Ethanol Extract of Limonium bicolor Improves Dextran Sulfate Sodium-Induced Ulcerative Colitis by Alleviating Inflammation and Restoring Gut Microbiota Dysbiosis in Mice
Abstract
1. Introduction
2. Results
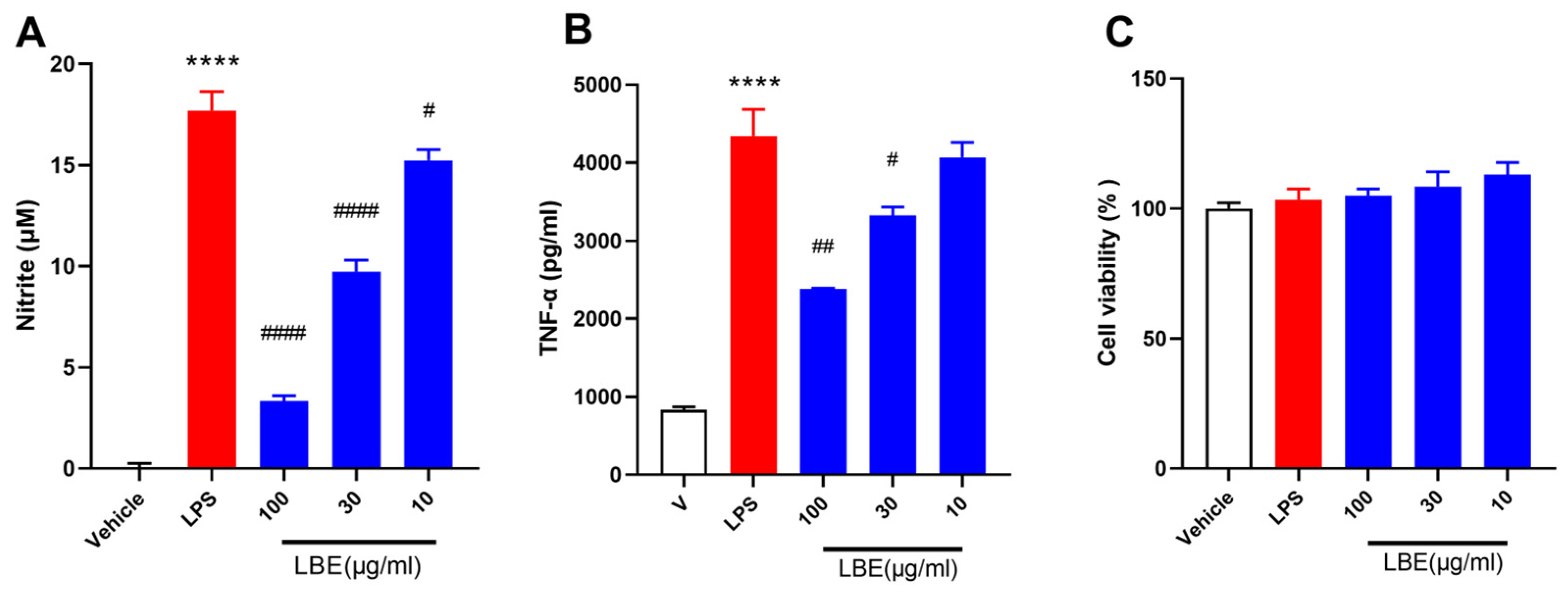
2.1. LBE Inhibited the Expression of Pro-Inflammatory Cytokines in LPS-Stimulated RAW264.7 Cells
2.2. LBE Attenuated DSS-Induced Colon Tissue Damage and Decreased DAI Scores
2.3. LBE Decreased the Levels of Pro-Inflammatory Cytokines
2.4. Identification of the Chemical Components of LBE
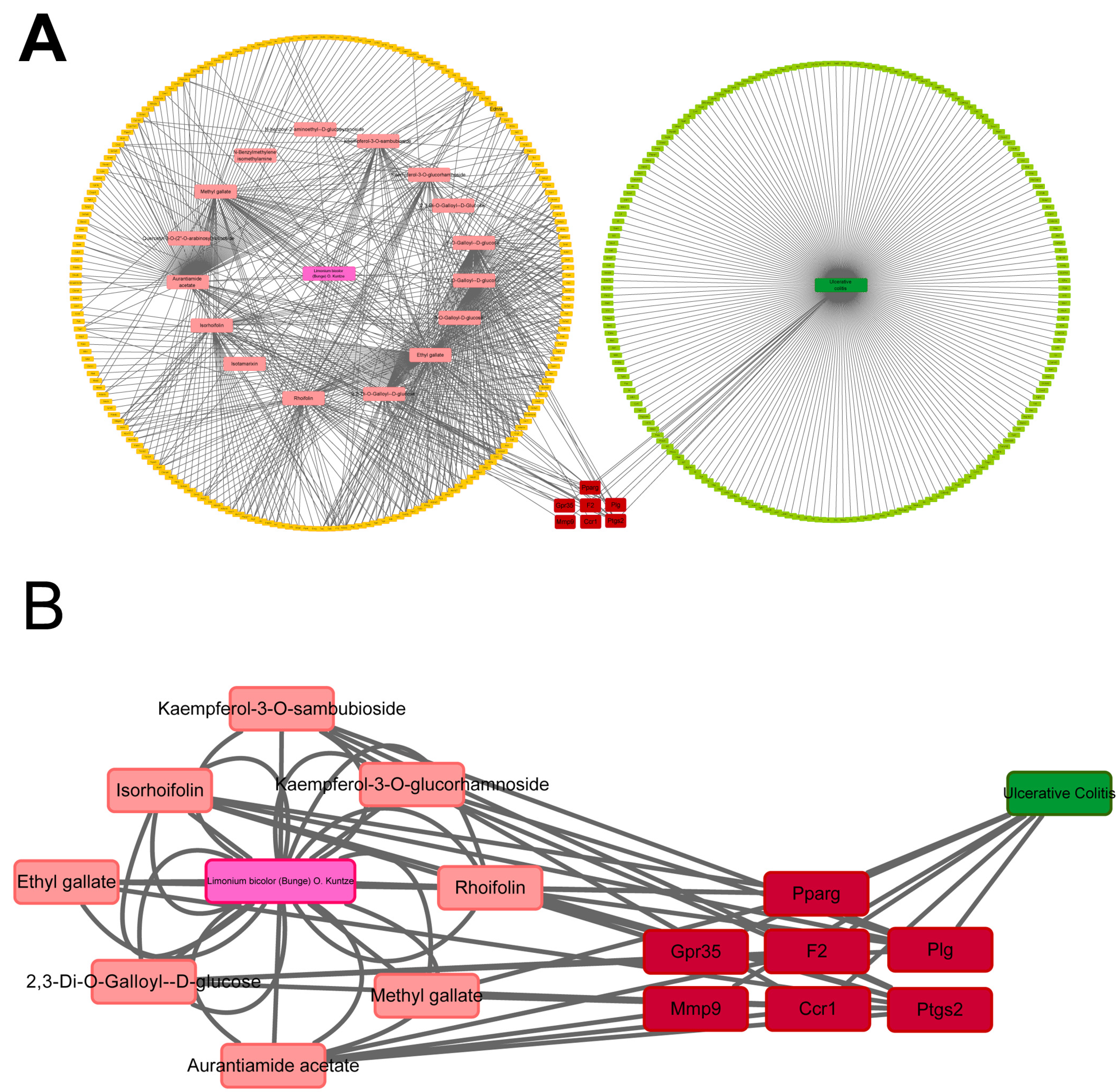
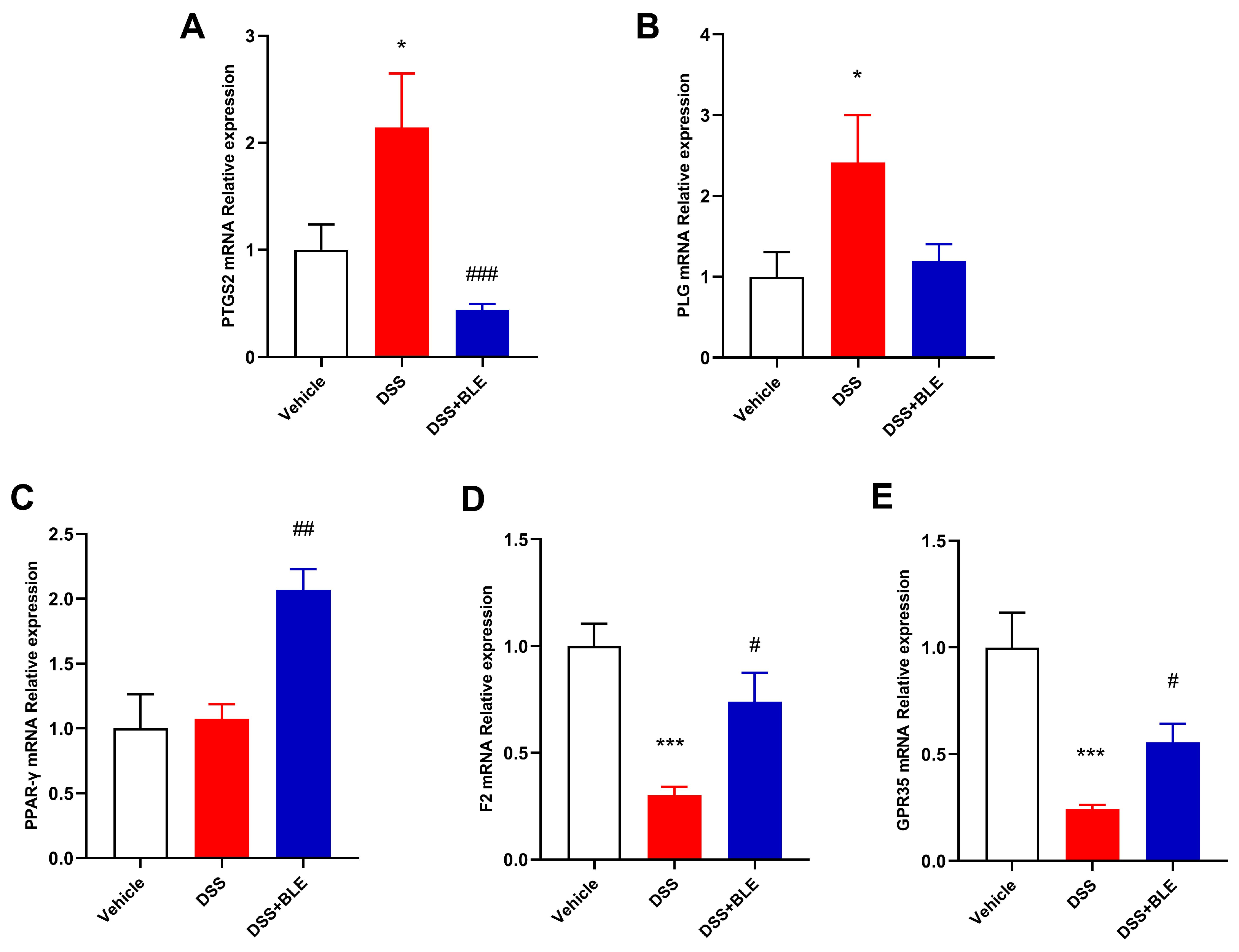
2.5. Potential Targets of LBE Predicted via Network Pharmacological Analysis
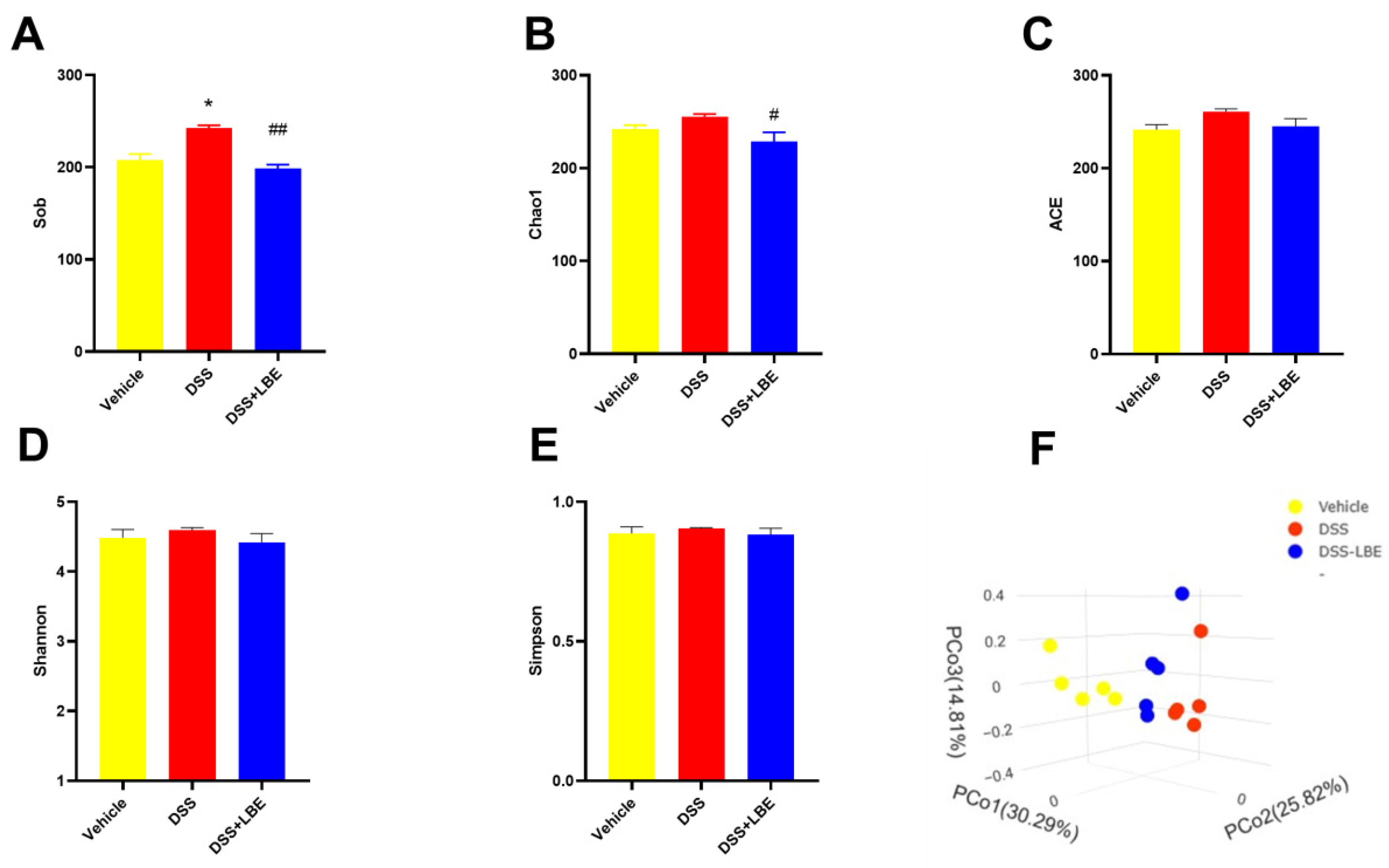
2.6. Effects of LBE on the Gut Microbiota of Mice with DSS-Induced UC
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Preparation of LBE
4.3. Cell Culture
4.4. NO Inhibition Assay
4.5. Cell Viability Assay
4.6. Animals and Treatment
4.7. Macroscopic Scoring and Histochemical Analysis
4.8. Detection of Pro-Inflammatory Cytokines in Serum
4.9. qRT-PCR
4.10. Identification of Major Chemical Components of LBE via LC-MS/MS
4.11. Network Pharmacology Analysis
4.12. 16S rRNA Sequencing of Gut Microbiota
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef]
- Conrad, K.; Roggenbuck, D.; Laass, M.W. Diagnosis and classification of ulcerative colitis. Autoimmun. Rev. 2014, 13, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Magro, F.; Langner, C.; Driessen, A.; Ensari, A.; Geboes, K.; Mantzaris, G.J.; Villanacci, V.; Becheanu, G.; Nunes, P.B.; Cathomas, G.; et al. European consensus on the histopathology of inflammatory bowel disease. J. Crohns Colitis 2013, 7, 827–851. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.X.; Shao, M.J.; Qi, Q.; Xu, Y.S.; Yang, X.Q.; Zhu, F.H.; He, S.J.; He, P.L.; Feng, C.L.; Wu, Y.W.; et al. Artemisinin analogue SM934 ameliorates DSS-induced mouse ulcerative colitis via suppressing neutrophils and macrophages. Acta Pharmacol. Sin. 2018, 39, 1633–1644. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 898. [Google Scholar] [CrossRef]
