceae, and Corynebacteriaceae. Noticeably, the Acidimicrobiia clade (in green) mainly has three paralogous groups, which are respectively associated with homolog(s) belonging to the Actinomycetes clade, suggesting their close relationship during evolution. Additionally, DtxR family regulators from Magnoliopsida (in orange) and Myxococcia (in dark green) are clustered with DtxRso/IdeR in the Actinomycetes clade, suggesting a possible horizontal transfer of the DtxRso homologs across the bacterial world. Considering the massive cryptic secondary metabolite BGCs and the wide distribution of DtxR family regulators in Actinomycetes, the genetic manipulation of these regulators would be an alternative approach to unlock silent or cryptic gene clusters.
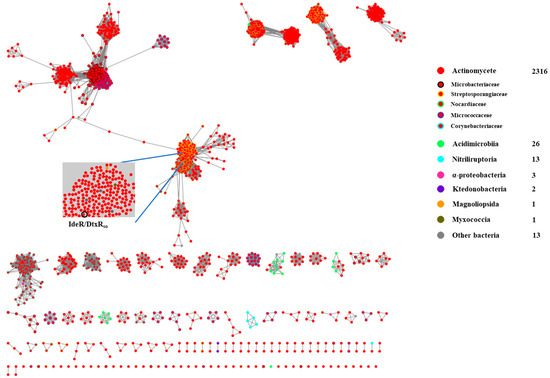
Figure 3.
Distribution of DtxRso homologs in bacteria. SSN analysis of DtxRso homologs in diverse bacteria is based on cutoff E value of 10−85 and visualized by Cytoscape [14].
3. Conclusions
In this study, three new compounds, olenamidonins A-C (1–3), and two known compounds (4 and 5) were obtained from the ΔdtxRso mutant of deepsea-derived S. olivaceus SCSIO 1071. Compounds 1–5 showed different degrees of growth inhibitory activities against E. faecalis CCARM 5172 and E. faecium CCARM 5203. The comparison of their structures and antibacterial activities suggested that the length of the fatty acyl chain probably plays an important role in antibacterial activities. Feeding lipids with longer chains may contribute to the generation of olenamidonin derivatives with better activities. The activation of olenamidonin production by knocking out dtxRso is very intriguing, and the underlying regulatory mechanism is worthy of further investigation. Changing the fermentation conditions of ΔdtxRso could serve to investigate the impacts of the dtxRso deletion on the productions of other metabolites. Given the wide distribution of DtxRso homologs, they could serve as alternative targets for the activation of silent BGCs in actinomycetes strains.
4. Materials and Methods
4.1. General Materials and Methods
Bacterial strains and plasmids used in this study are listed in Table S1; primers are listed in Table S2. Streptomyces olivaceus SCSIO 1071 was isolated from a deepsea mud sample from the South China Sea. The NMR spectra were recorded on a Bruker Avance III 600. Chemical shifts were referenced to the residual DMSO-d6 signal (ΔH 2.50 and ΔC 39.5 ppm for DMSO-d6). HRESIMS data were obtained on a Q-TOF Ultima Global GAA076 MS spectrometer. DNA isolation was carried out according to established protocols [15]. Plasmid was extracted using commercial kits (OMEGA).
4.2. Bioinformatic Analysis
The ole biosynthetic gene cluster was detected and analyzed using online antiSMASH 6.0.0 alpha software (http://antismash.secondarymetabolites.org/, 18 April 2024). Sequence comparisons and database searches were accomplished with BLAST programs (https://blast.ncbi.nlm.nih.gov/Blast.cgi, 18 April 2024).
The EFI-Enzyme Similarity Tool (EFI-EST) was used to generate sequence similarity networks (SSNs) [16]. For the distribution of DtxRso homologs, the DtxRso sequence was used as the query for searching a nonredundant protein sequence database using PSI-BLAST (position-specific iterated BLAST). A total of 2316 DtxRso homologs were used to build the SSNs. The 10−80 SSN was generated by applying an E value cutoff of 10−80 to the full network. Each node in the network represents a single sequence, and each edge represents the pairwise connection between two sequences for which the BLASTP E value was lower than the cutoff value. SSNs were visualized and colored by Cytoscape (v3.7.2) [14].
