αO-Conotoxin GeXIVA[1,2] Reduced Neuropathic Pain and Changed Gene Expression in Chronic Oxaliplatin-Induced Neuropathy Mice Model
Abstract
1. Introduction
2. Results
2.1. GeXIVA[1,2] Treatment Alleviated Oxaliplatin-Induced Mechanical Allodynia
2.2. Chronic GeXIVA[1,2] Injection Did Not Affect Mice Behaviors in OFT and ELPM
2.3. Overview of RNA-Seq Data
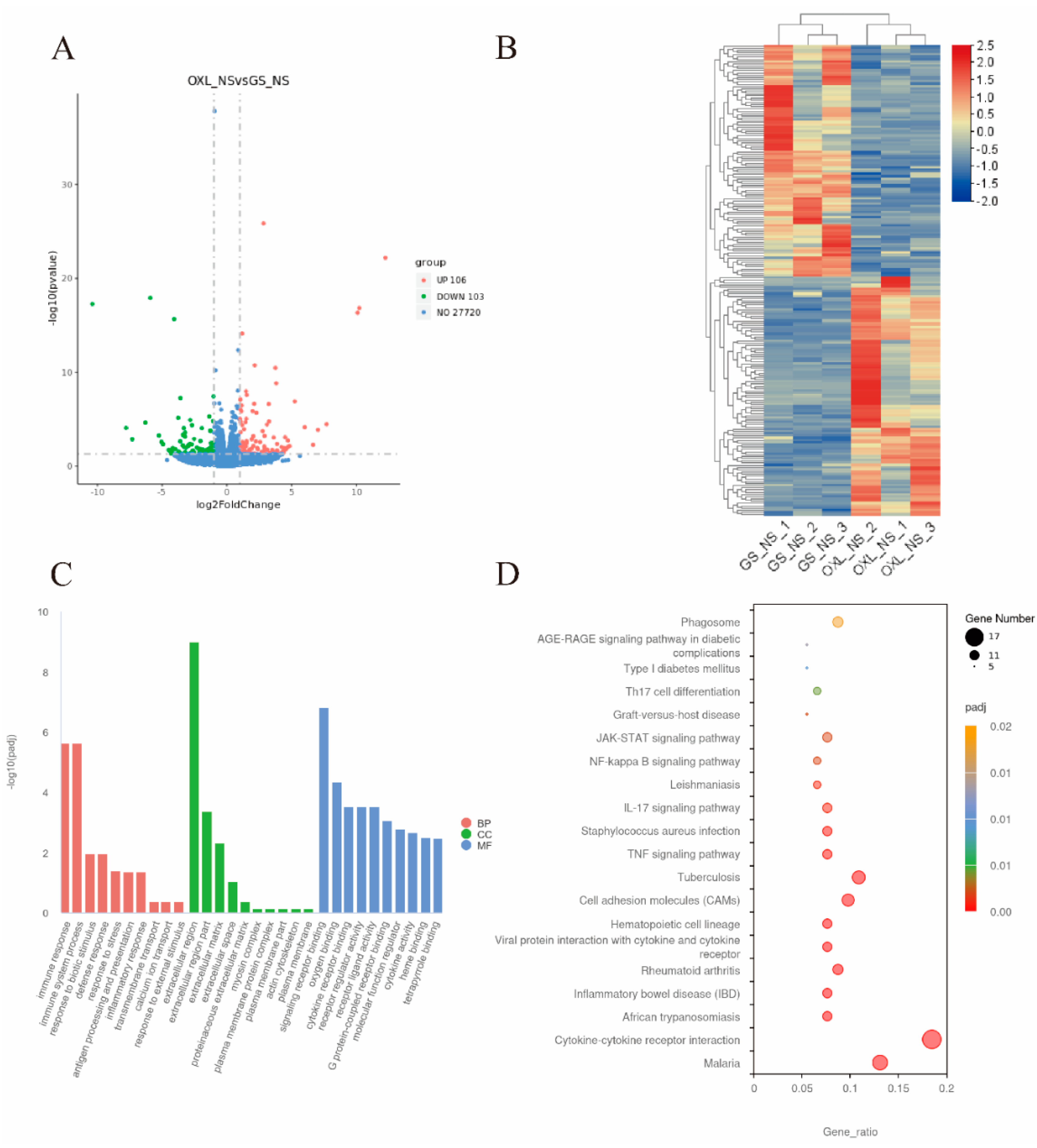
2.4. Differential Gene Expression in the Spinal Cord of CIPN Mice
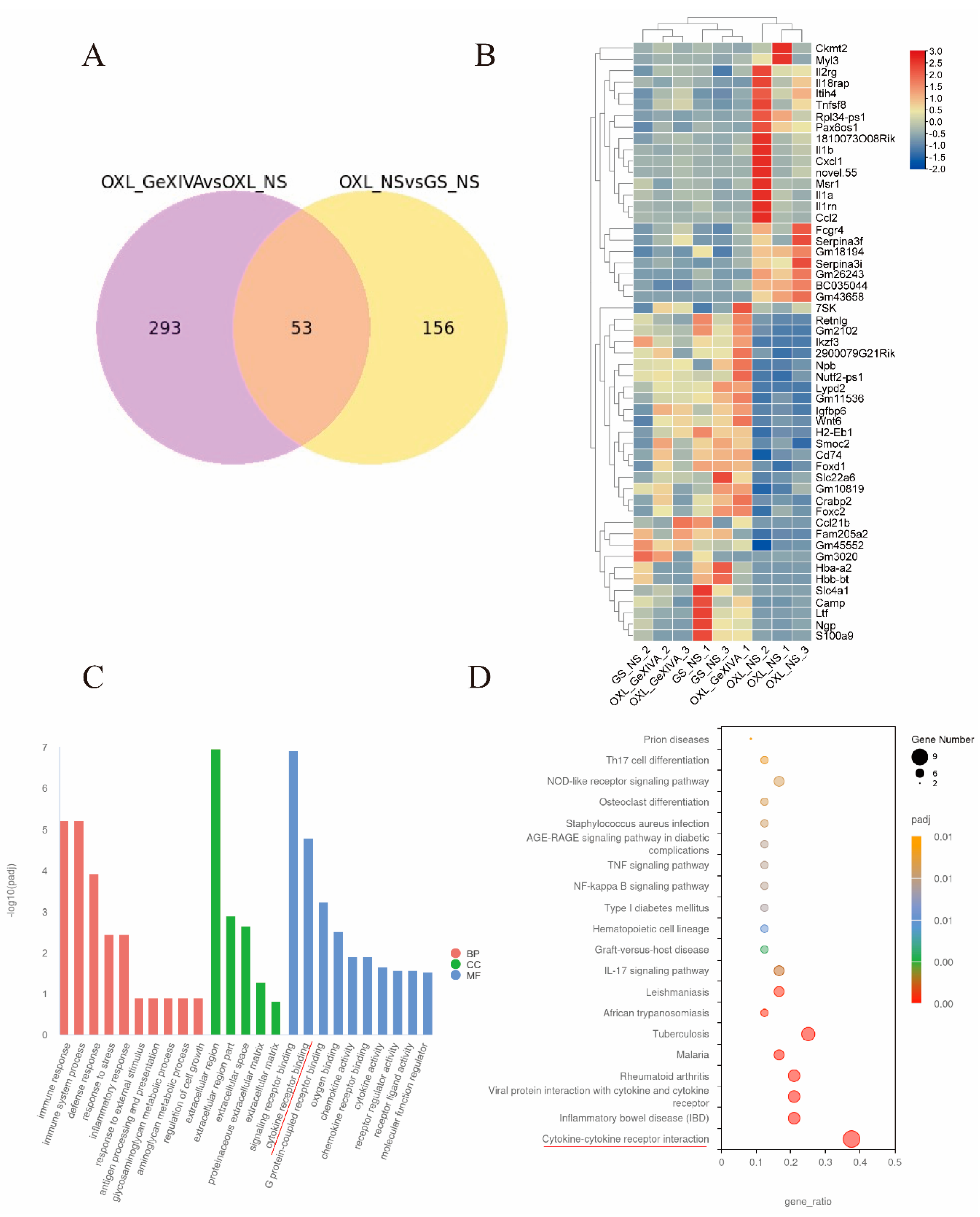
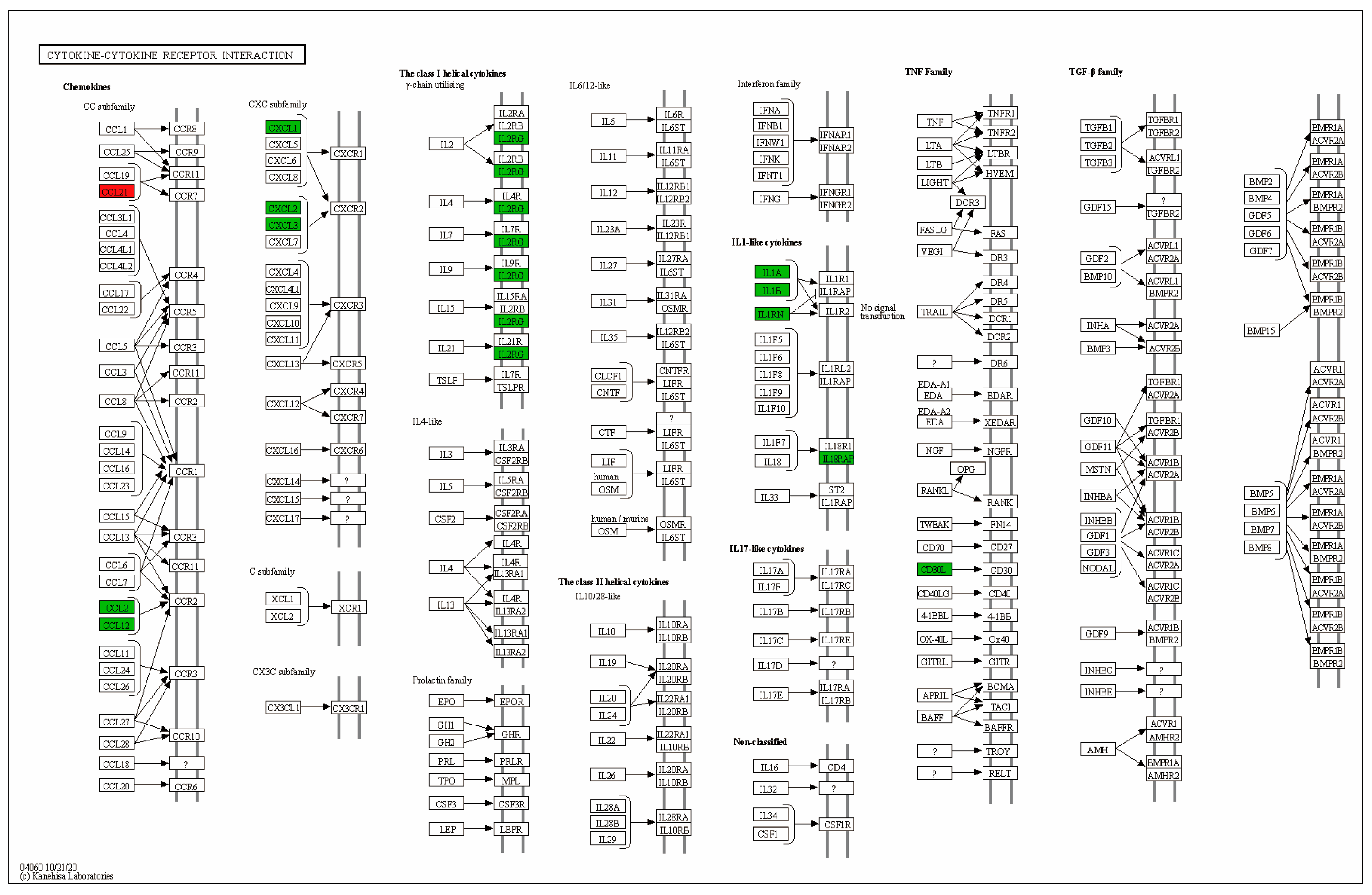
2.5. GeXIVA[1,2] Treatment Altered DEGs Identified in CIPN Model
2.6. GeXIVA[1,2] Changed Gene Expression in Spinal Cord of Normal Mice
2.7. qRT-PCR Validation
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Peptide Synthesis
4.3. Oxaliplatin Treatment and Experimental Schedule
4.4. Behavioral Tests
4.4.1. Mechanical Allodynia Testing
4.4.2. Open Field Test
4.4.3. Elevated Plus Maze Test
4.5. RNA Sequencing and qRT-PCR
4.6. Bioinformatic Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Chamberlain, M.C. Neurotoxicity of cancer treatment. Curr. Oncol. Rep. 2010, 12, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Brozou, V.; Vadalouca, A.; Zis, P. Pain in Platin-Induced Neuropathies: A Systematic Review and Meta-Analysis. Pain Ther. 2018, 7, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Navarro, M.; Maldonado, R.; Baños, J.E. Why mu-opioid agonists have less analgesic efficacy in neuropathic pain? Eur. J. Pain 2019, 23, 435–454. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Dougherty, P.M.; Abdi, S. Basic science and clinical management of painful and non-painful chemotherapy-related neuropathy. Gynecol. Oncol. 2015, 136, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Hone, A.J.; Servent, D.; McIntosh, J.M. α9-containing nicotinic acetylcholine receptors and the modulation of pain. Br. J. Pharmacol. 2018, 175, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tae, H.S.; Chu, Y.; Jiang, T.; Adams, D.J.; Yu, R. Medicinal chemistry, pharmacology, and therapeutic potential of α-conotoxins antagonizing the α9α10 nicotinic acetylcholine receptor. Pharmacol. Ther. 2021, 222, 107792. [Google Scholar] [CrossRef] [PubMed]