- Burri, E.; Maillard, M.H.; Schoepfer, A.M.; Seibold, F.; Van Assche, G.; Rivière, P.; Laharie, D.; Manz, M.; IBDnet, S.; Gastroenterology, S.S. Treatment Algorithm for Mild and Moderate-to-Severe Ulcerative Colitis: An Update. Digestion 2020, 101, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Pithadia, A.B.; Jain, S. Treatment of inflammatory bowel disease (IBD). Pharmacol. Rep. 2011, 63, 629–642. [Google Scholar] [CrossRef]
- Liu, Y.; Li, B.G.; Su, Y.H.; Zhao, R.X.; Song, P.; Li, H.; Cui, X.H.; Gao, H.M.; Zhai, R.X.; Fu, X.J.; et al. Potential activity of Traditional Chinese Medicine against Ulcerative colitis: A review. J. Ethnopharmacol. 2022, 289, 115084. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.S.; Fu, X.J.; Wu, Q.M.; Wang, C.Y.; Wang, Y.; Jiang, D.Z. Elementary exploration of the origin and development of marine Chinese materia medica. Zhonghua Yi Shi Za Zhi 2009, 39, 168–172. (In Chinese) [Google Scholar] [PubMed]
- Chen, J.; Teng, J.H.; Ma, L.; Tong, H.Y.; Ren, B.R.; Wang, L.S.; Li, W.L. Flavonoids Isolated from the Flowers of and their In vitro Antitumor Evaluation. Pharmacogn. Mag. 2017, 13, 222–225. [Google Scholar] [CrossRef]
- Kim, J.J.; Shajib, M.S.; Manocha, M.M.; Khan, W.I. Investigating Intestinal Inflammation in DSS-induced Model of IBD. JOVE J. Vis. Exp. 2012, 60, e3678. [Google Scholar] [CrossRef]
- Wu, M.N.; Li, P.Z.; An, Y.Y.; Ren, J.; Yan, D.; Cui, J.Z.; Li, D.; Li, M.; Wang, M.Y.; Zhong, G.S. Phloretin ameliorates dextran sulfate sodium-induced ulcerative colitis in mice by regulating the gut microbiota. Pharmacol. Res. 2019, 150. [Google Scholar] [CrossRef] [PubMed]
