Secondary Metabolites with Antifungal Activities from Mangrove Derived Fungus Monascus purpureus WMD2424
Abstract
1. Introduction
2. Results
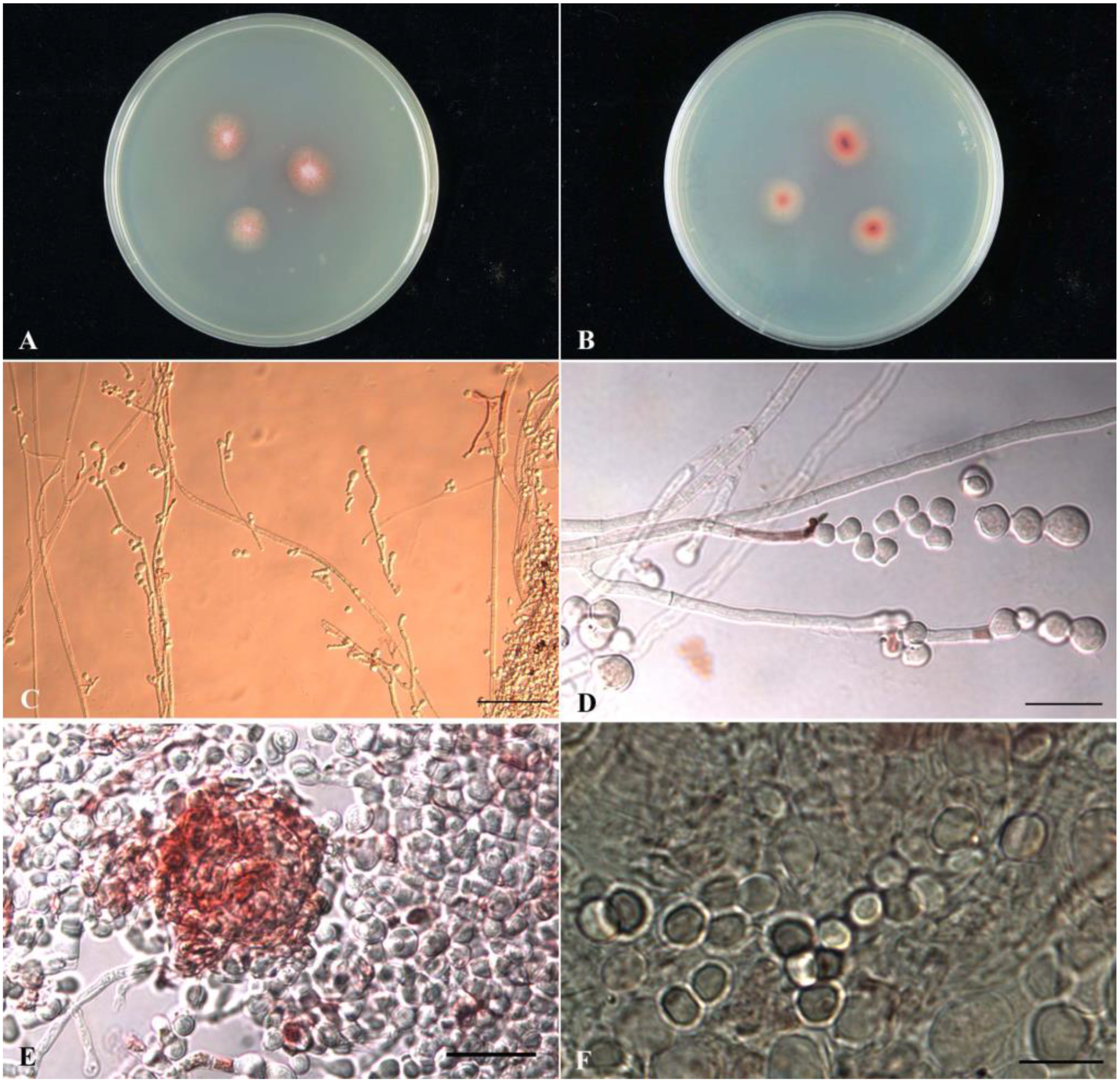
2.1. Taxonomic Identification (Phenotypic and Genotypic Data) of Monascus purpureus wmd2424
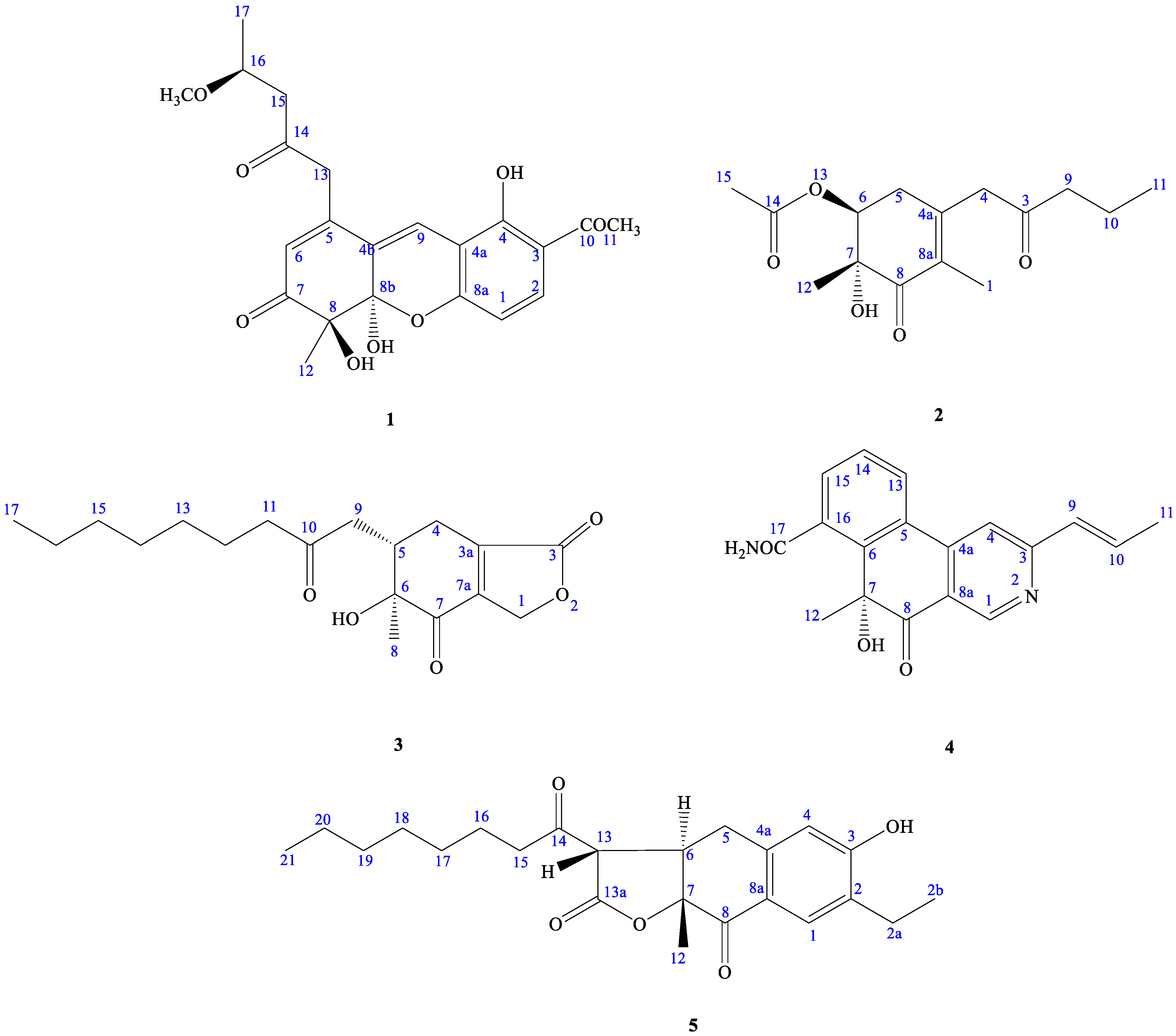
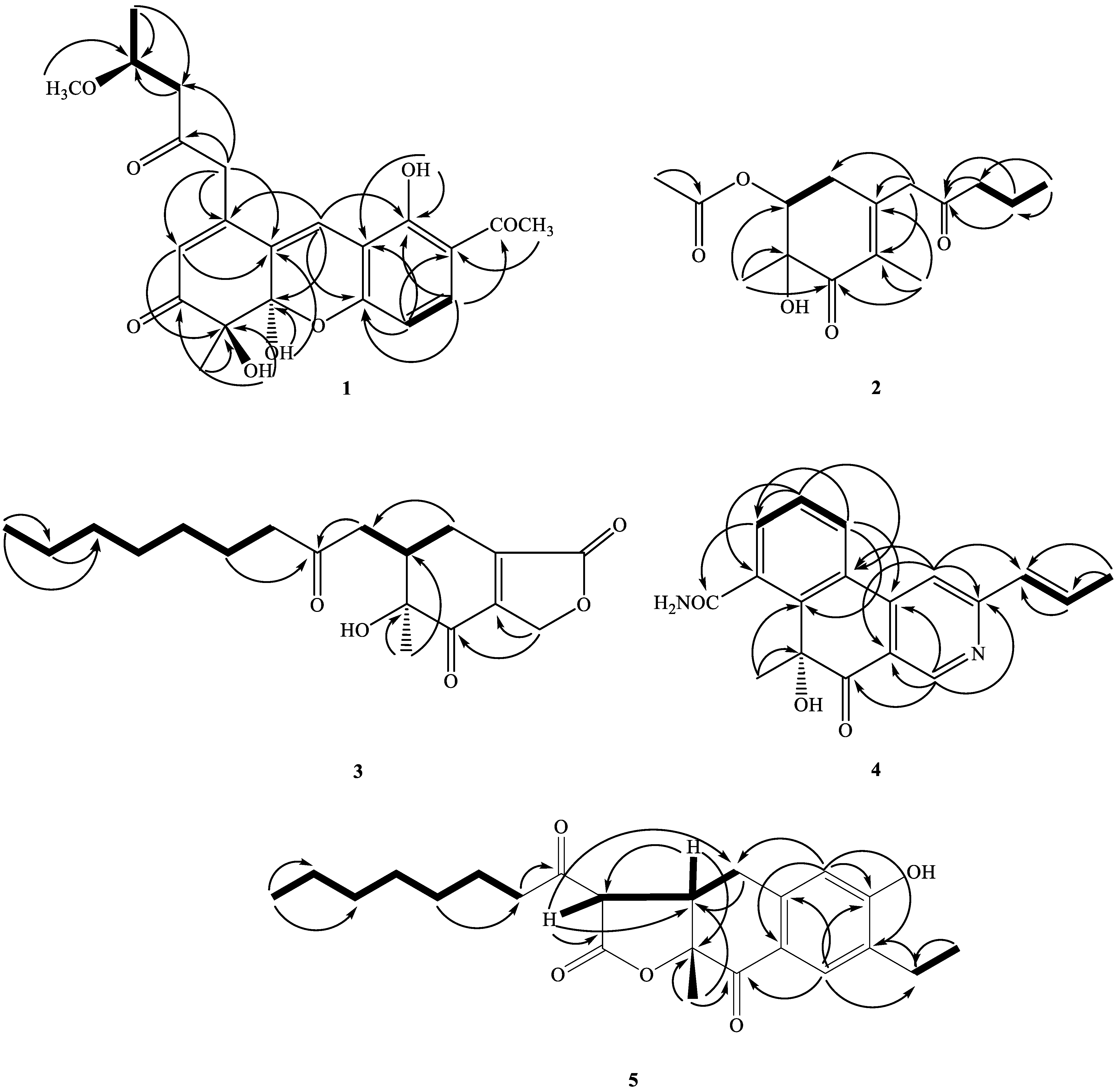
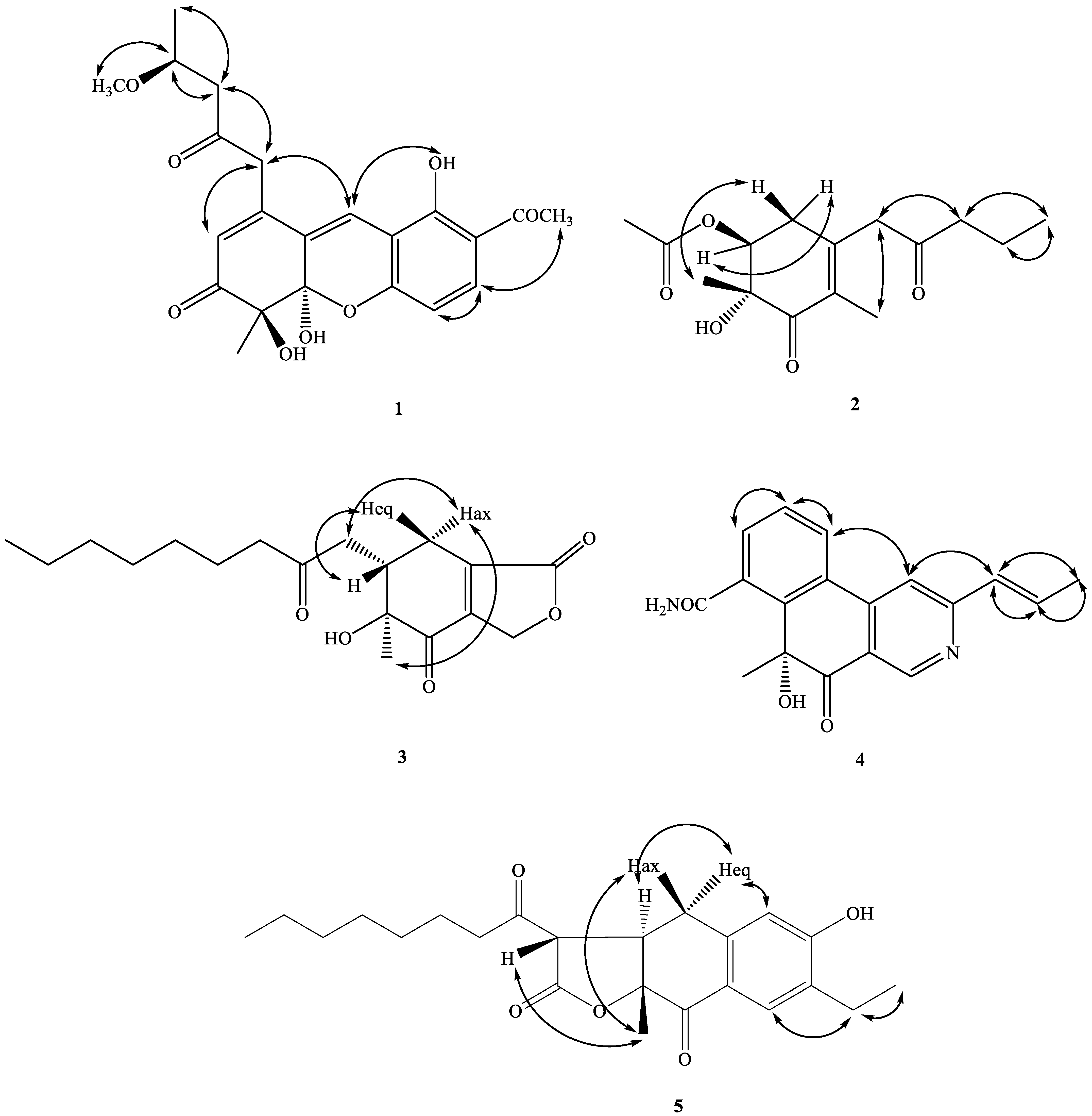
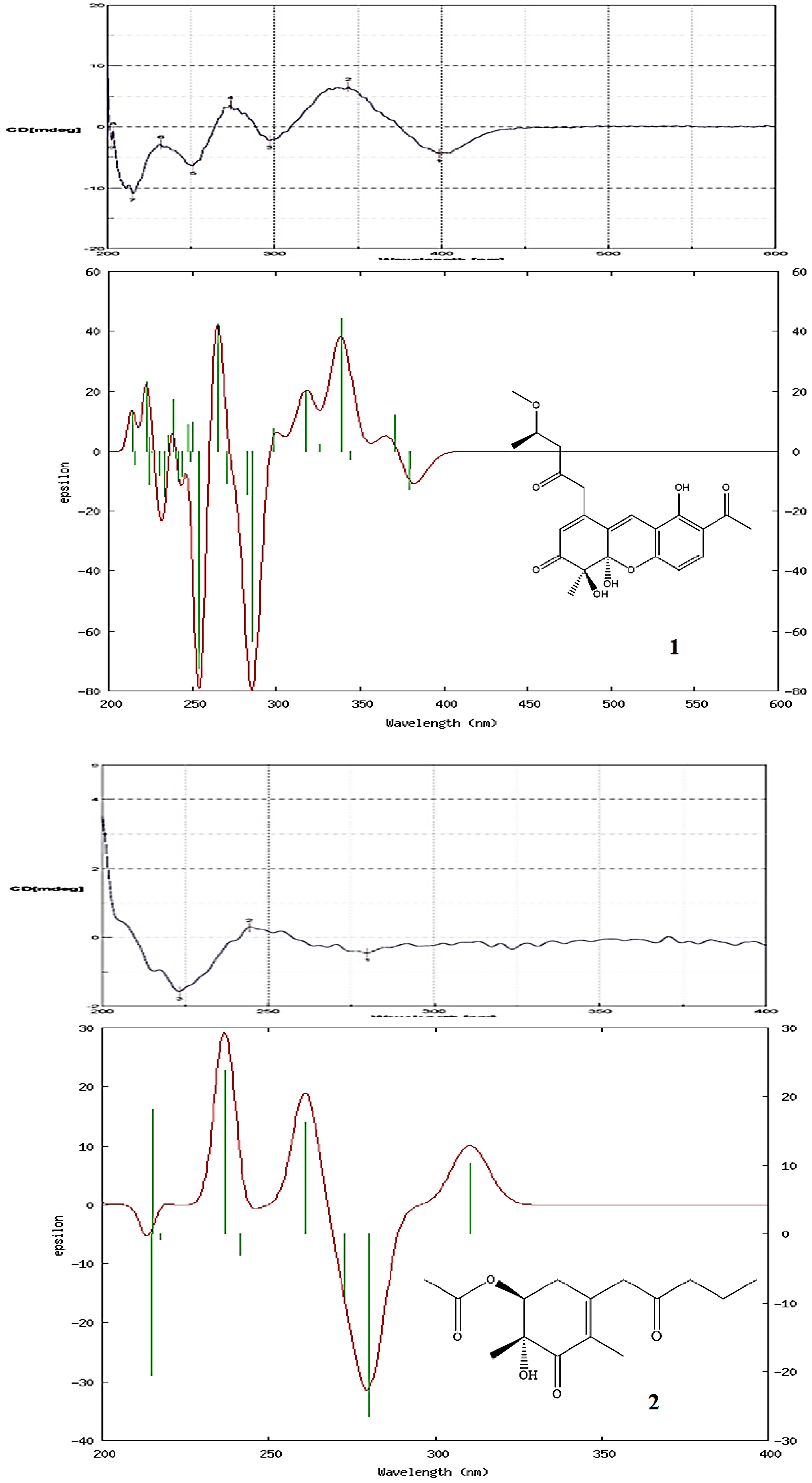
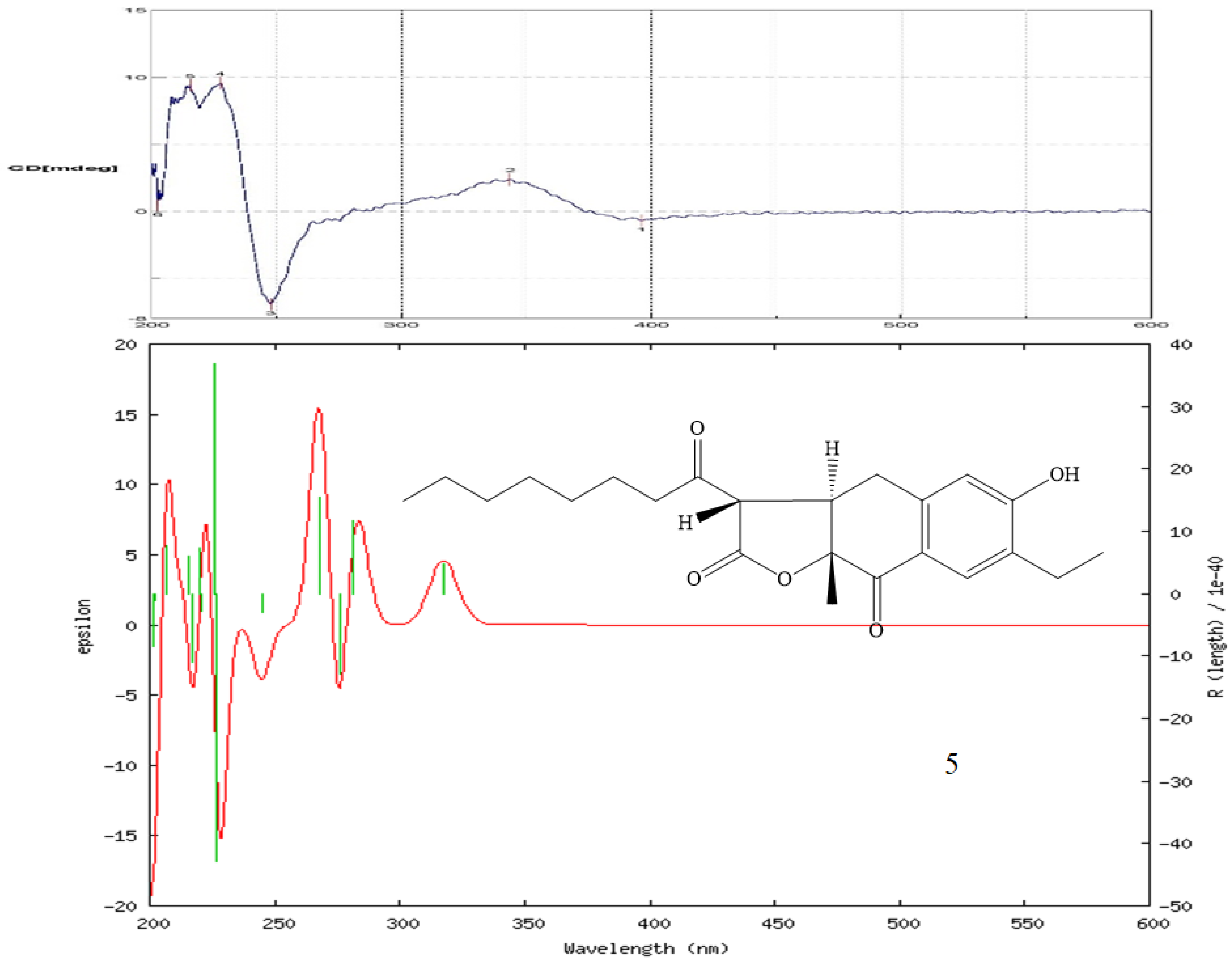
2.2. Structure Elucidation of Compounds
3. Discussion
Biological Studies
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Microorganism, Cultivation, and Preparation of the Strain
4.3. Isolation and Characterization of Secondary Metabolites
Computational Methods
4.4. Antifungal Activity Assays
4.4.1. Via Disk Diffusion Assay
4.4.2. Via Broth Dilution Assay
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nout, M.J.R.; Aidoo, K.E. Asian fungal fermented food. In Industrial Applications; Hofrichter, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 29–58. [Google Scholar]
- Liu, B.H.; Wu, T.S.; Su, M.C.; Chung, C.P.; Yu, F.Y. Evaluation of citrinin occurrence and cytotoxicity in Monascus fermentation products. J. Agric. Food Chem. 2005, 53, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, Y.; Ye, Q.; Li, J.; Hua, Y.; Ju, D.; Zhang, D.; Cooper, R.; Chang, M. Constituents of red yeast rice, a traditional Chinese food and medicine. J. Agric. Food Chem. 2000, 48, 5220–5225. [Google Scholar] [CrossRef] [PubMed]
- Endo, A. Monacolin K, a new hypocholesterolemic agent produced by a Monascus species. J. Antibiot. 1979, 32, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Endo, A. A historical perspective on the discovery of statins. P. Jpn. Acad. B-PHYS. 2010, 86, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.L.; Tsai, T.Y.; Wang, J.J.; Pan, T.M. In vivo hypolipidemic effects and safety of low dosage Monascus powder in a hamster model of hyperlipidemia. Appl. Microbiol. Biotechnol. 2006, 70, 533–540. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.; Duan, Z.; Guo, S. HMG-CoA reductase inhibitors from Monascus-fermented rice. J. Chem. 2013, 2013, 6. [Google Scholar] [CrossRef]