- Hone, A.J.; McIntosh, J.M. Nicotinic acetylcholine receptors: Therapeutic targets for novel ligands to treat pain and inflammation. Pharmacol. Res. 2023, 190, 106715. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Harvey, P.J.; Kaas, Q.; Wu, Y.; Zhu, X.; Hu, Y.; Li, X.; Tsetlin, V.I.; Christensen, S.; et al. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sci. USA 2015, 112, E4026–E4035. [Google Scholar] [CrossRef]
- Zhangsun, D.; Zhu, X.; Kaas, Q.; Wu, Y.; Craik, D.J.; McIntosh, J.M.; Luo, S. αO-Conotoxin GeXIVA disulfide bond isomers exhibit differential sensitivity for various nicotinic acetylcholine receptors but retain potency and selectivity for the human α9α10 subtype. Neuropharmacology 2017, 127, 243–252. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Zhangsun, D.; Yu, G.; Su, R.; Luo, S. The α9α10 Nicotinic Acetylcholine Receptor Antagonist αO-Conotoxin GeXIVA[1,2] Alleviates and Reverses Chemotherapy-Induced Neuropathic Pain. Mar. Drugs 2019, 17, 265. [Google Scholar] [CrossRef]
- Li, Z.; Han, X.; Hong, X.; Li, X.; Gao, J.; Zhang, H.; Zheng, A. Lyophilization Serves as an Effective Strategy for Drug Development of the α9α10 Nicotinic Acetylcholine Receptor Antagonist α-Conotoxin GeXIVA[1,2]. Mar. Drugs 2021, 19, 121. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, G.; Monza, L.; Cavaletti, G.; Rigolio, R.; Meregalli, C. Neuroinflammatory Process Involved in Different Preclinical Models of Chemotherapy-Induced Peripheral Neuropathy. Front. Immunol. 2020, 11, 626687. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.B.; Hone, A.J.; Roux, I.; Kniazeff, J.; Pin, J.P.; Upert, G.; Servent, D.; Glowatzki, E.; McIntosh, J.M. RgIA4 Potently Blocks Mouse α9α10 nAChRs and Provides Long Lasting Protection against Oxaliplatin-Induced Cold Allodynia. Front. Cell. Neurosci. 2017, 11, 219. [Google Scholar] [CrossRef]