- Sands, B.E.; Kaplan, G.G. The role of TNFα in ulcerative colitis. J. Clin. Pharmacol. 2007, 47, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.X.; Kirschning, C.J.; Mancinelli, R.; Xu, X.P.; Jin, Y.P.; Faure, E.; Mantovani, A.; Rothe, M.; Muzio, M.; Arditi, M. Bacterial lipopolysaccharide activates nuclear factor-κB through interleukin-1 signaling mediators in cultured human dermal endothelial cells and mononuclear phagocytes. J. Biol. Chem. 1999, 274, 7611–7614. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, K.; Sugimura, K.; Anezaki, K.; Bannai, H.; Ishizuka, K.; Asakura, H. Spectrum of cytokine gene expression in intestinal mucosal lesions of Crohn’s disease and ulcerative colitis. Digestion 1998, 59, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.S. Role and regulation of cyclooxygenase-2 during inflammation. Am. J. Med. 1999, 106, 37S–42S. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.Y.; Jiang, J.G.; Huang, C.L.; Zhu, W.; Zheng, C.Y. Polyphenols from Blossoms of L. var. Engl. Show Significant Anti-Complement and Anti-Inflammatory Effects. J. Agr. Food Chem. 2017, 65, 9061–9068. [Google Scholar] [CrossRef] [PubMed]
- Keragala, C.B.; Medcalf, R.L. Plasminogen: An enigmatic zymogen. Blood 2021, 137, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Yun, L.; Xu, R.; Li, G.H.; Yao, Y.C.; Li, J.M.; Yan, S.H. MTHFR C677T gene mutation affect the level of plasma homocysteine but do not related to early renal damage in hypertensive patients. J. Am. Coll. Cardiol. 2014, 64, C100–C101. [Google Scholar] [CrossRef]
- De Giovanni, M.; Tam, H.; Valet, C.; Xu, Y.; Looney, M.R.; Cyster, J.G. GPR35 promotes neutrophil recruitment in response to serotonin metabolite 5-HIAA. Cell 2022, 185, 1103–1104. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Baldassano, R.N.; Griffiths, A.; Russell, R.K.; Annese, V.; Dubinsky, M.; Kugathasan, S.; Bradfield, J.P.; Walters, T.D.; Sleiman, P.; et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009, 41, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; Folseraas, T.; Holm, K.; Ellinghaus, E.; Melum, E.; Balschun, T.; Laerdahl, J.K.; Shiryaev, A.; Gotthardt, D.N.; Weismüller, T.J.; et al. Genome-Wide Association Analysis in Primary Sclerosing Cholangitis and Ulcerative Colitis Identifies Risk Loci at and. Hepatology 2013, 58, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Kaya, B.; Melhem, H.; Niess, J.H. GPR35 in Intestinal Diseases: From Risk Gene to Function. Front. Immunol. 2021, 12, 717392. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, D.; Cabral, J.M.; Soares-da-Silva, P.; Magro, F. Role of epithelial ion transports in inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G460–G476. [Google Scholar] [CrossRef] [PubMed]
- Anbazhagan, A.N.; Priyamvada, S.; Alrefai, W.A.; Dudeja, P.K. Pathophysiology of IBD associated diarrhea. Tissue Barriers 2018, 6, e1463897. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Liu, Q.F.; Yuan, Q.N.; Ji, Y.J.; Zhu, S.N.; Tan, Y.X.; He, X.H.; Xu, Y.W.; Shi, J.J.; Cheng, X.; et al. Insights into divalent cation regulation and G-coupling of orphan receptor GPR35. Cell Discov. 2022, 8, 135. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cao, H.; Huang, Q.; Xiao, J.; Teng, H. Absorption, metabolism and bioavailability of flavonoids: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 7730–7742. [Google Scholar] [CrossRef] [PubMed]
- Hostetler, G.L.; Ralston, R.A.; Schwartz, S.J. Flavones: Food Sources, Bioavailability, Metabolism, and Bioactivity. Adv. Nutr. 2017, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Derrien, M.; Isolauri, E.; de Vos, W.M.; Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007, 73, 7767–7770. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Collado, M.C.; Ben-Amor, K.; Salminen, S.; de Vos, W.M. The mucin degrader is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 2008, 74, 1646–1648. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Wu, W.; Yang, L.; Lv, L.; Wang, Q.; Li, Y.; Ye, J.; Fang, D.; Wu, J.; Jiang, X.; et al. Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front. Microbiol. 2019, 10, 2259. [Google Scholar] [CrossRef]