- Shi, Y.C.; Pan, T.M. Anti-diabetic effects of Monascus purpureus NTU 568 fermented products on streptozotocin-induced diabetic rats. J. Agric. Food Chem. 2010, 58, 7634–7640. [Google Scholar] [CrossRef]
- Akihisa, T.; Tokuda, H.; Yasukawa, K.; Ukiya, M.; Kiyota, A.; Sakamoto, N.; Suzuki, T.; Tanabe, N.; Nishino, H. Azaphilones, furanoisophthalides, and amino acids from the extracts of Monascus pilosus-fermented rice (red-mold rice) and their chemopreventive effects. J. Agric. Food Chem. 2005, 53, 562–565. [Google Scholar] [CrossRef]
- Hsu, L.C.; Liang, Y.H.; Hsu, Y.W.; Kuo, Y.H.; Pan, T.M. Anti-inflammatory properties of yellow and orange pigments from Monascus purpureus NTU 568. J. Agric. Food Chem. 2013, 61, 2796–2802. [Google Scholar] [CrossRef]
- Yasukawa, K.; Takahashi, M.; Natori, S.; Kawai, K.I.; Yamazaki, M.; Takeuchi, M.; Takido, M. Azaphilones inhibit tumor promotion by 12-O-tetradecanoylphorbol-13-acetate in two-stage carcinogenesis in mice. Oncology 1994, 51, 108–112. [Google Scholar] [CrossRef]
- Cottone, S.; Lorito, M.; Riccobene, R.; Nardi, E.; Mulè, G.; Buscemi, S.; Geraci, C.; Guarneri, M.; Arsena, R.; Cerasola, G. Oxidative stress, inflammation and cardiovascular disease in chronic renal failure. J. Nephrol. 2008, 21, 175–179. [Google Scholar]
- Hong, M.Y.; Seeram, N.P.; Zhang, Y.; Heber, D. Anticancer effects of Chinese red yeast rice versus monacolin K alone on colon cancer cells. J. Nutr. Biochem. 2008, 19, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yau, L.F.; Lu, J.G.; Zhu, G.Y.; Wang, J.R.; Han, Q.B.; Hsiao, W.L.; Jiang, Z.H. Cytotoxic dehydromonacolins from red yeast rice. J. Agric. Food Chem. 2012, 60, 934–939. [Google Scholar] [CrossRef]
