Abstract
The venom of marine cone snails is mainly composed of peptide toxins called conopeptides, among which conotoxins represent those that are disulfide-rich. Publications on conopeptides frequently state that conopeptides attract considerable interest for their potent and selective activity, but there has been no analysis yet that formally quantifies the popularity of the field. We fill this gap here by providing a bibliometric analysis of the literature on cone snail toxins from 2000 to 2022. Our analysis of 3028 research articles and 393 reviews revealed that research in the conopeptide field is indeed prolific, with an average of 130 research articles per year. The data show that the research is typically carried out collaboratively and worldwide, and that discoveries are truly a community-based effort. An analysis of the keywords provided with each article revealed research trends, their evolution over the studied period, and important milestones. The most employed keywords are related to pharmacology and medicinal chemistry. In 2004, the trend in keywords changed, with the pivotal event of that year being the approval by the FDA of the first peptide toxin drug, ziconotide, a conopeptide, for the treatment of intractable pain. The corresponding research article is among the top ten most cited articles in the conopeptide literature. From the time of that article, medicinal chemistry aiming at engineering conopeptides to treat neuropathic pain ramped up, as seen by an increased focus on topological modifications (e.g., cyclization), electrophysiology, and structural biology.
1. Introduction
The origin of conopeptides contributes to their popularity because it involves the fascinating biology of the extraordinary organisms that produce them, the marine cone snails [1,2,3]. These carnivorous snails from the Conus genus live in tropical and subtropical seas and lie in ambush under the sand or in crevices waiting for their prey to pass by. Like their vegetarian cousins, these snails are slow paced. Nevertheless, they can catch fast-moving fish, or indeed slower moving mollusks or worms, by relying on peculiar hunting strategies [2]. They shoot hollow radular teeth filled with poisonous peptides, i.e., the conopeptides, through an elongated organ called the proboscis, and these teeth act as hypodermic needles to inject a deadly cocktail into their victim [2]. Some cone snails alternatively diffuse conopeptides in the water to anaesthetize a fish or a whole school of fish, enabling the snail to come in the open and engulf their unconscious prey in their mouth before stinging them while captive [2,4].
There are approximately 1000 cone snail species, each producing a mostly distinct set of several hundreds of conopeptides, which are diverse in terms of amino acid sequences, peptide folds, and pharmacological properties [5,6]. Most conopeptides are neuroactive and act on ligand-gated or voltage-gated ion channels with both high potency and selectivity; some of them display picomolar activities and most only target a narrow range of channel subtypes or even a single subtype [1]. This selectivity enables the nervous system of the prey to be subdued by over-activating specific pathways to disrupt cognition while simultaneously inhibiting other pathways related to flight [1]. Because of the large homology between ion channels in the animal kingdoms, conopeptides are often active at human targets. Some of them are considered drug leads, and one is an approved drug, as will be detailed in the Section 3 [5,6]. Conopeptides are also employed as molecular probes in neuroscience to study the involvement of certain ion channels in the nervous system [1,7]. This selectivity is contributed by numerous types of post-translational modifications, including C-terminal amidation, prolyl-hydroxylation, bromination of tryptophan, or phosphorylation, and these modifications considerably expand the chemical space [6,8]. The most common modification is the formation of disulfide bonds, which are important for the stability of peptides. More than 30 disulfide bond scaffolds have been identified so far in conopeptides, and most of those that have been structurally characterized adopt distinct folds [5]. Conopeptides are therefore numerous and diverse in terms of sequence, structure, and chemistry. The marine origin of this natural, highly diverse combinatorial library of ultra-potent and selective peptides appeals to the imaginary and has been qualified as “bounty”, “treasure house”, or “pharmaceutical treasure” [9,10,11].
Shortly after the physiological description of the cone snail envenomation apparatus by Kohn et al. in 1960 [12], Whyte and Endean evidenced the activity of the venom on the nervous system in 1962 [13], but it was only 20 years later that conopeptides started to be individually characterized with the pioneering investigations of Baldomero Olivera, who effectively founded the field [14]. In 1981, he published the first complete amino acid sequences of conopeptides that inhibited muscle contraction [15]. In two following seminal publications in 1985 and 1990 in the journal Science, he revealed that conopeptides are a vast natural peptide family displaying an unparalleled level of diversity and breadth of molecular targets [16,17], seeding an interest that has not faded 30–40 years later. Articles on conopeptides frequently state that conopeptides “attract considerable interest” but there is no definitive analysis to quantify that claim. To fill this gap, we have carried out a bibliometric analysis of the literature on conopeptides from the year 2000 to 2022, providing an unbiased estimate of the breadth of activity in the field, also identifying the main contributors in terms of geographic region, institutions, and investigators, and finally analyzing the major trends of the field over that period through an analysis of publication keywords.