- Qu, S.; Fan, L.; Qi, Y.; Xu, C.; Hu, Y.; Chen, S.; Liu, W.; Liu, W.; Si, J. Akkermansia muciniphila Alleviates Dextran Sulfate Sodium (DSS)-Induced Acute Colitis by NLRP3 Activation. Microbiol. Spectr. 2021, 9, e0073021. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Lee, Y.J.; Yoo, H.J.; Kim, M.; Chang, Y.; Lee, D.S.; Lee, J.H. Consumption of Dairy Yogurt Containing Lactobacillus paracasei ssp. paracasei, Bifidobacterium animalis ssp. lactis and Heat-Treated Lactobacillus plantarum Improves Immune Function Including Natural Killer Cell Activity. Nutrients 2017, 9, 558. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Vilchez-Padial, L.M.; Gil, A. Evidence of the Anti-Inflammatory Effects of Probiotics and Synbiotics in Intestinal Chronic Diseases. Nutrients 2017, 9, 555. [Google Scholar] [CrossRef] [PubMed]
- Aximujiang, K.; Kaheman, K.; Wushouer, X.; Wu, G.; Ahemaiti, A.; Yunusi, K. Lactobacillus acidophilus and HKL Suspension Alleviates Ulcerative Colitis in Rats by Regulating Gut Microbiota, Suppressing TLR9, and Promoting Metabolism. Front. Pharmacol. 2022, 13, 859628. [Google Scholar] [CrossRef] [PubMed]
- Ahl, D.; Liu, H.; Schreiber, O.; Roos, S.; Phillipson, M.; Holm, L. Lactobacillus reuteri increases mucus thickness and ameliorates dextran sulphate sodium-induced colitis in mice. Acta Physiol. 2016, 217, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ning, Y.; Hu, J.; Wang, Z.; Chen, X.; Zhao, X. The Preventive Effect of Lactobacillus plantarum ZS62 on DSS-Induced IBD by Regulating Oxidative Stress and the Immune Response. Oxid. Med. Cell Longev. 2021, 2021, 9416794. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Zhang, X.; Hao, H.; Liu, Q.; Zhou, Z.; Liang, X.; Liu, T.; Gong, P.; Zhang, L.; Zhai, Z.; et al. Lactobacillus rhamnosus GG Derived Extracellular Vesicles Modulate Gut Microbiota and Attenuate Inflammatory in DSS-Induced Colitis Mice. Nutrients 2021, 13, 3319. [Google Scholar] [CrossRef] [PubMed]