- Chang, W.T.; Chuang, C.H.; Lee, W.J.; Huang, C.S. Extract of Monascus purpureus CWT715 fermented from sorghum liquor biowaste inhibits migration and invasion of SK-Hep-1 human hepatocarcinoma cells. Molecules 2016, 21, 1691. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.W.; Chen, M.H.; Fang, W.H.; Hung, C.M.; Chen, Y.L.; Wu, M.D.; Yuan, G.F.; Wu, M.J.; Wang, Y.J. Preventive effects of Monascus on androgen-related diseases: Androgenetic alopecia, benign prostatic hyperplasia, and prostate cancer. J. Agric. Food Chem. 2013, 61, 4379–4386. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, H.C.; Park, S.W.; So, H.S.; Woo, C.Y.; Choi, J.H.; Kim, S.H.; Kim, S.J.; Oh, Y.M. A case of isolated pulmonary mucormycosis in an immunocompetent host. Tuberc. Respir. Dis. 2013, 74, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Hotamisligil, G.S. Inflammation, stress, and diabetes. J. Clin. Invest. 2005, 115, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Leonie, K.H.; Lesley, V.C. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr. Pharm. Des. 2008, 14, 1225–1230. [Google Scholar]
- Kamer, A.R.; Craig, R.G.; Dasanayake, A.P.; Brys, M.; Glodzik-Sobanska, L.; de Leon, M.J. Inflammation and Alzheimer’s disease: Possible role of periodontal diseases. Alzheimers Dement. 2008, 4, 242–250. [Google Scholar] [CrossRef]
- Vasto, S.; Candore, G.; Listì, F.; Balistreri, C.R.; Colonna-Romano, G.; Malavolta, M.; Lio, D.; Nuzzo, D.; Mocchegiani, E.; Di Bona, D.; et al. Inflammation, genes and zinc in Alzheimer’s disease. Brain Res. Rev. 2008, 58, 96–105. [Google Scholar] [CrossRef]
- Martínková, L.; J°zlová, P.; Veselý, D. Biological activity of polyketide pigments produced by the fungus Monascus. J. Appl. Microbiol. 1995, 79, 609–616. [Google Scholar]