- Dyachenko, I.A.; Palikova, Y.A.; Palikov, V.A.; Korolkova, Y.V.; Kazakov, V.A.; Egorova, N.S.; Garifulina, A.I.; Utkin, Y.N.; Tsetlin, V.I.; Kryukova, E.V. α-Conotoxin RgIA and oligoarginine R8 in the mice model alleviate long-term oxaliplatin induced neuropathy. Biochimie 2022, 194, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Huynh, P.N.; Christensen, S.B.; McIntosh, J.M. RgIA4 Prevention of Acute Oxaliplatin-Induced Cold Allodynia Requires α9-Containing Nicotinic Acetylcholine Receptors and CD3(+) T-Cells. Cells 2022, 11, 3561. [Google Scholar] [CrossRef]
- Pacini, A.; Micheli, L.; Maresca, M.; Branca, J.J.; McIntosh, J.M.; Ghelardini, C.; Di Cesare Mannelli, L. The α9α10 nicotinic receptor antagonist α-conotoxin RgIA prevents neuropathic pain induced by oxaliplatin treatment. Exp. Neurol. 2016, 282, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Romero, H.K.; Christensen, S.B.; Di Cesare Mannelli, L.; Gajewiak, J.; Ramachandra, R.; Elmslie, K.S.; Vetter, D.E.; Ghelardini, C.; Iadonato, S.P.; Mercado, J.L.; et al. Inhibition of α9α10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci. USA 2017, 114, E1825–E1832. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y.; Wu, Y.; Huang, Y.; Yu, S.; Ding, Q.; Zhangsun, D.; Luo, S. Anti-hypersensitive effect of intramuscular administration of αO-conotoxin GeXIVA[1,2] and GeXIVA[1,4] in rats of neuropathic pain. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2016, 66, 112–119. [Google Scholar] [CrossRef]
- Mohammadi, S.A.; Burton, T.J.; Christie, M.J. α9-nAChR knockout mice exhibit dysregulation of stress responses, affect and reward-related behaviour. Behav. Brain Res. 2017, 328, 105–114. [Google Scholar] [CrossRef]
- Chang, H.H.V.; Morley, B.J.; Cullen, K.E. Loss of α-9 Nicotinic Acetylcholine Receptor Subunit Predominantly Results in Impaired Postural Stability Rather Than Gaze Stability. Front. Cell. Neurosci. 2021, 15, 799752. [Google Scholar] [CrossRef]