- Cannon, A.R.; Shim, E.H.; Kuprys, P.V.; Choudhry, M.A. IL-22 and Lactobacillus delbrueckii mitigate alcohol-induced exacerbation of DSS-induced colitis. J. Leukoc. Biol. 2022, 112, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Slingerland, A.E.; Schwabkey, Z.; Wiesnoski, D.H.; Jenq, R.R. Clinical Evidence for the Microbiome in Inflammatory Diseases. Front. Immunol. 2017, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.D.; Walker, A.W.; Churcher, C.; Clark, S.K.; Tekkis, P.P.; Johnson, M.W.; Parkhill, J.; Ciclitira, P.J.; Dougan, G.; Nicholls, R.J.; et al. The bacteriology of pouchitis: A molecular phylogenetic analysis using 16S rRNA gene cloning and sequencing. Ann. Surg. 2010, 252, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Cope, J.L.; Nagy-Szakal, D.; Dowd, S.; Versalovic, J.; Hollister, E.B.; Kellermayer, R. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes 2016, 7, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Ando, M.; Kamada, N.; Nagano, Y.; Narushima, S.; Suda, W.; Imaoka, A.; Setoyama, H.; Nagamori, T.; et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 2015, 163, 367–380. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef]
- Rajilic-Stojanovic, M.; Biagi, E.; Heilig, H.G.; Kajander, K.; Kekkonen, R.A.; Tims, S.; de Vos, W.M. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Konikoff, T.; Gophna, U. Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, R.P.D.; Machado, A.; Galvez, J.; Cazarin, C.B.B.; Marostica Junior, M.R. Ulcerative colitis: Gut microbiota, immunopathogenesis and application of natural products in animal models. Life Sci. 2020, 258, 118129. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]




| No. | Ionization Model | Compounds | RT (min) | Relative Content |
|---|---|---|---|---|