- Martínková, L.; Patáková-Jůzlová, P.; Krent, V.; Kucerová, Z.; Havlícek, V.; Olsovský, P.; Hovorka, O.; Ríhová, B.; Veselý, D.; Veselá, D.; et al. Biological activities of oligoketide pigments of Monascus purpureus. Food Addit. Contam. 2001, 16, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Jung, H.; Kim, J.H.; Shin, C.S. Effect of monascus pigment derivatives on the electrophoretic mobility of bacteria, and the cell adsorption and antibacterial activities of pigments. Colloids Surf. B 2006, 47, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.P.; Li, Y.Q.; Yang, J.; Cui, K.Y. Antibacterial characteristics of orange pigment extracted from Monascus pigments against Escherichia coli. Czech J. Food Sci. 2016, 34, 197–203. [Google Scholar] [CrossRef]
- Kim, C.; Jung, H.; Kim, Y.O.; Shin, C.S. Antimicrobial activities of amino acid derivatives of monascus pigments. FEMS Microbiol. Lett. 2006, 264, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Jůzlová, P.; Martínková, L.; Křen, V. Secondary metabolites of the fungus Monascus: A review. J. Ind. Microbiol. 1996, 16, 163–170. [Google Scholar] [CrossRef]
- Wong, H.C.; Koehler, P.E. Production and isolation of an antibiotic from Monascus purpureus and its relationship to pigment production. J. Food Sci. 1981, 46, 589–592. [Google Scholar] [CrossRef]
- Fabric, C.E.; Santerre, A.L.; Loret, M.O.; Baberian, R.; Pareilleux, A.; Goma, G.; Blanc, P.J. Production and food applications of the red pigments of Monascus ruber. J. Food Sci. 1993, 58, 1099–1102. [Google Scholar]
- Sun, J.M.; Kim, S.J.; Kim, G.W.; Rhee, J.K.; Kim, N.D.; Jung, H.; Jeun, J.; Lee, S.H.; Han, S.H.; Shin, C.S.; et al. Inhibition of hepatitis C virus replication by Monascus pigment derivatives that interfere with viral RNA polymerase activity and the mevalonate biosynthesis pathway. J. Antimicrob. Chemother. 2011, 67, 49–58. [Google Scholar] [CrossRef]
- Hsu, Y.W.; Hsu, L.C.; Liang, Y.H.; Kuo, Y.H.; Pan, T.M. Monaphilones A−C, three new antiproliferative azaphilone derivatives from Monascus purpureus NTU 568. J. Agric. Food Chem. 2010, 58, 8211–8216. [Google Scholar] [CrossRef]