- Wala, E.P.; Crooks, P.A.; McIntosh, J.M.; Holtman, J.R., Jr. Novel small molecule α9α10 nicotinic receptor antagonist prevents and reverses chemotherapy-evoked neuropathic pain in rats. Anesth. Analg. 2012, 115, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.R.; Dougherty, P.M. Spinal astrocyte gap junction and glutamate transporter expression contributes to a rat model of bortezomib-induced peripheral neuropathy. Neuroscience 2015, 285, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.; Chen, Z.; Muscoli, C.; Bryant, L.; Esposito, E.; Cuzzocrea, S.; Dagostino, C.; Ryerse, J.; Rausaria, S.; Kamadulski, A.; et al. Targeting the overproduction of peroxynitrite for the prevention and reversal of paclitaxel-induced neuropathic pain. J. Neurosci. Off. J. Soc. Neurosci. 2012, 32, 6149–6160. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Li, Y.; Liu, X.; Gong, D.; Zheng, W. Notch activation enhances microglial CX3CR1/P38 MAPK pathway in rats model of vincristine-induced peripheral neuropathy. Neurosci. Lett. 2020, 715, 134624. [Google Scholar] [CrossRef] [PubMed]
- Navia-Pelaez, J.M.; Choi, S.H.; Dos Santos Aggum Capettini, L.; Xia, Y.; Gonen, A.; Agatisa-Boyle, C.; Delay, L.; Gonçalves Dos Santos, G.; Catroli, G.F.; Kim, J.; et al. Normalization of cholesterol metabolism in spinal microglia alleviates neuropathic pain. J. Exp. Med. 2021, 218, e20202059. [Google Scholar] [CrossRef] [PubMed]
- Brandolini, L.; d’Angelo, M.; Antonosante, A.; Allegretti, M.; Cimini, A. Chemokine Signaling in Chemotherapy-Induced Neuropathic Pain. Int. J. Mol. Sci. 2019, 20, 2904. [Google Scholar] [CrossRef]
- Vichaya, E.G.; Chiu, G.S.; Krukowski, K.; Lacourt, T.E.; Kavelaars, A.; Dantzer, R.; Heijnen, C.J.; Walker, A.K. Mechanisms of chemotherapy-induced behavioral toxicities. Front. Neurosci. 2015, 9, 131. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, W. The Role of Satellite Glial Cells, Astrocytes, and Microglia in Oxaliplatin-Induced Neuropathic Pain. Biomedicines 2020, 8, 324. [Google Scholar] [CrossRef]