| 1 | [M − H]− | 3-O-galloyl-d-glucose | 1.54 | 4.43 × 107 |
| 2 | [M − H]− | 1-O-galloyl-β-d-glucose | 1.6 | 4.52 × 107 |
| 3 | [M − H]− | 6-O-galloyl-β-d-glucose | 1.61 | 4.29 × 107 |
| 4 | [M + H]+ | N-benzylmethylene isomethylamine | 1.92 | 2.70 × 107 |
| 5 | [M + H]+ | N-benzoyl-2-aminoethyl-β-d-glucopyranoside | 1.99 | 3.10 × 107 |
| 6 | [M − H]− | 2,3-di-O-galloyl-β-d-glucose | 2.26 | 5.39 × 107 |
| 7 | [M − H]− | Methyl gallate | 3.17 | 3.23 × 107 |
| 8 | [M + H]+ | Quercetin-3-O-(2″-O-arabinosyl) rutinoside | 3.43 | 3.08 × 107 |
| 9 | [M + H]+ | Isotamarixin | 3.54 | 2.83 × 107 |
| 10 | [M + H]+ | Kaempferol-3-O-glucorhamnoside | 3.92 | 2.30 × 107 |
| 11 | [M − H]− | Gallic acid ethyl ester | 3.94 | 4.92 × 107 |
| 12 | [M + H]+ | Kaempferol-3-O-sambubioside | 3.96 | 4.81 × 107 |
| 13 | [M + H]+ | Isorhoifolin | 3.96 | 2.93 × 107 |
| 14 | [M + H]+ | Rhoifolin | 4.12 | 2.65 × 107 |
| 15 | [M + H]+ | Aurantiamide acetate | 7.62 | 2.84 × 107 |
| Gene | Name | Degree |
|---|---|---|
| PTGS2 | Prostaglandin endoperoxide synthase 2 | 13 |
| PLG | Plasminogen | 9 |
| PPAR-γ | Peroxisome proliferator-activated receptor gamma | 6 |
| F2 | Coagulation factor II, thrombin | 6 |
| GPR35 | G protein-coupled receptor 35 | 5 |
| ABCB1 | ATP-binding cassette subfamily B member 1 | 3 |
| MPO | Myeloperoxidase | 3 |
| SYK | Spleen-associated tyrosine kinase | 3 |
| IL2 | Interleukin-2 | 3 |
| AKT1 | Akt serine/threonine kinase 1 | 3 |
| MET | Met proto-oncogene, receptor tyrosine kinase | 3 |
| MMP9 | Matrix metallopeptidase 9 | 3 |
| CCR1 | C-C motif chemokine receptor 1 | 3 |
| MMP3 | Matrix metallopeptidase 3 | 3 |
| TNF | Tumor necrosis factor | 2 |
| RELA | Rela proto-oncogene, NF-kB subunit | 2 |
| CHEK2 | Checkpoint kinase 2 | 2 |
| TLR9 | Toll-like receptor 9 | 2 |
| Gene | Forward Primers | Reverse Primers |
|---|---|---|
| GAPDH | GGTGAAGGTCGGTGTGAACG | CTCGCTCCTGGAAGATGGTG |
| TNF-α | AATGGCCTCCCTCTCATCAGTTCT | TGAGATAGCAAATCGGCTGACGGT |