- Jongrungruangchok, S.; Kittakoop, P.; Yongsmith, B.; Bavovada, R.; Tanasupawat, S.; Lartpornmatulee, N.; Thebtaranonth, Y. Azaphilone pigments from a yellow mutant of the fungus Monascus kaoliang. Phytochemistry 2004, 65, 2569–2575. [Google Scholar] [CrossRef] [PubMed]
- Loret, M.-O.; Morel, S. Isolation and structural characterization of two new metabolites from Monascus. J. Agric. Food Chem. 2010, 58, 1800–1803. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Ku, S. Beneficial effects of Monascus sp. KCCM 10093 pigments and derivatives: A mini review. Molecules 2018, 23, 98. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Chen, Y.F.; Cheng, M.J.; Wu, M.D.; Chen, Y.L.; Chang, H.S. Investigations into Chemical Components from Monascus purpureus with Photoprotective and Anti-Melanogenic Activities. J. Fungi 2021, 7, 619. [Google Scholar] [CrossRef]
- Diedrich, C.; Grimme, S. Systematic investigation of modern quantum chemical methods to predict electronic circular dichroism spectra. J. Phys. Chem. A 2003, 107, 2524–2539. [Google Scholar] [CrossRef]
- Ma, M.Z.; Ge, H.J.; Yi, W.W.; Wu, B.; Zhang, Z.Z. Bioactive drimane sesquiterpenoids and isocoumarins from the marine-derived fungus Penicillium minioluteum ZZ1657. Tetrahedron Lett. 2020, 61, 151504. [Google Scholar] [CrossRef]
- Knecht, A.; Cramer, B.; Humpf, H.-U. New Monascus metabolites: Structure elucidation and toxicological properties studied with immortalized human kidney epithelial cells. Mol. Nutr. Food. Res. 2006, 50, 314–321. [Google Scholar] [CrossRef]
- Takikawa, H.; Hachisu, Y.; Bode, J.W.; Suzuki, K. Catalytic enantioselective crossed aldehyde-ketone benzoin cyclization. Angew. Chem. Int. Ed. Engl. 2006, 45, 3492–3494. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts, Approved Guideline, CLSI Document M44-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2004. [Google Scholar]