- Stephens, K.E.; Chen, Z.; Sivanesan, E.; Raja, S.N.; Linderoth, B.; Taverna, S.D.; Guan, Y. RNA-seq of spinal cord from nerve-injured rats after spinal cord stimulation. Mol. Pain 2018, 14, 1744806918817429. [Google Scholar] [CrossRef]
- Echeverria-Villalobos, M.; Tortorici, V.; Brito, B.E.; Ryskamp, D.; Uribe, A.; Weaver, T. The role of neuroinflammation in the transition of acute to chronic pain and the opioid-induced hyperalgesia and tolerance. Front. Pharmacol. 2023, 14, 1297931. [Google Scholar] [CrossRef]
- Grau, V.; Richter, K.; Hone, A.J.; McIntosh, J.M. Conopeptides [V11L;V16D]ArIB and RgIA4: Powerful Tools for the Identification of Novel Nicotinic Acetylcholine Receptors in Monocytes. Front. Pharmacol. 2018, 9, 1499. [Google Scholar] [CrossRef] [PubMed]
- Tsetlin, V.; Haufe, Y.; Safronova, V.; Serov, D.; Shadamarshan, P.; Son, L.; Shelukhina, I.; Kudryavtsev, D.; Kryukova, E.; Kasheverov, I.; et al. Interaction of α9α10 Nicotinic Receptors With Peptides and Proteins From Animal Venoms. Front. Cell. Neurosci. 2021, 15, 765541. [Google Scholar] [CrossRef] [PubMed]
- AlSharari, S.D.; Toma, W.; Mahmood, H.M.; Michael McIntosh, J.; Imad Damaj, M. The α9α10 nicotinic acetylcholine receptors antagonist α-conotoxin RgIA reverses colitis signs in murine dextran sodium sulfate model. Eur. J. Pharmacol. 2020, 883, 173320. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, M.; Whiteaker, P.; Shi, F.D.; Morley, B.J.; Lukas, R.J. Attenuation in Nicotinic Acetylcholine Receptor α9 and α10 Subunit Double Knock-Out Mice of Experimental Autoimmune Encephalomyelitis. Biomolecules 2019, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lei, B.; Yuan, Y.; Zhang, L.; Hu, L.; Jin, S.; Kang, B.; Liao, X.; Sun, W.; Xu, F.; et al. Brain control of humoral immune responses amenable to behavioural modulation. Nature 2020, 581, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Herz, S.M.; Stokes, C.; Damaj, M.I.; Grau, V.; Papke, R.L. Pharmacological profiles and anti-inflammatory activity of pCN-diEPP and mCN-diEPP, new α9α10 nicotinic receptor ligands. Neuropharmacology 2023, 240, 109717. [Google Scholar] [CrossRef]
