Abstract
Bacillus spp. could be one of the most suitable substitutes for the control and prevention of aquatic diseases. The occurrence of species population, antimicrobial character, and virulence diversity in Bacillus spp. recovered from the mariculture system in China between 2009 and 2021 were investigated, screening for probiotic Bacillus strains with good biological safety that can inhibit Vibrio parahaemolyticus, V. alginolyticus, V. harveyi, V. owensii, V. campbellii. The results showed that 116 Bacillus isolates were divided into 24 species, and the top three species were B. subtilis (37/116), B. velezensis (28/116), and B. amyloliquefaciens (10/116). Among the 116 Bacillus isolates, 32.8% were effective against V. parahaemolyticus, 30.1% for V. alginolyticus, 60.3% for V. harveyi, 69.8% for V. owensii and 74.1% for V. campbellii. More than 62% of Bacillus isolates were susceptible to florfenicol, doxycycline and tetracycline, etc., and 26/116 Bacillus isolates were found to be multiple-antibiotic-resistant (MAR), with MARI values ranging from 0 to 0.06. Eighteen kinds of antibiotic resistance genes were tested; only tetB, blaTEM, and blaZ were detected. And 9 isolates in 2 Bacillus species were excluded by 6/10 kinds of Bacillus-related toxin gene (hblA, hblC, nheB, nheC, entFM, cykK). Bio-safety testing indicated that three kinds of probiotics were good probiotic candidates to prevent Vibriosis. These results provide comprehensive genetic diversity, potential risks, and probiotic characteristics of Bacillus in the mariculture system in China, and provide basic support for green and healthy development of aquatic industry.
1. Introduction
Aquatic products play an important role as a component of healthy and indispensable supplements to nutritional diets. The aquaculture output reach to 80 million tons in recent years [1]. However, the occurrence of diseases is gradually becoming a major issue for the aquaculture industry [2]. For instance, approximately 60% of the reduced production in shrimp aquaculture can be attributed to diseases, 20% of which are caused by bacterial diseases [3]. Vibrio spp. is a Gram-negative bacteria commonly found in ocean and estuarine environments. Vibriosis is regarded as the bacterial disease caused by Vibrio spp. in shellfish and finfish aquaculture and is also a main cause of mortality in cultured shrimp throughout the world [4]. Pathogenic Vibrio spp. has been reported to cause foodborne diseases associated with seafood consumption. Meanwhile, Vibriosis in shrimps has been reported by many researchers, and at least 14 Vibrio species have been deemed responsible, including V. parahaemolyticus, V. alginolyticus, V. harveyi, V. anguillarum, V. splendidus, V. mimicus, V. damsella, V. vulnificus, V. fischeri, V. cambellii, V. ordalli, V. mediterrani, V. orientalis and V. logei [5,6,7].
The beneficial microbial strains referred to as probiotics are known to be “live microorganisms, when administered in adequate amounts, confer a health benefit on the host” [8]. Probiotics hold potential for use as effective drugs against aquaculture pathogens. Bacillus spp. is a commonly used probiotics in aquaculture. The application of Bacillus spp. could provide the most suitable substitute for the control and prevention of animal diseases. The majority of Bacillus species used as probiotics in aquaculture include B. subtilis, B. licheniformis, B. pumilus, B. amyloliquefaciens, etc. [6]. After Bacillus supplementation, increased resistance has been recorded against V. parahaemolyticus, Edwardsiella tarda [9], Aeromonas salmonicida, Lactococcus garvieae, Streptococcus iniae [10], A. hydrophila [11], Acinetobacter spp. [12]. Furthermore, improved disease resistance through the administration of dietary Bacillus has been reported in various aquatic species, such as rainbow trout [13], tilapia [14], and white shrimp [15].
On the other hand, certain Bacillus may cause aquatic diseases; for example, B. cereus is known as an important foodborne pathogen that can cause distinct forms of illness [16]. The occurrence of bacterial white spot syndrome (BWSS) may be associated with the unconventional use of probiotics containing B. subtilis in shrimp ponds [17]. Not only that, the probiotic bacterial species may present resistant strains and carry the risk of becoming conduits for the spread of toxin genes and antibiotic-resistance genes [18]. Therefore, only antibiotic-resistance and toxin-free candidate species should be considered for inclusion in probiotics [19].
At present, various genetic diversity studies have been conducted regarding the use of Bacillus in ecology, biotechnology, and industry [20,21]. However, poor-quality Bacillus is found in aquaculture, and systematic research on the ecological risk assessment of Bacillus is almost completely lacking. Bacillus leads to the release of pathogenic Bacillus, increasing environmental ecological risks and risks to the health of farmed animals. Based on this, Bacillus spp. isolates were collected from different aquatic environments in China between 2009 and 2021 for this study, and the genetic diversity, antibacterial and antibiotic properties, virulence, and antibiotics resistance genotype of the Bacillus spp. isolates were evaluated. Then, candidate strains with multi-pathogen antagonism and environmental friendliness were used for bio-safety testing. The results will provide a basic theoretical basis for the healthy use of Bacillus in mariculture production and to provide high-quality products for the aquaculture industry. This will promote the healthy, green, and sustainable development of the aquaculture industry.
2. Results
2.1. Bacillus Isolates Information
A total of 116 Bacillus isolates were activated and purified from the mariculture system. The collected Bacillus were identified on the basis of morphological, physiological, and biochemical characterizations as well as 16S rDNA and recA sequencing. Sequencing analysis classified 116 Bacillus isolates into 24 different species (Table 1), including B. subtilis (37/116), B. velezensis (28/116), B. amyloliquefaciens(10/116), B. stercoris (6/116), B. cereus (6/116), B. thuringiensis (3/116), B. megaterium (3/116), B. flexus (3/116), B. nealsonii (2/116), B. altitudinis (2/116), B. aryabhattai (2/116), B. atrophaeus (2/116), B. tequilensis (1/116), B. inaquosorum (1/116), B. stratosphericus (1/116), B. koreensis (1/116), B. lehensis (1/116), B. gibsonii (1/116), B. methylotrophicus (1/116), B. licheniformis (1/116), B. pumilus (1/116), B. haikouensis (1/116), B. circulans (1/116), B. marisflavi (1/116).

Table 1.
Type and quantity of Bacillus isolates.
Strains were photographed for colony morphology characteristics, and different morphotypes were discovered in 24 Bacillus species. Six Bacillus species showed that the colony was obtained in dried spore form with irregular edges, including B. subtilis, B. velezensis, B. amyloliquefaciens, B. tequilensis, B. haikouensis. Eighteen Bacillus species showed a moist and smooth morphology with regular edges, including B.stercoris, B. cereus, B. thuringiensis, B. megaterium, B. flexus, B. nealsonii, B. altitudinis, B. aryabhattai, B. atrophaeus, B.inaquosorum, B. stratosphericus, B. koreensis, B. lehensis, B. gibsonii, B. methylotrophicus, B. pumilus, B. circulans, B. marisflavi. Single-colony photographs of 24 Bacillus species are shown in Figure S1.