4.3. The Construction of the dtxRso Mutant Strain
A PCR-targeting strategy was adopted to obtain the dtxRso mutant strain [17,18]. Briefly, the amplified aac(3)IV-oriT resistance gene from pIJ773 was transformed into E. coli BW25113/pIJ790/pWLI615 to replace the dtxRso gene, resulting in the mutant cosmid pWLI1002 (ΔdtxRso). The mutant cosmid was passed through E. coli ET12567/pUZ8002 and then introduced into S. olivaceus SCSIO 1071 via conjugation [19]. The dtxRso mutant strain was selected through apramycin-resistant and neomycin-sensitive phenotype screening, followed by PCR confirmation.
4.4. Production and Purification of Olenamidonins
The fermentation of S. olivaceus SCSIO 1071/pWLI1002 in a total volume of 35 L was performed by growing cultures in 250 mL baffled Erlenmeyer flasks each containing 50 mL of AF/MS medium (glucose 20 g, yeast extract 2 g, soya flour 8 g, CaCO3 4 g, NaCl 1 g, distilled H2O 1000 mL, pH = 7.3), incubated at 30 °C on a rotary shaker at 220 rpm for 7 days. The cells were extracted with acetone by sonication. The combined organic extracts were concentrated and then partitioned between 90% MeOH and n-hexane to yield two residues. The aqueous MeOH layer (1.05 g) was subjected to a reversed-phase (C18) open column chromatography with 20−100% MeOH to afford 6 fractions. Compounds 1–5 were obtained by a further purification of fraction 4 on reversed-phase HPLC (YMC-Pack ODS-A column 250 mm × 10 mm, i.d. 5 μm; wavelength at 260 nm) eluting with 49% CH3CN + 0.1% HCOOH (v/v) (1.5 mL/min).
Olenamidonin A (1): colorless oil; [α= +10.4 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 206 (3.03), 266 (3.04) nm; 1H and 13C NMR data, Table 1; HRESIMS m/z 654.3994 [M + H]+ (calcd for C34H51N7O6, 654.3979).
Olenamidonin B (2): colorless oil; [α= +34.6 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 206 (3.02), 266 (3.01) nm; 1H and 13C NMR data, Table 1; HRESIMS m/z 654.3991 [M + H]+ (calcd for C34H51N7O6, 654.3979).
Olenamidonin C (3): colorless oil; [α= +6.6 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 206 (3.42), 266 (3.40) nm; 1H and 13C NMR data, Table 1; HRESIMS m/z 626.3675 [M + H]+ (calcd for C32H47N7O6, 626.3666).
Autucedine A (4): colorless oil; [α= +22.4 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 208 (3.16), 266 (3.20) nm; 1H and 13C NMR data, Table S3; HRESIMS m/z 612.3522 [M + H]+ (calcd for C31H45N7O6, 612.3510).
Autucedine D (5): colorless oil; [α= +9 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 208 (3.22), 266 (3.27) nm; 1H and 13C NMR data, Table S3; HRESIMS m/z 640.3840 [M + H]+ (calcd for C33H49N7O6, 640.3823).
4.5. Antibacterial Assay
The MDR bacterial strains M. luteus ML01, E. faecalis CCARM 5172, E. faecium CCARM 5203, E. coli CCARM 1009, and P. aeruginosa 15690 were grown overnight at 37 °C in liquid LB medium, then diluted with the LB broth to 106 CFU/mL. The sample solutions were diluted with MeOH to make a series of concentrations. After that, 20 μL of the sample solutions with different concentrations was dispensed into 180 μL of the bacterial suspension in 96-well plates and incubated at 37 °C for 18 h. The growth of MDR strains was measured on a microplate reader at a wavelength of 620 nm. Tetracycline (for M. luteus ML01, E. faecium CCARM 5203, E. coli CCARM 1009, and P. aeruginosa 15690) or ciprofloxacin (for E. faecalis CCARM 5172) was used as a positive control, methanol was used as a negative control, and LB broth was used as a blank.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/md22060262/s1, Figure S1: Neighbor-joining phylogenetic tree of S. olivaceus 1071 SCSIO; Figures S2 and S3: The HPLC chromatogram data of D-FDAA derivatives of the amino acid units in olenamidonin A (1); Figures S4–S37: HR-ESI-MS, 1H NMR, 13C NMR and, HSQC, HMBC, 1H-1H COSY, NOESY, ECD spectra of compounds 1–5. Tables S1 and S2: 1490 nt 16S rRNA and dtxRso sequence of S. olivaceus SCSIO 1071; Tables S3 and S4: Bacteria, plasmids and primer pairs used in this study; Table S5: 1H (600 MHz) and 13C (150 MHz) NMR data of 4 and 5; Table S6: Proposed functions of proteins encoded by the ole biosynthetic gene cluster in S. olivaceus SCSIO 1071. Reference [20] are cited in the Supplementary Materials.