- Zhu, X.; Yuan, M.; Wang, H.; Zhangsun, D.; Yu, G.; Che, J.; Luo, S. Novel αO-conotoxin GeXIVA[1,2] Nonaddictive Analgesic with Pharmacokinetic Modelling-Based Mechanistic Assessment. Pharmaceutics 2022, 14, 1789. [Google Scholar] [CrossRef] [PubMed]
- Milligan, E.D.; Watkins, L.R. Pathological and protective roles of glia in chronic pain. Nat. Rev. Neurosci. 2009, 10, 23–36. [Google Scholar] [CrossRef]
- Di Cesare Mannelli, L.; Pacini, A.; Micheli, L.; Tani, A.; Zanardelli, M.; Ghelardini, C. Glial role in oxaliplatin-induced neuropathic pain. Exp. Neurol. 2014, 261, 22–33. [Google Scholar] [CrossRef]
- Di Cesare Mannelli, L.; Pacini, A.; Bonaccini, L.; Zanardelli, M.; Mello, T.; Ghelardini, C. Morphologic features and glial activation in rat oxaliplatin-dependent neuropathic pain. J. Pain 2013, 14, 1585–1600. [Google Scholar] [CrossRef]
- Di Cesare Mannelli, L.; Cinci, L.; Micheli, L.; Zanardelli, M.; Pacini, A.; McIntosh, J.M.; Ghelardini, C. α-conotoxin RgIA protects against the development of nerve injury-induced chronic pain and prevents both neuronal and glial derangement. Pain 2014, 155, 1986–1995. [Google Scholar] [CrossRef] [PubMed]
- Janes, K.; Wahlman, C.; Little, J.W.; Doyle, T.; Tosh, D.K.; Jacobson, K.A.; Salvemini, D. Spinal neuroimmune activation is independent of T-cell infiltration and attenuated by A3 adenosine receptor agonists in a model of oxaliplatin-induced peripheral neuropathy. Brain Behav. Immun. 2015, 44, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Elgoyhen, A.B. The α9α10 acetylcholine receptor: A non-neuronal nicotinic receptor. Pharmacol. Res. 2023, 190, 106735. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M. Ethical guidelines for investigations of experimental pain in conscious animals. Pain 1983, 16, 109–110. [Google Scholar] [CrossRef] [PubMed]