2.2. Antibacterial Activity Characterization of Bacillus Isolates against 5 Species of Vibrio
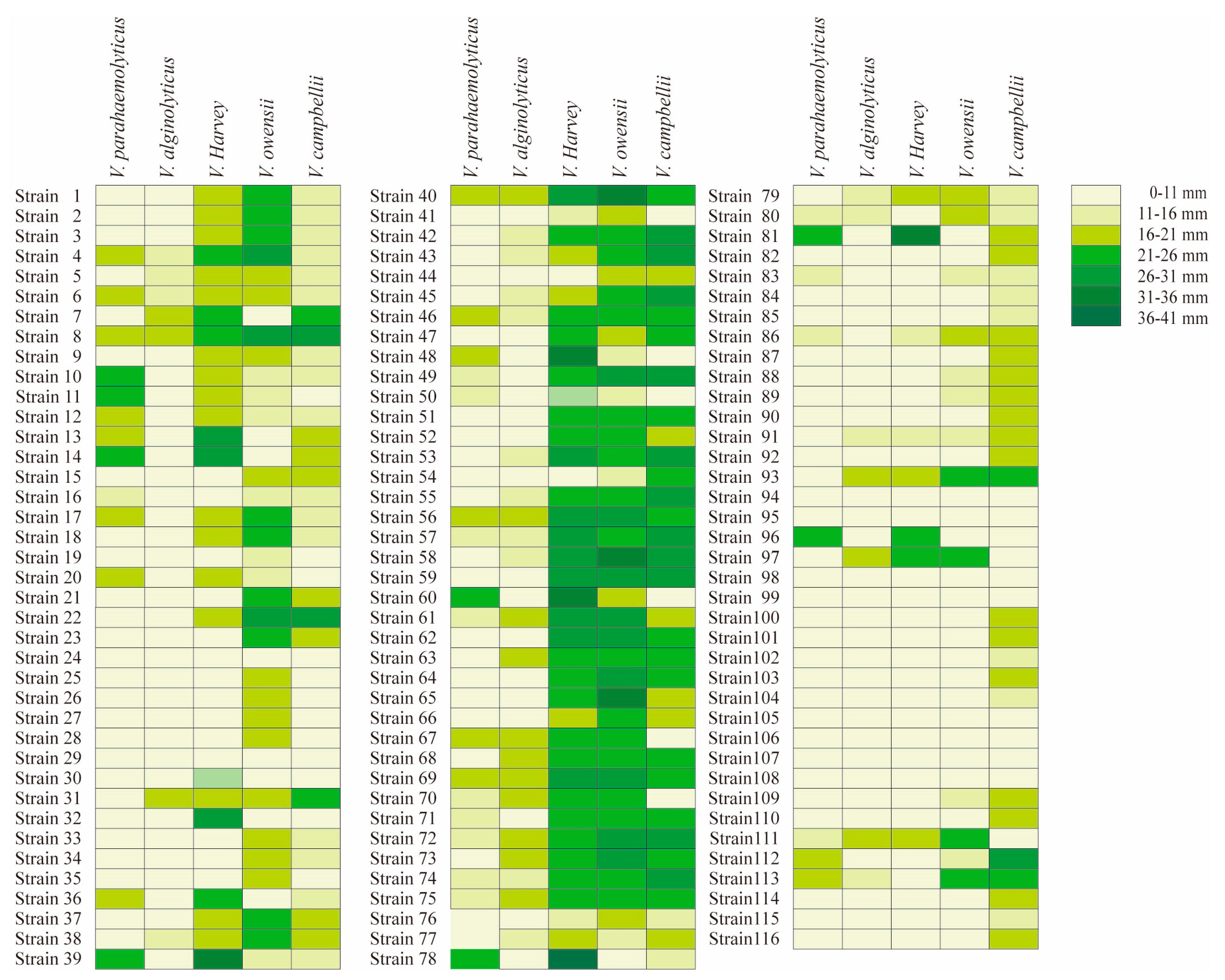
Bacillus isolates were tested on 5 species of Vibrio, and displayed typically different antibacterial ability among Vibrio species (Figure 1). The criterion of antibacterial activity was the presence of an inhibition zone, and the antibacterial ability showed typical differences among Bacillus isolates. Some probiotics had good antibacterial effects, and obvious inhibition zones can be observed. Among the 116 Bacillus isolates, 38 isolates were effective against V. parahaemolyticus, 35 isolates against V. alginolyticus, 70 isolates against V. harveyi, 81 isolates against V. owensii and 86 isolates against V. campbellii (Table 2).
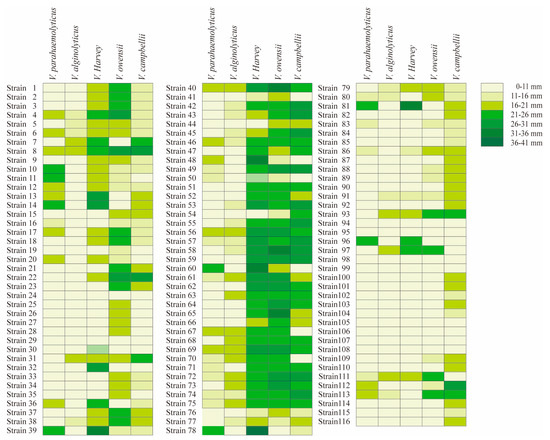
Figure 1.
Inhibition zone of 116 Bacillus isolates under five kinds of Vibrio. The left is the strain number, and the right is the antibacterial diameter character. The colors from light to dark represent 0–11 mm, 11–16 mm, 16–21 mm, 21–26 mm, 26–31 mm, 31–36 mm and 36–41 mm repectively. Strains 1–37 were B. subtilis, strains 38–65 were B. velezens, strains 66–75 were B.amyloliquefaciens, strains 76–81 were B. stercoris, strains 82–87 were B. cereus, strains 88–90 were B. thuringiensis, strain 91 was B. tequilensis, strains 92–93 were B. atrophaeus, strains 94–95 were B.nealsonii, strain 96 was B. inaquosorum, strain 97 was B. stratosphericus, strains 98–100 were B. flexus, strains 101–103 were B. megaterium, strain 104 was B. koreensis, strains 105–106 were B. aryabhattai, strain 107 was B. lehensis, strain 108 was B.gibsonii, strain 109 was B. methylotrophicus, strain 110 was B. licheniformis, strain 111 was B. pumilus, strains 112–113 were B. altitudinis, strain 114 was B. haikouensis, strain 115 was B. circulans, strain 116 was B. marisflavi.

Table 2.
Cluster of the Bacillus isolates that inhibit 5 pathogenic bacteria.
Furthermore, in terms of Vibrio species, the antibacterial ability of Bacillus isolates showed differences among species and isolates. For the 38 isolates that were effective against V. parahaemolyticus, there were 12 B. subtilis isolates, 10 B. velezensis isolates, 7 B. amyloliquefaciens isolates, 3 B. stercoris isolates, 2 B. altitudinis isolates, 2 B. cereus isolates, 1 B. pumilus isolates, and 1 B. inaquosorum isolates. For the 35 isolates that were effective against V. alginolyticus, there were 6 B. subtilis isolates, 13 B. velezensis isolates, 8 B. amyloliquefaciens isolates, 3 B. stercoris isolates, 1 B. tequilensis, B. atrophaeus, B. stratosphericus, B. pumilus, and B. altitudinis isolates.
Among the 70 strains against V. harveyi, there were 23 B. subtilis isolates, 26 B. velezensis isolates, 10 B. amyloliquefaciens isolates, 1 B. cereus, B. tequilensis, B. atrophaeus, B. stratosphericus, B. pumilus and B. inaquosorum isolates. Among the 81 strains that against V. owensii, there were 37 B. subtilis isolates, 28 B. velezensis isolates, 10 B. amyloliquefaciens isolates, 3 B. stercoris isolates, 2 B.cereus and B. altitudinis isolates, 1 B. tequilensis, B. atrophaeus, B. stratosphericus, B. pumilus, B. thuringiensis and B. methylotrophicus isolates. Among the 81 strains that against V. campbellii, there were 37 B. subtilis isolates, 28 B. velezensis isolates, 8 B. amyloliquefaciens isolates, 6 B. stercoris isolates, 6 B. cereus isolates, 3 B. megaterium isolates, 2 B. thuringiensis, B. altitudinis and B. atrophaeus isolates, 1 B. tequilensis, B. flexa, B. haikouensis, B. licheniformis and B. methylotrophicus, B. circulans, B. marisflavi and B. koreensis isolates.
In addition, for the homogeneous Bacillus species, different effects were observed for the same type of Vibrio species. Among the 37 B. subtilis isolates, 25 isolates had no antibacterial effect on V. parahaemolyticus, and the zones of the remaining 12 isolates were 13.2–22 mm; 31 isolates had no antibacterial effect on V. alginolyticus, and the inhibition zone of the remaining 6 isolates was 14–20.9 mm; 14 isolates had no antibacterial effect on V. harveyi, and the inhibition zone of remaining 23 isolates was16.4–27.4 mm; 8 isolates had no antibacterial effect on V. owensii, and the inhibition zone of the remaining 29 isolates was 14.3–29.1 mm; 12 isolates had no antibacterial effect on V. campbellii, and the inhibition zone of the remaining 25 isolates was 14.3–29.6 mm.
For B. velezensis, among the 28 B. velezensis, 18 isolates had no effect on V. parahaemolyticus, and the inhibition zones of the remaining 10 isolates ranged from 13.8 to 21.7 mm; 15 strains had no effect on V. alginolyticus, and the diameter range of remaining 13 isolates ranged from 14.3 to 18.5 mm; for the V. harveyi, only 2 strains had no effective, remining isolates. The maximum diameter could reach 33.8 mm, while 14.7 is the minimum diameter. All 28 isolates were effective on V. owensii, from 11.7 to 33.4 mm; 4 isolates had no effect on V. campbellii, the diameter of the remaining 24 isolates ranged from 14.2 to 29.2 mm.