Author Contributions
Q.S. performed chemical and bioactive experiments, analyzed the data, and wrote the manuscript. D.Y. measured the NMR spectra. X.Z. formal analysis. F.X. and W.L. designed the experiments and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Shandong Provincial Natural Science Foundation (ZR2021ZD28), the National Natural Science Foundation of China (32070054, U22A20582, 81991525 and 32100051), and the Fundamental Research Funds for the Central Universities (202172002).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data are included in the article and the Supplementary Materials.
Acknowledgments
We would like to thank Jianhua Ju (Shandong University of China) for providing us with the Streptomyces strain used in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; He, J.; Wei, X.; Ju, J.; Ma, J. Exploration and genome mining of natural products from marine Streptomyces. Appl. Microbiol. Biotechnol. 2020, 104, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Boyd, J.; Oza, M.N.; Murphy, J.R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 1990, 87, 5968–5972. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yang, R.; Lyu, M.; Wang, S.; Liu, X.; Wen, Y.; Song, Y.; Li, J.; Chen, Z. IdeR, a DtxR family iron response regulator, controls iron homeostasis, morphological differentiation, secondary metabolism, and the oxidative stress response in Streptomyces avermitilis. Appl. Environ. Microbiol. 2018, 84, e01503-18. [Google Scholar] [PubMed]
- Deng, Y.; Zhang, X. DtxR, an iron-dependent transcriptional repressor that regulates the expression of siderophore gene clusters in Thermobifida fusca. FEMS Microbiol. Lett. 2015, 362, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hantke, K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001, 4, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Flores, F.J.; Martín, J.F. Iron-regulatory proteins DmdR1 and DmdR2 of Streptomyces coelicolor form two different DNA-protein complexes with iron boxes. Biochem. J. 2004, 380, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Flores, F.J.; Barreiro, C.; Coque, J.J.; Martín, J.F. Functional analysis of two divalent metal-dependent regulatory genes dmdR1 and dmdR2 in Streptomyces coelicolor and proteome changes in deletion mutants. FEBS J. 2005, 272, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Namatame, I.; Tomoda, H.; Matsuda, D.; Tabata, N.; Kobayashi, S.; Omura, S. K97-0239A and B, new inhibitors of macrophage foam cell formation, produced by Streptomyces sp. K97-0239. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2006, 78, 45–50. [Google Scholar] [CrossRef]
- Ohshiro, T.; Rudel, L.L.; Omura, S.; Tomoda, H. Selectivity of microbial acyl-CoA: Cholesterol acyltransferase inhibitors toward isozymes. J. Antibiot. 2007, 60, 43–51. [Google Scholar] [CrossRef]
- Son, S.; Ko, S.K.; Kim, S.M.; Kim, E.; Kim, G.S.; Lee, B.; Ryoo, I.J.; Kim, W.G.; Lee, J.S.; Hong, Y.S.; et al. Antibacterial cyclic lipopeptide enamidonins with an enamide-linked acyl chain from a Streptomyces Species. J. Nat. Prod. 2018, 81, 2462–2469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, C.; Li, Q.; Ma, J.; Ju, J. Metabolic blockade-based genome mining reveals lipochain-linked dihydro-β-alanine synthetases involved in autucedine biosynthesis. Org. Lett. 2022, 24, 5535–5540. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor: New York, NY, USA, 2001; pp. 895–909. [Google Scholar]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Chandra, G.; Jakimowicz, D.; Yuqing, T.; Bruton, C.J.; Chater, K.F. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 2004, 54, 107–128. [Google Scholar] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A.; Charter, K.; Bib, M.J.; Bipp, M.; Keiser, T.; Butner, M. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Macneil, D.J.; Occi, J.L.; Gewain, K.M.; Macneil, T.; Gibbons, P.H.; Ruby, C.L.; Danis, S.J. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 1992, 115, 119–125. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).