- Dowell, C.; Olivera, B.M.; Garrett, J.E.; Staheli, S.T.; Watkins, M.; Kuryatov, A.; Yoshikami, D.; Lindstrom, J.M.; McIntosh, J.M. α-conotoxin PIA is selective for α6 subunit-containing nicotinic acetylcholine receptors. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 8445–8452. [Google Scholar] [CrossRef]
- Dixon, W.J. Efficient analysis of experimental observations. Annu. Rev. Pharmacol. Toxicol. 1980, 20, 441–462. [Google Scholar] [CrossRef] [PubMed]
- Deuis, J.R.; Dvorakova, L.S.; Vetter, I. Methods Used to Evaluate Pain Behaviors in Rodents. Front. Mol. Neurosci. 2017, 10, 284. [Google Scholar] [CrossRef]
- Li, X.; Xiong, J.; Zhang, B.; Zhangsun, D.; Luo, S. α-Conotoxin TxIB Inhibits Development of Morphine-Induced Conditioned Place Preference in Mice via Blocking α6β2* Nicotinic Acetylcholine Receptors. Front. Pharmacol. 2021, 12, 772990. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mao, K.; Li, X.; Chen, Z.; Dong, X.; Zhangsun, D.; Zhu, X.; Luo, S. α-Conotoxin TxIB Improved Behavioral Abnormality and Changed Gene Expression in Zebrafish (Danio rerio) Induced by Alcohol Withdrawal. Front. Pharmacol. 2022, 13, 802917. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]




| Sample | Raw_Reads | Clean_Reads | Clean_Bases | Total_Map | Q20 | Q30 |
|---|---|---|---|---|---|---|
| GS_NS_1 | 46716272 | 45655274 | 6.85 G | 44223518 (96.86%) | 98.11 | 94.62 |
| GS_NS_2 | 44480262 | 42952690 | 6.44 G | 41313746 (96.18%) | 98.02 | 94.45 |
| GS_NS_3 | 46572300 | 43424244 | 6.51 G | 41201614 (94.88%) | 97.72 | 93.79 |
| GS_GeXIVA_1 | 44574590 | 42223054 | 6.33 G | 40322767 (95.5%) | 97.83 | 94.1 |
| GS_GeXIVA_2 | 46047054 | 44190706 | 6.63 G | 42161490 (95.41%) | 97.79 | 93.97 |
| GS_GeXIVA_3 | 48388854 | 46555380 | 6.98 G | 44491735 (95.57%) | 98.1 | 94.7 |
| OXL_GeXIVA_1 | 52600066 | 48849060 | 7.33 G | 46288111 (94.76%) | 97.85 | 94.09 |
| OXL_GeXIVA_2 | 47023926 | 44871854 | 6.73 G | 42765248 (95.31%) | 97.65 | 93.65 |
| OXL_GeXIVA_3 | 47806906 | 47702712 | 7.16 G | 45096434 (94.54%) | 97.9 | 94.27 |
| OXL_NS_1 | 45368676 | 43316100 | 6.5 G | 41569301 (95.97%) | 97.37 | 92.89 |