- Clinical and Laboratory Standards Institute. Zone Diameter Interpretive Standards, Corresponding Minimal Inhibitory Concentration (MIC) Interpretive Breakpoints, and Quality Control Limits for Antifungal Disk Diffusion Susceptibility Testing of Yeasts, Informational Supplement, CLSI Document M44-S2, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. Method for Antifungal Disk Diffusion Susceptibility Testing of Filamentous Fungi, Proposed Guideline, CLSI Document M51-P; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, Approved Standard, CLSI Document M27-A3, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, Informational Supplement, CLSI Document M27-S3, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, Approved Standard, CLSI Document M38-A2, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Velíšek, J.; Davídek, J.; Cejpek, K. Biosynthesis of food constituents: Natural pigments. Part 2—A review. Czech J. Food Sci. 2008, 26, 73–98. [Google Scholar] [CrossRef]

| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 6.68 (1H, dd, J = 8.8) | 1.76 (3H, q, J = 1.2) | 4.89 (1H, dd, J = 18.0, 4.5) 5.05 (1H, dd, J =18.0, 3.3) | 9.05 (1H, s) | 7.91 (1H, s) |
| 2 | 7.71 (1H, d, J = 8.8) | ||||
| 2a | 2.64 (3H, q, J = 7.2) | ||||
| 2b | 1.26 (3H, J = 7.2) | ||||
| 4 | 3.33 (1H, d, J = 16.8) 3.48 (1H, d, J = 16.8) | 2.10–2.12 (1H, m) 2.95, d (1H, dd, J = 19.0, 4.5, 3.3) | 7.59 (1H, s) | 6.63 (1H, s) | |
| 5 | 2.49 (1H, ddd, J = 18.0, 10.7, 1.2 Hz, Hax-5) 2.53 (1H, ddd, J = 18.0, 6.2, 1.2 Hz, Heq-5) | 2.80–2.82 (1H, m) | 3.15 (1H, dd, J =16.0, 4.2, H-eq) 2.92 (1H, dt, J = 16.0, 12.3, H-ax) | ||
| 6 | 5.94 (1H, s) | 4.83 (1H, dd, J = 10.7, 6.2) | 3.34 (1H, td, J = 12.6, 4.2) | ||
| 8 | 1.24 (3H, s) | ||||
| 9 | 7.53 (1H, d, J = 8.8) | 2.45 (2H, t, J = 7.8) | 2.49–2.52 (1H, m) 3.03 (1H, dd, J = 18.0, 3.2) | 6.65 (1H dq, J = 15.6, 1.8) | |