For B. amyloliquefaciens, among the 10 B. amyloliquefaciens, 3 strains had no effect on V. parahaemolyticus, and the inhibition zones of the remaining 7 isolates ranged from 13 to 19.7 mm; 2 strains had no effect on V. alginolyticus, and the inhibition zone of the remining 8 isolates was 14.7–19.7 mm; 10 strains had effective on V. harveyi, from 20.3–26.8 mm; 10 strains were effective on V. owensii, ranging from 22.2 to 27.9 mm; only 2 strains were effective for V. campbellii. The inhibition zone of the remining eight isolates was 20.3–27.1 mm.
2.3. Antibiotic Susceptibility Characterization of Different Kinds of Bacillus Isolates
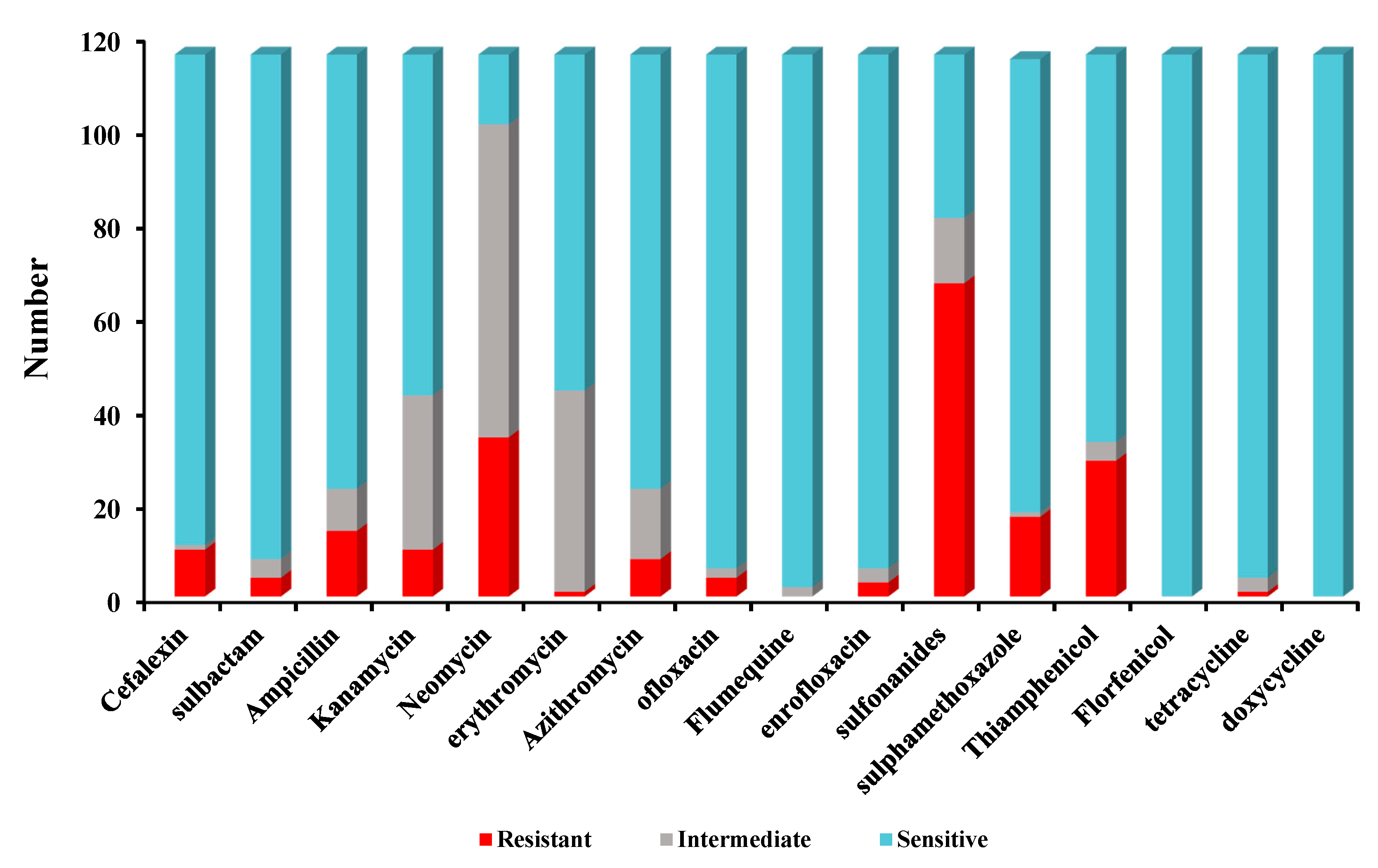
Antimicrobial susceptibility tests indicated that high sulfadiazine resistance (67/116) was observed in 116 isolates. However, high susceptibility was observed to florfenicol (116/116), doxycycline (116/116), sulfamethoxazole-trimethoprim (114/116), tetracycline (112/116), ofloxacin (110/116), enrofloxacin (110/116), sulbactam (108/116), cefalexin (105/116), sulfamethoxazole (97/116), ampicillin (93/116), azithromycin (93/116), thiamphenicol (83/116). Low susceptibility to kanamycin (73/116), erythromycin (72/116), neomycin (15/116) was observed among 116 Bacillus isolates. The antimicrobial susceptibility for 116 Bacillus species against 16 antimicrobial agents are listed in Figure 2.
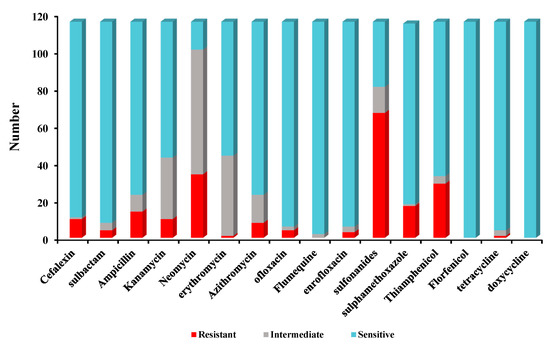
Figure 2.
The antimicrobial susceptibility of 116 Bacillus isolates for 16 antibiotics.
Furthermore, 26/116 Bacillus isolates were found to be multiple antibiotic resistant (MAR) and displaying resistance to at least three of the antibiotics tested in this study. The MARI of all the isolates ranged from 0 to 0.06. None of the tested isolates were resistant to all antibiotics at the same time. Two isolates (strain 7, strain 23) were resistant to seven types of antibiotics. The MAR of isolates resistant to 3 types of antibiotics was the most concentrated, and the proportion of isolates resistant to 4–6 types of drugs was lower (Table 3).

Table 3.
Multiple antibiotic resistance (MAR) index of Bacillus isolates.