| OXL_NS_2 | 44891890 | 43867380 | 6.58 G | 42222855 (96.25%) | 97.65 | 93.49 |
| OXL_NS_3 | 42498354 | 41571208 | 6.24 G | 39995953 (96.21%) | 97.71 | 93.7 |
| Gene Symbol | Full Name | Forward Primer (5′~3′) | Reverse Primer (5′~3′) |
|---|---|---|---|
| Lcn2 | Lipocalin 2 | TGGCCCTGAGTGTCATGTG | CTCTTGTAGCTCATAGATGGTGC |
| Scgb3a1 | Secretoglobin, family 3A, member 1 | ACCACCACCTTTCTAGTGCTC | GGCTTAATGGTAGGCTAGGCA |
| Ch25h | Cholesterol 25-hydroxylase | TGCTACAACGGTTCGGAGC | AGAAGCCCACGTAAGTGATGAT |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 | TGAGCAACTATTCCAAACCAGC | GCACGTAGTCTTCGATCACTATC |
| Lrg1 | Leucine-rich alpha-2-glycoprotein 1 | TTGGCAGCATCAAGGAAGC | CAGATGGACAGTGTCGGCA |
| Neb | Nebulin | CTCCTGCCGACATGCTGAG | CAGCTCGATGTTCATTGCGTC |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, X.; Qiao, Y.; Wang, M.; Wang, W.; McIntosh, J.M.; Zhangsun, D.; Luo, S. αO-Conotoxin GeXIVA[1,2] Reduced Neuropathic Pain and Changed Gene Expression in Chronic Oxaliplatin-Induced Neuropathy Mice Model. Mar. Drugs 2024, 22, 49. https://doi.org/10.3390/md22010049
Wang H, Li X, Qiao Y, Wang M, Wang W, McIntosh JM, Zhangsun D, Luo S. αO-Conotoxin GeXIVA[1,2] Reduced Neuropathic Pain and Changed Gene Expression in Chronic Oxaliplatin-Induced Neuropathy Mice Model. Marine Drugs. 2024; 22(1):49. https://doi.org/10.3390/md22010049
Chicago/Turabian StyleWang, Huanbai, Xiaodan Li, Yamin Qiao, Meiting Wang, Wen Wang, J. Michael McIntosh, Dongting Zhangsun, and Sulan Luo. 2024. "αO-Conotoxin GeXIVA[1,2] Reduced Neuropathic Pain and Changed Gene Expression in Chronic Oxaliplatin-Induced Neuropathy Mice Model" Marine Drugs 22, no. 1: 49. https://doi.org/10.3390/md22010049
APA StyleWang, H., Li, X., Qiao, Y., Wang, M., Wang, W., McIntosh, J. M., Zhangsun, D., & Luo, S. (2024). αO-Conotoxin GeXIVA[1,2] Reduced Neuropathic Pain and Changed Gene Expression in Chronic Oxaliplatin-Induced Neuropathy Mice Model. Marine Drugs, 22(1), 49. https://doi.org/10.3390/md22010049