| 10 | 1.61 (2H, sextet, J = 7.8) | 7.13 (1H, dd, J = 15.6, 6.8) | |||
| 11 | 2.60 (3H, s) | 0.92 (3H, t, J = 7.8) | 2.44–2.47 (2H, m) | 2.05 (3H, dd, J = 6.8, 1.8) | |
| 12 | 1.48 (3H, s) | 1.38 (3H, s) | 1.55–1.60 (2H, m) | 1.85 (3H, s) | 1.47 (3H, s) |
| 13 | 3.72 (1H, d-like, J = 17.0), 3.77 (1H, d-like, J = 17.0) | 1.20–1.35 (2H, m) | 8.04 (1H, dd, J = 7.8) | 3.72 (3H, d, J = 12.6) | |
| 14 | 1.20–1.35 (2H, m) | 7.70 (1H, t, J = 7.8) | |||
| 15 | 2.72 (1H, d, J = 16.2), 2.75 (1H, d, J = 16.2) | 2.09 (3H, s) | 1.20–1.35 (2H, m) | 7.90 (1H, dd, J = 7.8, 0.6) | 2.65/3.03 (each 1H, dt, J = 18.0, 7.2) |
| 16 | 4.25 (1H, m) | 1.20–1.35 (2H, m) | 1.64 (2H, pentet, J = 7.2) | ||
| 17 | 1.28 (3H, t, J = 6.4) | 0.90 (3H, t, J = 7.2) | 1.30–1.33 (2H, m) | ||
| 18 | 1.30–1.33 (2H, m) | ||||
| 19 | 1.30–1.33 (2H, m) | ||||
| 20 | 1.30–1.33 (2H, m) | ||||
| 21 | 0.91 (3H, t, J =7.2) | ||||
| OH-3 | 5.42 (1H, br s) | ||||
| OCH3–16 | 3.21 (3H, s) | ||||
| OH-4 | 13.4 (1H, s) | ||||
| OH-8 | 3.50 (1H, br s)/4.15 (1H, br s) | ||||
| OH-8b | 4.15 (1H, br s)/3.50 (1H, br s) |
| No. | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 108.8 | 12.3 | 67.2 | 149.8 | 130.4 |
| 2 | 134.0 | 130.0 | |||
| 2a | 22.0 | ||||
| 2b | 14.2 | ||||
| 3 | 114.8 | 205.4 | 170.9 | 161.8 | 159.0 |
| 3a | 144.5 | ||||
| 4 | 161.2 | 48.8 | 25.9 | 114.0 | 115.8 |
| 4a | 110.2 | 146.8 | 143.5 | 140.9 | |
| 4b | 125.3 | ||||
| 5 | 149.2 | 37.9 | 40.7 | 126.7 | 30.2 |
| 6 | 123.5 | 67.9 | 63.2 | 151.0 | 43.1 |
| 7 | 198.4 | 85.1 | 198.5 | 84.9 | 84.2 |
| 7a | 148.9 | ||||
| 8 | 79.8 | 195.6 | 19.2 | 192.8 | 192.1 |
| 8a | 157.3 | 132.0 | 122.7 | 124.7 | |
| 8b | 97.2 | ||||
| 9 | 122.7 | 45.0 | 41.8 | 131.8 | |
| 10 | 203.1 | 17.2 | 209.1 | 137.2 | |
| 11 | 26.6 | 13.6 | 43.4 | 18.8 | |
| 12 | 23.0 | 16.2 | 23.5 | 27.3 | 17.4 |
| 13 | 48.9 | 29.0 | 129.1 | 54.9 | |
| 13a | 29.0 | 170.9 | |||
| 14 | 206.1 | 170.2 | 29.0 | 132.3 | 203.9 |