2.4. Distribution of Antimicrobial Resistance Genes among Bacillus Isolates
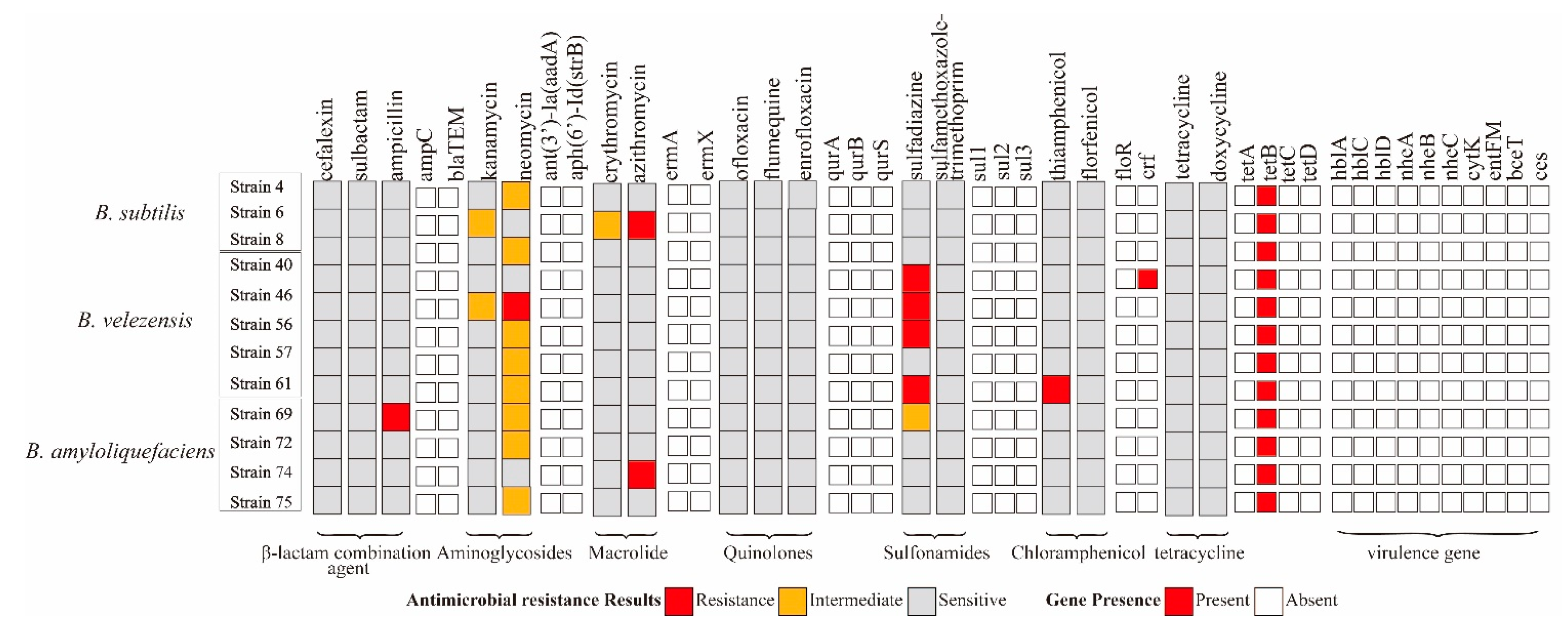
A total of 18 kinds of antimicrobial resistance genes were tested, but only 3 genes were detected (Figure S1), including the tetracycline gene tetB, which was detected in 116 isolates (100%), the chloramphenicol gene cfr, which was detected in 10/116 isolates (8.62%), and the β-lactams gene blaTEM, which was detected in 3/116 isolates (2.59%). The tetracycline resistance genes tetA, tetC, tetD; aminoglycoside resistance genes ant(3′)-Ia(aadA) and aph(6′)-Id(strB); quinolones genes qnrA, qnrB, qnrS, chloramphenicol genes flor and cfr; Sulfonamides genes sul1, sul2, sul3; and macrolides genes ermA, and ermX were not detected in any isolates.
Under 67 sulfonamides resistant isolates, none of the selected sulfonamides antimicrobial resistance genes were identified. All isolates were sensitive to tetracycline, but tetB was detected in 116 isolates. The blaTEM and cfr was detected some isolates that susceptibility to cefalexin, sulbactam, thiamphenicol and florfenicol.
2.5. Distribution of Virulence Genes among Bacillus Isolates
Based on the reported Bacillus-associated virulence genes, 10 kinds of virulence genes were detected in the Bacillus isolates. Only 9 of 116 isolates (7.8%) carried virulence-associated genes, of which 9/116 carried entFM, 7/116 carried hblA and hblC, 6/116 carried nheB, 5/116 carried nheC and cytK. The hblD, nheA, and ces genes were not detected in any strains. The positive, uniformly sized PCR products were sequenced and reconfirmed based on the NCBI database.
In addition, all virulence genes were detected in 2 Bacillus species (6 B. cereus isolates and 3 B. thuringiensis isolates); the entFM genes were found in 9 isolates (Figure S2); the detection rate was 100%; the hblA and hblC were detected in 7 of 9 isolates; the nheB was found in 6/9 isolates; nheC, cytK and bceT were found in 5 of 9 isolates. The largest number of virulence-associated genes belonged to the hblA+hblC+nheC+cytK+entFM+bceT and hblA+hblC+nheB+nheC+cytK+entFM pattern.
2.6. Holistic Assessment of Potential Probiotics
Among the 116 Bacillus isolates, 12 Bacillus isolates were shown to have an inhibitory effect on 5 Vibrio species and can be used as potential probiotics to inhibit Vibriosis (Table 4), including 3 B. subtilis isolates, 5 B. velezensis isolates, and 4 B. amyloliquefaciens isolates. The largest inhibition zone for V. parahaemolyticus was 20.1 mm, and those for V. alginolyticus, V. harveyi, V. owensii, V. campbellii were 19.7 mm, 28.7 mm, 33.4 mm, and 28.7 mm, respectively.

Table 4.
Antibacterial character of 12 potential probiotics Bacillus isolates against 5 Vibrio.
For antibiotic susceptibility characterization, B. subtilis strain 4, 6 was resistant to neomycin and azithromycin, and B. velezens strain 46, 61 was resistant to ampicillin and thiamphenicol. B. velezens strain 40, 46, 50, 61 was resistant to sulfonamides. Only B. subtilis strain 4 carried chloramphenicol gene cfr. The other 11 isolates contain 1 resistance gene tetB but were susceptible to tetracycline. All virulence genes were not detected in these strains (Figure 3).
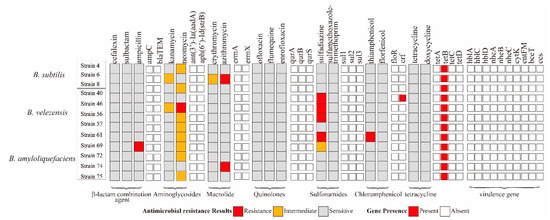
Figure 3.
Gene presence and antibiotic susceptibility characteristics of 12 potential probiotics Bacillus isolates.
2.7. Safety Testing of Potential Probiotics Bacillus Isolates in L. vannamei
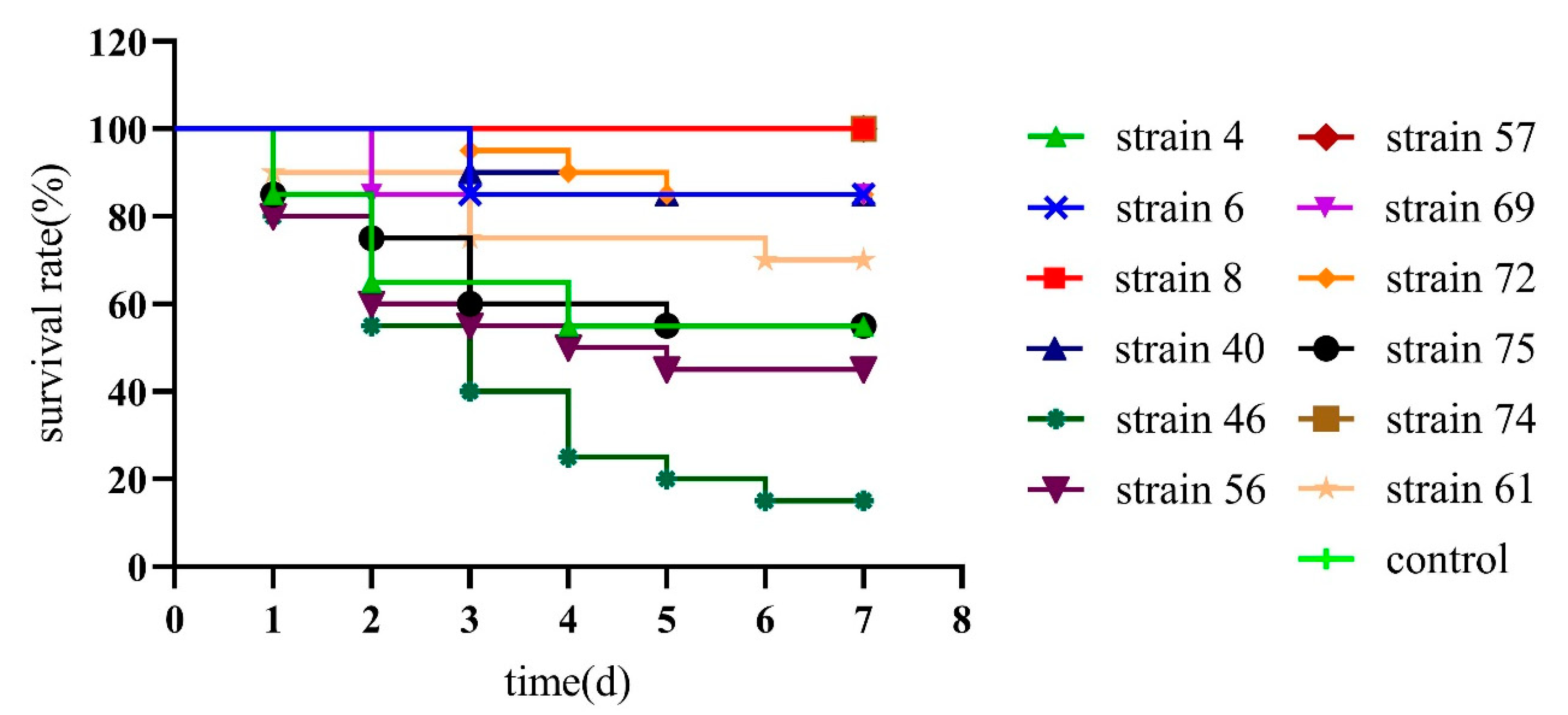
The 12 potential probiotics Bacillus suspension of 108 CFU/mL was injected into the muscle of the second abdominal segment of L. vannamei and observed for 7 days to confirm the safety of the strains. The results showed that B. subtilis strain 4, 6, 40, B. velezens 46, 56, 61, and B. amyloliquefaciens 69, 72, 75 had pathogenic and lethal effects. B. velezens strain 46 mortality rates reached 85%, and the mortality rate of the remaining strains was between 15% and 55%. For the death experimental groups. Some minor mortality groups may be caused by the deterioration of the culture environment, rather than the pathogenicity of the strain. And the absolutely safety Bacillus isolates for L. vannamei were selected, 3 strains with obvious non-lethality were B. subtilis strain 8, B. velezens strain 57, and B. amyloliquefaciens strain 74 were a safe strain without death, and can be applied as a potential probiotic in L. vannamei (Figure 4).