| IL-6 | AATTAAGCCTCCGACTTGTGAAG | CTTCCATCCAGTTGCCTTCTTG |
| iNOS | CCCGTCCACAGTATGTGAGGAT | CATTACCTAGAGCCGCCAGTGA |
| Time (min) | Solvent A (%) | Solvent B (%) |
|---|---|---|
| 0 | 95 | 5 |
| 9 | 5 | 95 |
| 10 | 5 | 95 |
| 11 | 95 | 5 |
| 14 | 95 | 5 |
| Gene | Forward Primers | Reverse Primers |
|---|---|---|
| Ptgs2 | TGCACTATGGTTACAAAAGCTGG | TCAGGAAGCTCCTTATTTCCCTT |
| PLG | TCCCAATGAGGGACTAGAAGAG | CGGATCTGTAGTGTAGCACCA |
| PPAR-γ | GCCCTTTGGTGACTTTATGGA | GCAGCAAGGTTGTCTTGGATG |
| F2 | TCCGGTAGAACCAGGCTTTTC | GGGCAAACCAATCACAAACAC |
| GPR35 | GGTCACCTCCCTGGTAGTG | AGCGGCAGGTAGAATCCCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, W.; Yu, S.; Liu, X.; Le, Q.; He, X.; Yu, L.; He, J.; Yang, L.; Gao, H. Ethanol Extract of Limonium bicolor Improves Dextran Sulfate Sodium-Induced Ulcerative Colitis by Alleviating Inflammation and Restoring Gut Microbiota Dysbiosis in Mice. Mar. Drugs 2024, 22, 175. https://doi.org/10.3390/md22040175
Jia W, Yu S, Liu X, Le Q, He X, Yu L, He J, Yang L, Gao H. Ethanol Extract of Limonium bicolor Improves Dextran Sulfate Sodium-Induced Ulcerative Colitis by Alleviating Inflammation and Restoring Gut Microbiota Dysbiosis in Mice. Marine Drugs. 2024; 22(4):175. https://doi.org/10.3390/md22040175
Chicago/Turabian StyleJia, Wei, Siyu Yu, Xi Liu, Qingqing Le, Xiwen He, Lutao Yu, Jianlin He, Longhe Yang, and Huiyuan Gao. 2024. "Ethanol Extract of Limonium bicolor Improves Dextran Sulfate Sodium-Induced Ulcerative Colitis by Alleviating Inflammation and Restoring Gut Microbiota Dysbiosis in Mice" Marine Drugs 22, no. 4: 175. https://doi.org/10.3390/md22040175
APA StyleJia, W., Yu, S., Liu, X., Le, Q., He, X., Yu, L., He, J., Yang, L., & Gao, H. (2024). Ethanol Extract of Limonium bicolor Improves Dextran Sulfate Sodium-Induced Ulcerative Colitis by Alleviating Inflammation and Restoring Gut Microbiota Dysbiosis in Mice. Marine Drugs, 22(4), 175. https://doi.org/10.3390/md22040175