| 15 | 50.3 | 21.3 | 31.4 | 127.8 | 42.8 |
| 16 | 73.4 | 22.4 | 125.8 | 23.5 | |
| 17 | 23.0 | 13.9 | 168.5 | 29.1 | |
| 18 | 29.1 | ||||
| 19 | 31.7 | ||||
| 20 | 22.8 | ||||
| 21 | 13.9 |
| Test Microorganism | Isolated Compounds | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Ketoconazole | |
| A. niger | 15.4 ± 0.7 | 29.1 ± 3.5 | 29.3 ± 1.9 | 32.0 ± 1.8 | 27.5 ± 2.8 | 34.2 ± 1.8 |
| P. italicum | 17.8 ± 1.2 | 28.5 ± 2.1 | 29.4 ± 1.4 | 28.3 ± 3.1 | 17.5 ± 2.2 | 35.9 ± 2.3 |
| C. albicans | 16.2 ± 5.4 | 27.6 ± 3.9 | 36.2 ± 3.6 | 31.2 ± 3.5 | 28.0 ± 3.1 | 39.3 ± 3.1 |
| S. cerevisiae | 12.9 ± 1.1 | 30.1 ± 4.0 | 21.9 ± 2.5 | 28.2 ± 2.8 | 27.3 ± 1.4 | 34.2 ± 1.1 |
| Compounds | A. niger | P. italicum | C. albicans | S. cerevisiae |
|---|---|---|---|---|
| 2 | >100 | >100 | >100 | 43.45 ± 2.33 a |
| 3 | >100 | >100 | 32.87 ± 2.19 a | >100 |
| 4 | 29.65 ± 3.54 a | >100 | 58.43 ± 1.51 a | >100 |
| 5 | >100 | >100 | >100 | >100 |
| Ketoconazole | 4.10 ± 0.84 a | 5.34 ± 2.56 a | 10.88 ± 5.67 a | 3.57 ± 0.98 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.-D.; Chen, J.-J.; Cheng, M.-J. Secondary Metabolites with Antifungal Activities from Mangrove Derived Fungus Monascus purpureus WMD2424. Mar. Drugs 2023, 21, 200. https://doi.org/10.3390/md21040200
Wu M-D, Chen J-J, Cheng M-J. Secondary Metabolites with Antifungal Activities from Mangrove Derived Fungus Monascus purpureus WMD2424. Marine Drugs. 2023; 21(4):200. https://doi.org/10.3390/md21040200
Chicago/Turabian StyleWu, Ming-Der, Jih-Jung Chen, and Ming-Jen Cheng. 2023. "Secondary Metabolites with Antifungal Activities from Mangrove Derived Fungus Monascus purpureus WMD2424" Marine Drugs 21, no. 4: 200. https://doi.org/10.3390/md21040200
APA StyleWu, M.-D., Chen, J.-J., & Cheng, M.-J. (2023). Secondary Metabolites with Antifungal Activities from Mangrove Derived Fungus Monascus purpureus WMD2424. Marine Drugs, 21(4), 200. https://doi.org/10.3390/md21040200