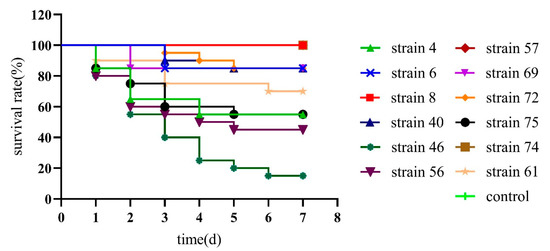
Figure 4.
Survival curve of Litopenaeus vannamei after injected infection with 12 potential probiotics Bacillus isolates.
3. Discussion
Biological antagonism aims to achieve the effect of “treating bacteria with bacteria” under natural conditions. Since Pseudomonas bromoutilis was isolated as antagonistic bacteria for the first time in 1966 [22], more and more antagonistic bacteria have been utilized. Probiotic bacteria isolated from the culture medium might exhibit antagonistic effects on present pathogens. Bacillus isolated from the marine environment can improve disease-resistant activity against bacterial infection [23]. It is a priority and inherently advantageous to use B. pumilus in the marine environment as a biocontrol agent or probiotic in aquaculture [24]. To form the basis of morphological characterizations and gene sequencing, the study reflected the diversity of Bacillus in our aquaculture systems. A total of 116 Bacillus isolates were categorized into 24 different species, including B. subtilis, B. velezensis, B. amyloliquefaciens, and B. stercoris, in this study. At present, Vibriosis has become an economically important disease in marine culture and adversely affected many cultured animals [25]. In the shrimp aquaculture system, V. harveyi, V. alginolyticus and V. parahaemolyticus are most frequently isolated [26]. V. campbellii, V. owensii were also pathogens of shrimp acute hepatopancreatic necrosis disease [27,28]. In this study, different Bacillus isolates had a variety of effects and also showed interspecific differences in pathogen inhibition effects. In addition, for the homogeneous Bacillus species, different effects were shown on the same type of Vibrio isolate. In this regard, the isolation of Bacillus from the mariculture system may have provided these isolates with an advantage in the inhibition of pathogens isolated from the marine environment.
According to the guidelines suggested by the European Food Safety Authority, this is essential for the absence of virulence factors in probiotic strains [29]. The presence of toxin genes in probiotic strains could potentially contribute to the prevalence of toxin genes among bacteria through horizontal transfer in some cases. To ensure the safe use of probiotic Bacillus, a safety evaluation must be carried out, common virulence genes must be screened, and the use of strains carrying virulence genes should be reduced as much as possible. In recent years, studies have been focused on the toxin genes character in probiotic Bacillus spp. Fu et al. [19] found that surveillance of anthrax toxin revealed that cya was detected in 8 of 31 farms. Almost half of the isolates could produce enterotoxins and various cytotoxic surfactin-like toxins in Cui et al. [30,31] report, and they summarized the state of current knowledge about the toxins of B, cereus sensu lato to be considered for safety assessment of probiotic candidates. Among the 116 Bacillus spp. isolates, very few isolates detected the presence of virulence genes, and 9 isolates of Bacillus can be excluded by virulence genes’ testing. Five isolates (4.3%) were tested as positive for hemolytic cytotoxin K (cytK), which is responsible for severe food poisoning. None of the species were positive for the emetic toxin gene ces, and the prevalence of nheABC and hblCDA was shown to be rare in the comparison [30,32]. Hemolytic activity assays are considered to be an important screening process. Hemolysin is a very common virulence factor, which frequently causes anemia and edema in the host; hence, hemolytic strains are not recommended for use as feed additives. The non-hemolytic strains would be preferable for probiotic use [33]. Generally, nheABC and hblCDA genes were widely distributed among B. cereus isolates from various origins, except for probiotic origin [30]. Deng et al. [34] suggested that nheABC and hblCDA were gradually prevalent in probiotic Bacillus spp. isolates. The current observations suggest that hblC and nheA were detected in B. cereus and B. thuringiensis isolates. No virulence genes were detected in 12 potential probiotics isolates.
Antibiotic resistance is one of the imminent challenges to global health with the current trend of escalating environmental contaminants; furthermore, the natural environment is a potential repository for the spread of antibiotic resistance [35]. Antibiotic susceptibility is one of the most important features of probiotic bacteria [36]. Reacher Yaylaci et al. [37] reported that B. pumilus was susceptible to furazolidone, erythromycin, ampicillin, oxytetracycline, rifampicin, and ciprofoxacin. Sorokulova et al. [38] previously reported that Bacillus spp. may be resistant to chloramphenicol. Our study is not similar in terms of results: 116 strains of Bacillus isolates showed low resistance to antibiotics, and more than 62% isolates were susceptible to florfenicol, doxycycline, sulfamethoxazole-trimethoprim, tetracycline, ofloxacin, enrofloxacin, sulbactam, cefalexin, sulfamethoxazole, ampicillin, azithromycin, thiamphenicol, kanamycin and erythromycin. The MARI reflects the degree of the environmental pollution caused by antibiotics that may be dangerous to human health [39]. A value of higher than 0.2 indicates a high antibiotic exposure risk, while a value lower than 0.2 indicates a low antibiotic exposure risk [40]. The MARI values of the 116 Bacillus isolates ranged from 0 to 0.06, which indicates a low antibiotic exposure risk.
Furthermore, the antibiotic resistance burden has serious implications for human health owing to the potential transfer of antibiotic resistance genes between bacteria, thereby impairing the efficacy of antibiotic treatment and compromising public health [41]. A total of 18 kinds of antibiotic resistance genes were tested, and only tetB, blaTEM, and blaZ were detected in this study. The previous studies indicated that the majority of Bacillus spp. for probiotics exhibited resistance to tetracycline, which mainly resulted in mobile tetracycline-resistance genes such as tetB and tet45 [19,31,32]. In this study, 112 Bacillus isolates showed an antibiotics phenotype that was sensitive to tetracycline, but the tetB detection rate was 100%. Many researchers have observed discrepancies between antimicrobial resistance phenotypes and the prevalence of antimicrobial resistance genes [42,43]. This might be due to the antimicrobial phenotypes that can be expressed upon the stimulation of many different genetic determinants [44]. The macrolide-resistance genes present on extrachromosomal elements have been identified in mobile elements, such as the plasmid-encoded ermC from B. subtilis [45]. Tetracycline-resistance determinants have also been found in mobile elements, including the plasmid-encoded tetL gene from B. subtilis [46], and the tetM contained within the conjugative transposon Tn5397 of B. subtilis [47]. Other tetracycline resistance genes, such as tetK, have been observed in some Bacillus isolates [48]. The detection of chloramphenicol gene cfr (8.62%) and β-lactams gene blaTEM (2.59%) was low. Chloramphenicol resistance can be attributed to the enzymatic inhibition of the drug facilitated by chloramphenicol acetyl-transferases [49]. Beta lactams are the most broadly used class of antimicrobial agents, characterized by minimal toxicity and employed in the treatment of various bacterial ailments, including those attributed with different Vibrio species [50]. Apparently, resistance among the beta-lactam and chloramphenicol drugs was low.
4. Materials and Methods
4.1. Bacterial Strains and Growth Condition
A total of 116 Bacillus isolates were collected from a mariculture system in China with geographic and chronological differences between 2009 and 2021. All samples were cultured on tryptone soybean broth (TSB) agar medium at 28 °C for 36 h. Single colonies were picked and inoculated on TSB medium to obtain purified bacteria. Then single colony was picked up with a tip under sterile conditions, transferred to 100 μL ultrapure water and repeatedly pipetted and, after 12 min in a metal bath at 99 °C, centrifuged for 5 min at 4 °C and 12,000 rpm with a high-speed centrifuge, the supernatant was taken. The DNA concentration was determined in the supernatant using a spectrophotometer (Nano Drop 1000) as template DNA in PCR. The PCR system consists of 1 μL of DNA template, 25 μL of Taq Master Mix (Vazyme, Nanjing, China), 1 μL of forward primers and 1 μL of reverse primers, and double-distilled water for a total volume of 50 μL. PCR amplification conditions were: 94 °C, 4 min; 30 cycles: 94 °C, 30 s, 55 °C, 30 s, 72 °C, 1.5 min; store at 72 °C, 10 min, 4 °C. PCR products were sequenced after detection by 1% agarose gel electrophoresis. Primer synthesis and gene sequencing were completed by Sangon Biotech (Shanghai, China). Colonies were preliminarily identified by morphology and 16S rDNA sequencing. BLAST was used for nucleotide comparison to obtain percentage similarity. The sequence analysis was carried out based on recA sequence to more accurately identify the dominant bacteria, as described by Mohkam.
Five common pathogenic Vibrio spp. of Litopenaeus vannamei, including V. parahaemolyticus, V. alginolyticus, V. harvey, V. owensii, and V. campbellii, were tested to observe the antibacterial effect of Bacillus isolates. These strains were activated and cultured at 28 °C for 24 h in TSB for activation.
4.2. Antibacterial Ability of Bacillus Isolates against Vibrio spp.
Antibacterial activity was assessed using the agar well diffusion assay method. The Bacillus and Vibrio isolates were cultured in TSB media at 28 °C, 180 rpm for 24 h. Then, the bacteria suspension was adjusted to 1 × 109 CFU/mL for Bacillus, 1 × 106 CFU/mL for Vibrio. 100 μL Vibrio suspension was smeared on the surface of TSB medium and 100 μL of Bacillus suspension was added into the hole and cultured at 28 °C. After 36 h, checks were carried out for the appearance of inhibition haloes surrounding the putative antagonists’ spots and the diameter of the inhibition zone was measured with Scan 1200 inhibition zone reader function (Interscience, Saint-Nom-la-Bretèche, France) to screen the beneficial strains with good antibacterial effect. All the experiments were performed in triplicate.
4.3. Antibiotic Susceptibility Test
The antimicrobial resistance of 116 Bacillus isolates was determined using the Kirby–Bauer disk diffusion method on TSB agar medium according to the guidelines by the National Committee for Clinical Laboratory Standards with minor changes [51]. A total of 16 kinds of antibiotics were used in this study, including thiamphenicol, erythromycin, kanamycin, ofloxacin, florfenicol, azithromycin, cefalexin, flumequine, sulfamethoxazole–trimethoprim, enrofloxacin, tetracycline, sulfadiazine, neomycin, sulbactam, ampicillin, and doxycycline. Escherichia coli ATCC 25922 was used for quality control. The plates were incubated at 28 °C for 24 h. All the experiments were performed in triplicate. Then, the antibiotic susceptibility was reported as resistant (R), intermediate (I) and sensitive (S) based on the inhibitory zone diameter according to the CLSI standards.
Furthermore, the isolates were considered multiple-antibiotic resistant (MAR) if the isolate was resistant to three or more separate antimicrobial classes [52]. The MAR and multiple antibiotic resistance index (MARI) were measured for all isolates according to the CLSI protocol [53].
4.4. Antimicrobial Resistance Genes Detection among Bacillus Isolates
18 antimicrobial resistance genes belong to 7 categories were studied using PCR amplification, including tetracycline resistance (tetA, tetB, tetE), extended-spectrum β-lactamase (ESBL) (blaTEM, ampC, blaZ), aminoglycoside resistance [ant(3)-Ia(aadA), aph(6′)-Ib(strB)], sulphonamide resistance (sul1, sul2 and sul3), macrolides (ermA, ermX), and phenicols (floR, cfr), as well as quinolones (qnrA, qnrB, qnrS) resistance genes. Table 1 describes the primers and protocols used. Each assay was composed of 1 μL of DNA material, 12.5 μL of Taq Master Mix (Vazyme, China), 1 μL of forward and 1 μL of reverse primers, and double-distilled water to obtain a total volume of 25 μL. The PCR product was analyzed by electrophoresis using 1% (w/v) agarose gel and then visualized under UV transilluminator. The positive, uniformly sized PCR products were sequenced and reconfirmed based on the NCBI database.
4.5. Molecular Detection of Potential Virulence Gene among Bacillus Isolates
Based on Fu et al. [54], the presence of 10 kinds of Bacillus-related toxin genes was detected in 116 Bacillus isolates, including hblA, hblC, hblD, nheA, nheB, nheC, cytK, entFM, bceT and ces. The primers and the expected size of the DNA product for each of the investigated genes are shown in Table 5.

Table 5.
PCR primers used to this study.
The primers were synthesized by Sangon Biotech (Qingdao, China). The DNA of the purified bacteria was extracted by the Beijing Tiangen biological bacterial genome DNA extraction kit. Extracted DNA material served as a template for PCR amplification for each tested sample. Each assay was composed of 1 μL of DNA material, 12.5 μL of Taq Master Mix (Vazyme, China), 1 μL of forward and 1 μL of reverse primers, and double-distilled water to obtain a total of 25 μL. The PCR product was analyzed by electrophoresis using a 1% (w/v) agarose gel and then visualized under a UV transilluminator. The positive bands’ sequence information was determined and compared to verify the accuracy of the sequence information.
4.6. Bio-Safety Testing in Litopenaeus vannamei
To evaluate the in vivo security of 12 potential probiotics Bacillus isolates, L. vannamei was used in this study. Bacillus were cultured in TSB and incubated with shaking for 36 h at 28 °C. Shrimp (5–8 cm) were randomly divided into 13 groups (12 groups for Bacillus infection and 1 group for control), with each group containing 20 shrimp. The shrimp were injected with 1.0 × 108 CFU/mL Bacillus suspension, and PBS was injected as control. Then, mortality was monitored and recorded for 7 days.
4.7. Statistical Analysis
Data were preliminary analyzed by excel 2016 software, and all statistical analyses were carried out using SPSS software (version 26.0). The results are expressed as mean ± SE of mean (SEM) with One-way ANOVA at 5%. The colony photographs were adjusted with Adobe Illustrator 2019 to best represent the colony morphology. Survival curves were made by GraphPad.
5. Conclusions
Our study provides comprehensive diversity and shows the potential risks of the Bacillus species collected from geographically distinct regions in the mariculture system in China for the first time. The phenotype and genetic diversity of different kinds of Bacillus spp. were investigated. An integral evaluation of the virulence and antimicrobial resistance risk of Bacillus isolates was carried out. Then, the potential probiotics were screened with multiple antibacterial activities for different Vibrio species and evaluated the probiotic character. Finally, three potential probiotic strains were found. These results provide basic support for microecological preparations in aquatic disease preservation and promote the healthy and green development of aquatic industry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21040228/s1, Figure S1: Morphological diversity of representative Bacillus species. Figure S2: Antimicrobial resistance genes gel electrophoresis.; Figure S3: Virulence genes gel electrophoresis.
Author Contributions
Conceptualization, Y.W. and Y.Y.; methodology, Y.Y., Z.Z. and Y.Z.; software, J.G. and B.L.; validation, C.W. and Z.Z.; formal analysis, Y.Z. and C.W.; investigation, Y.Z., M.L. and Y.Y.; resources, Y.Y. and Z.Z.; data curation, Y.Z. and J.W.; writing—original draft preparation, Y.Y. and Y.Z.; writing—review and editing, M.L. and X.R.; visualization, X.R. and Y.Y.; supervision, Y.W. and Z.Z.; project administration, Y.W. and Z.Z.; funding acquisition, Y.W. and Z.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Key Research and Development Program of Shandong Province (2021SFGC0701), Key technology research and industrialization demonstration project of Qingdao city (23-1-3-hygg-22-hy, 22-3-3-hygg-3-hy), and Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD40).
Institutional Review Board Statement
All animal experiments described in the present study were conducted at the Key Laboratory of Maricultural Organism Disease Control (Qingdao, China) in accordance with the recommendations of Directive YSFRI-2022006 on the protection of animals used for scientific purposes.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Food and Agriculture Organization. The State of World Fisheries and Aquaculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Abarike, E.D.; Jian, J.; Tang, J.; Cai, J.; Yu, H.; Lihua, C.; Jun, L. Influence of traditional Chinese medicine and Bacillus species (TCMBS) on growth, immune response and disease resistance in Nile tilapia, Oreochromis niloticus. Aquac. Res. 2018, 49, 2366–2375. [Google Scholar] [CrossRef]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Panda, S.K.; Luyten, W. Anti-vibrio and immune-enhancing activity of medicinal plants in shrimp: A comprehensive review. Fish Shellfish Immunol. 2021, 117, 192–210. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Akagawa-Matsushita, M.; Muroga, K.; Microbiology, E. Vibrio penaeicida sp. nov., a pathogen of kuruma prawns (Penaeus japonicus). Int. J. Syst. Bacteriol. 1995, 45, 134–138. [Google Scholar] [CrossRef]
- Yi, Y.; Zhang, Z.; Zhao, F.; Liu, H.; Yu, L.; Zha, J.; Wang, G. Probiotic potential of Bacillus velezensis JW: Antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol. 2018, 78, 322–330. [Google Scholar] [CrossRef]
- Gao, Y.; Han, G.; Qiang, L.; Zhang, L.; Tan, R.; Yu, Y. Hematological varieties, histological changes, and immune responses in the early stage of infection with Vibrio parahaemolyticus in Black rockfish Sebastes schlegelii. Aquacult Int. 2023, 31, 381–399. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Santos, R.A.; Monteiro, M.; Rangel, F.; Jerusik, R.; Saavedra, M.J.; Carvalho, A.P.; Oliva-Teles, A.; Serra, C.R. Bacillus spp. Inhibit Edwardsiella tarda Quorum-Sensing and Fish Infection. Mar. Drugs 2021, 19, 602. [Google Scholar] [CrossRef]
- Cha, J.-H.; Rahimnejad, S.; Yang, S.-Y.; Kim, K.-W.; Lee, K.-J. Evaluations of Bacillus spp. as dietary additives on growth performance, innate immunity and disease resistance of olive flounder (Paralichthys olivaceus) against Streptococcus iniae and as water additives. Aquaculture 2013, 402, 50–57. [Google Scholar] [CrossRef]
- Ramesh, D.; Souissi, S. Effects of potential probiotic Bacillus subtilis KADR1 and its subcellular components on immune responses and disease resistance in Labeo rohita. Aquac. Res. 2018, 49, 367–377. [Google Scholar] [CrossRef]
- Kavitha, M.; Raja, M.; Perumal, P. Evaluation of probiotic potential of Bacillus spp. isolated from the digestive tract of freshwater fish Labeo calbasu (Hamilton, 1822). Aquac. Rep. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Newaj-Fyzul, A.; Adesiyun, A.A.; Mutani, A.; Ramsubhag, A.; Brunt, J.; Austin, B. Bacillus subtilis AB1 controls Aeromonas infection in rainbow trout (Oncorhynchus mykiss, Walbaum). J. Appl. Microbiol. 2007, 103, 1699–1706. [Google Scholar] [CrossRef]
- Aly, S.M.; Abdel-Galil Ahmed, Y.; Abdel-Aziz Ghareeb, A.; Mohamed, M.F. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 2008, 25, 128–136. [Google Scholar] [CrossRef]
- Tseng, D.Y.; Ho, P.L.; Huang, S.Y.; Cheng, S.C.; Shiu, Y.L.; Chiu, C.S.; Liu, C.H. Enhancement of immunity and disease resistance in the white shrimp, Litopenaeus vannamei, by the probiotic, Bacillus subtilis E20. Fish Shellfish Immunol. 2009, 26, 339–344. [Google Scholar] [CrossRef]
- Stenfors Arnesen, L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef]
- Wang, Y.G.; Lee, K.L.; Najiah, M.; Shariff, M.; Hassan, M.D. A new bacterial white spot syndrome (BWSS) in cultured tiger shrimp Penaeus monodon and its comparison with white spot syndrome (WSS) caused by virus. Dis. Aquat. Org. 2000, 41, 9–18. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.; G de los Reyes-Gavilán, C.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef]
- Fu, S.; Yang, Q.; He, F.; Lan, R.; Hao, J.; Ni, P.; Liu, Y.; Li, R. National Safety Survey of Animal-use Commercial Probiotics and Their Spillover Effects From Farm to Humans: An Emerging Threat to Public Health. Clin. Infect. Dis. 2020, 70, 2386–2395. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, A.; Pratush, A. Molecular diversity and functional variability of environmental isolates of Bacillus species. Springerplus 2014, 3, 312. [Google Scholar] [CrossRef]
- Kanamoto, E.; Terashima, K.; Shiraki, Y.; Nishida, H. Diversity of Bacillus Isolates from the Sake Brewing Process at a Sake Brewery. Microorganisms 2021, 9, 1760. [Google Scholar] [CrossRef]
- Burkholder, P.R.; Pfister, R.M.; Leitz, F.H. Production of a pyrrole antibiotic by a marine bacterium. Appl. Microbiol. 1966, 14, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xia, Y.; Zhu, C.; Chu, W. Isolation of Marine Bacillus sp. with Antagonistic and Organic-Substances-Degrading Activities and Its Potential Application as a Fish Probiotic. Mar. Drugs 2018, 16, 196. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Y.; Liu, Y.; Miao, L.L.; Li, E.W.; Hou, T.T.; Liu, Z.P. Mechanism of anti-Vibrio activity of marine probiotic strain Bacillus pumilus H2, and characterization of the active substance. AMB Express 2017, 7, 23. [Google Scholar] [CrossRef]
- Uzun, E.; Ogut, H. The isolation frequency of bacterial pathogens from sea bass (Dicentrarchus labrax) in the Southeastern Black Sea. Aquaculture 2015, 437, 30–37. [Google Scholar] [CrossRef]
- Zhou, J.; Fang, W.; Yang, X.; Zhou, S.; Hu, L.; Li, X.; Qi, X.; Su, H.; Xie, L. A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease” of Litopenaeus vannamei shrimp. PLoS ONE 2012, 7, e29961. [Google Scholar] [CrossRef]
- Dong, X.; Wang, H.; Xie, G.; Zou, P.; Guo, C.; Liang, Y.; Huang, J. An isolate of Vibrio campbellii carrying the pir(VP) gene causes acute hepatopancreatic necrosis disease. Emerg. Microbes Infect. 2017, 6, e2. [Google Scholar] [CrossRef]
- Liu, L.Y.; Xiao, J.Z.; Zhang, M.M.; Zhu, W.Y.; Xia, X.M.; Dai, X.L.; Pan, Y.J.; Yan, S.L.; Wang, Y.J. A Vibrio owensii strain as the causative agent of AHPND in cultured shrimp, Litopenaeus vannamei. J. Invertebr. Pathol. 2018, 153, 156–164. [Google Scholar] [CrossRef]
- Azimonti, G.; Bampidis, V.; Bastos MD, L.; Christensen, H.; Dusemund, B.; Fašmon Durjava, M.; Kouba, M.; López-Alonso, M.; López Puente, S.; Marcon, F.; et al. EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). EFSA J. Eur. Food Saf. Auth. 2018, 20, e07268. [Google Scholar]
- Cui, Y.; Martlbauer, E.; Dietrich, R.; Luo, H.; Ding, S.; Zhu, K. Multifaceted toxin profile, an approach toward a better understanding of probiotic Bacillus cereus. Crit. Rev. Toxicol. 2019, 49, 342–356. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, S.; Ding, S.; Shen, J.; Zhu, K. Toxins and mobile antimicrobial resistance genes in Bacillus probiotics constitute a potential risk for One Health. J. Hazard. Mater. 2020, 382, 121266. [Google Scholar] [CrossRef]
- Zhu, K.; Holzel, C.S.; Cui, Y.; Mayer, R.; Wang, Y.; Dietrich, R.; Didier, A.; Bassitta, R.; Martlbauer, E.; Ding, S. Probiotic Bacillus cereus Strains, a Potential Risk for Public Health in China. Front. Microbiol. 2016, 7, 718. [Google Scholar] [CrossRef]
- Nandi, A.; Dan, S.K.; Banerjee, G.; Ghosh, P.; Ghosh, K.; Ringo, E.; Ray, A.K. Probiotic Potential of Autochthonous Bacteria Isolated from the Gastrointestinal Tract of Four Freshwater Teleosts. Probiotics Antimicro. 2017, 9, 12–21. [Google Scholar] [CrossRef]
- Deng, F.; Chen, Y.; Sun, T.; Wu, Y.; Su, Y.; Liu, C.; Zhou, J.; Deng, Y.; Wen, J. Antimicrobial resistance, virulence characteristics and genotypes of Bacillus spp. from probiotic products of diverse origins. Food Res. Int. 2021, 139, 109949. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martinez, J.L. Antibiotic Resistance: Moving From Individual Health Norms to Social Norms in One Health and Global Health. Front. Microbiol. 2020, 11, 1914. [Google Scholar] [CrossRef]
- Patel, A.K.; Ahire, J.J.; Pawar, S.P.; Chaudhari, B.L.; Chincholkar, S.B. Comparative accounts of probiotic characteristics of Bacillus spp. isolated from food wastes. Food Res. Int. 2009, 42, 505–510. [Google Scholar] [CrossRef]
- Yaylaci, E.U. Isolation and characterization of Bacillus spp. from aquaculture cage water and its inhibitory effect against selected Vibrio spp. Arch. Microbiol. 2021, 204, 26. [Google Scholar] [CrossRef]
- Sorokulova, I.B.; Pinchuk, I.V.; Denayrolles, M.; Osipova, I.G.; Huang, J.M.; Cutting, S.M.; Urdaci, M.C. The safety of two Bacillus probiotic strains for human use. Dig. Dis. Sci. 2008, 53, 954–963. [Google Scholar] [CrossRef]
- Pall, E.; Niculae, M.; Brudasca, G.F.; Ravilov, R.K.; Sandru, C.D.; Cerbu, C.; Olah, D.; Zablau, S.; Potarniche, A.V.; Spinu, M.; et al. Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania. Antibiotics 2021, 10, 333. [Google Scholar] [CrossRef]
- Mohamad, N.; Amal, M.N.A.; Saad, M.Z.; Yasin, I.S.M.; Zulkiply, N.A.; Mustafa, M.; Nasruddin, N.S. Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet. Res. 2019, 15, 176. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.G.; Singer, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- Dang, H.; Zhang, X.; Song, L.; Chang, Y.; Yang, G. Molecular characterizations of oxytetracycline resistant bacteria and their resistance genes from mariculture waters of China. Mar. Pollut. Bull. 2006, 52, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Yu, Y.; Liao, M.; Wang, Y.; Yang, G.; Zhang, Z.; Li, B.; Rong, X.; Wang, C. Physiology, metabolism, antibiotic resistance, and genetic diversity of Harveyi clade bacteria isolated from coastal mariculture system in China in the last two decades. Front. Mar. Sci. 2022, 9. [Google Scholar] [CrossRef]
- Van, C.N.; Zhang, L.J.; Thanh, T.V.T.; Son, H.P.H.; Ngoc, T.T.; Huang, Q.; Zhou, R. Association between the Phenotypes and Genotypes of Antimicrobial Resistance in Haemophilus parasuis Isolates from Swine in Quang Binh and Thua Thien Hue Provinces, Vietnam. Engineering 2020, 6, 40–48. [Google Scholar] [CrossRef]
- Monod, M.; Denoya, C.; Dubnau, D. Sequence and Properties of pIM13, a Macrolide-Lincosamide-Streptogramin B Resistance Plasmid from Bacillus subtilis. J. Bacteriol. 1986, 167, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Phelan, R.W.; Clarke, C.; Morrissey, J.P.; Dobson, A.D.; O’Gara, F.; Barbosa, T.M. Tetracycline resistance-encoding plasmid from Bacillus sp. strain #24, isolated from the marine sponge Haliclona simulans. Appl. Environ. Microbiol. 2011, 77, 327–329. [Google Scholar] [CrossRef]
- Roberts, A.P.; Pratten, J.; Wilson, M.; Mullany, P. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol. Lett. 1999, 177, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Neela, F.A.; Nonaka, L.; Rahman, M.H.; Suzuki, S. Transfer of the chromosomally encoded tetracycline resistance gene tet(M) from marine bacteria to Escherichia coli and Enterococcus faecalis. World J. Microbiol. Biotechnol. 2009, 25, 1095–1101. [Google Scholar] [CrossRef]
- Alvarez-Contreras, A.K.; Quinones-Ramirez, E.I.; Vazquez-Salinas, C. Prevalence, detection of virulence genes and antimicrobial susceptibility of pathogen Vibrio species isolated from different types of seafood samples at “La Nueva Viga” market in Mexico City. Antonie Leeuwenhoek 2021, 114, 1417–1429. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. beta-Lactams and beta-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Wayne, PA, USA, 2018; Volume M100. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Chon, J.W.; Kim, J.H.; Lee, S.J.; Hyeon, J.Y.; Song, K.Y.; Park, C.; Seo, K.H. Prevalence, phenotypic traits and molecular characterization of emetic toxin-producing Bacillus cereus strains isolated from human stools in Korea. J. Appl. Microbiol. 2012, 112, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Heini, N.; Stephan, R.; Ehling-Schulz, M.; Johler, S. Characterization of Bacillus cereus group isolates from powdered food products. Int. J. Food Microbiol. 2018, 283, 59–64. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Vukov, N.; Schulz, A.; Shaheen, R.; Andersson, M.; Martlbauer, E.; Scherer, S. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 2005, 71, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Sung, K.; Khan, S.A.; Khan, A.A.; Steele, R. Biochemical and molecular characterization of tetracycline-resistant Aeromonas veronii isolates from catfish. Appl. Environ. Microbiol. 2006, 72, 6461–6466. [Google Scholar] [CrossRef] [PubMed]
- Furlaneto-Maia, L.; Rocha, K.R.; Siqueira, V.L.; Furlaneto, M.C. Comparison between automated system and PCR-based method for identification and antimicrobial susceptibility profile of clinical Enterococcus spp. Rev. Inst. Med. Trop. São Paulo 2014, 56, 97–103. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef]
- Sandvang, D.; Aarestrup, F.M.; Jensen, L.B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. Microbiol. Lett. 1997, 157, 177–181. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Wu, X.; Huo, S. Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Poult. Sci. 2015, 94, 601–611. [Google Scholar] [CrossRef]
- Bochniarz, M.; Adaszek, L.; Dziegiel, B.; Nowaczek, A.; Wawron, W.; Dabrowski, R.; Szczubial, M.; Winiarczyk, S. Factors responsible for subclinical mastitis in cows caused by Staphylococcus chromogenes and its susceptibility to antibiotics based on bap, fnbA, eno, mecA, tetK, and ermA genes. J. Dairy Sci. 2016, 99, 9514–9520. [Google Scholar] [CrossRef]
- Letchumanan, V.; Yin, W.F.; Lee, L.H.; Chan, K.G. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 2015, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Gxalo, O.; Digban, T.O.; Igere, B.E.; Olapade, O.A.; Okoh, A.I.; Nwodo, U.U. Virulence and Antibiotic Resistance Characteristics of Vibrio Isolates From Rustic Environmental Freshwaters. Front. Cell. Infect. Microbiol. 2021, 11, 732001. [Google Scholar] [CrossRef] [PubMed]
- Mohkam, M.; Nezafat, N.; Berenjian, A.; Mobasher, M.A.; Ghasemi, Y. Identification of Bacillus Probiotics Isolated from Soil Rhizosphere Using 16S rRNA, recA, rpoB Gene Sequencing and RAPD-PCR. Probiotics Antimicrob. Proteins 2016, 8, 8–18. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).