2. Materials and Methods
2.1. Data Collection
The list of publications on cone snail toxins made over the twenty years from 2000 to 2022 was retrieved from Clarivate Analytics Web of Science (WoS; “https://www.webofscience.com (accessed on 13 January 2023)”). A request was made to the Web of Science Core Collection (WoSCC) bibliographic database using the query “TS = (con-ikot-ikot* OR conantokin* OR coninsulin* OR conkunitzin* OR conoCAP* OR conoGAY* OR conoNPY* OR conodipine* OR conohyal* OR conolysin* OR conomap* OR conomarphin* OR conopeptide* OR conophan* OR conophysin* OR conoporin* OR conopressin* OR conofamide* OR conorphin* OR conotoxin* OR contryphan* OR contulakin*)”. A total of 3451 research articles and reviews were downloaded in the “Web of Science Core Collection” format, which is standardized using a set of two-letter tags and could conveniently be analyzed computationally. We also retrieved the description made in NCBI PubMed (“https://pubmed.ncbi.nlm.nih.gov (accessed on 13 January 2023)”) for each article and review when it was possible. The publication data in PubMed were retrieved using the PubMed identifier provided by WoS. We reasoned that WoS displays a larger breadth of research journals, which should provide a more comprehensive snapshot of the literature than PubMed.
2.2. Data Post-Processing
The literature data were first curated where necessary to increase data consistency as well as correct mis- and alternative spellings. The corrections were carried out in four steps, which were implemented in three python scripts provided in the Supplementary Materials.
A manual inspection of the data revealed that several articles were wrongly classified as either reviews or original research. The first python script (“01-retype.py”) re-classified the articles as being either review or original research based on the description made in the database NCBI PubMed in the “PT” field, which we found to be reliable. During that analysis, we also discarded the most recent articles for which no publication date was yet provided (ahead of print articles), as identified using the “PY” fields of WoS.
A second script (“02-rekeywords.py”) was used to homogenize the keywords provided by the authors (“DE” field) and by WoS (“ID” field; known as “Keywords Plus®”), enabling the downstream analysis of keyword frequencies. The changes include using the plural of each term (e.g., “isoforms” instead of “isoform”), equivalent terms (e.g., “conopeptides” instead of “Conus peptides”), and renaming peptides because of alternative spelling (e.g., “mu-conotoxin kiiia” instead of “kiiia”) or alternative names (e.g., “omega-conotoxin mviia” instead of “ziconotide”). Several non-informative, albeit frequently used keywords were removed, such as “water” or “identification”. The list of the 594 keyword changes is provided in the Supplementary Materials.
A third script (“03-authors_replace.py”) was used to correct for inconsistent author first names, which are in the “AU” and “AF” fields of the WoS format. The corrections were proposed using a semi-automatic pre-analysis, which involves extracting the frequency of names + first names, names with additional first names, or nearly equivalent names (identical first letters of names). Alternative first names result from partial spelling of the first names, for example, “D”, “DJ”, “D J”, “David”, or “David J” were recorded for “David James Craik”. Occasionally, alternative spellings of first names translated from non-Latin scripts were also identified, such as “Maksim E Astashev” and “Maxim E Astashev”. Names were manually checked for co-occurrence with other names and institutions, resulting in a list of 1964 modifications.
The data used for analysis consisted of 3421 publications, which comprised 3028 original research articles and 393 reviews.
2.3. Data Analysis
Our methodology is similar to that described by Zhu et al. (2021) [18], which describes a bibliometric analysis of nicotinic acetylcholine receptors. VOSviewer 1.6.18 [19,20] was used to identify the most active authors, their institutions and countries, as well as the most cited articles and co-cited references. Keywords and trends were analyzed with CiteSpace 6.1.R6 (64-bit) Basic [21].
3. Results and Discussion
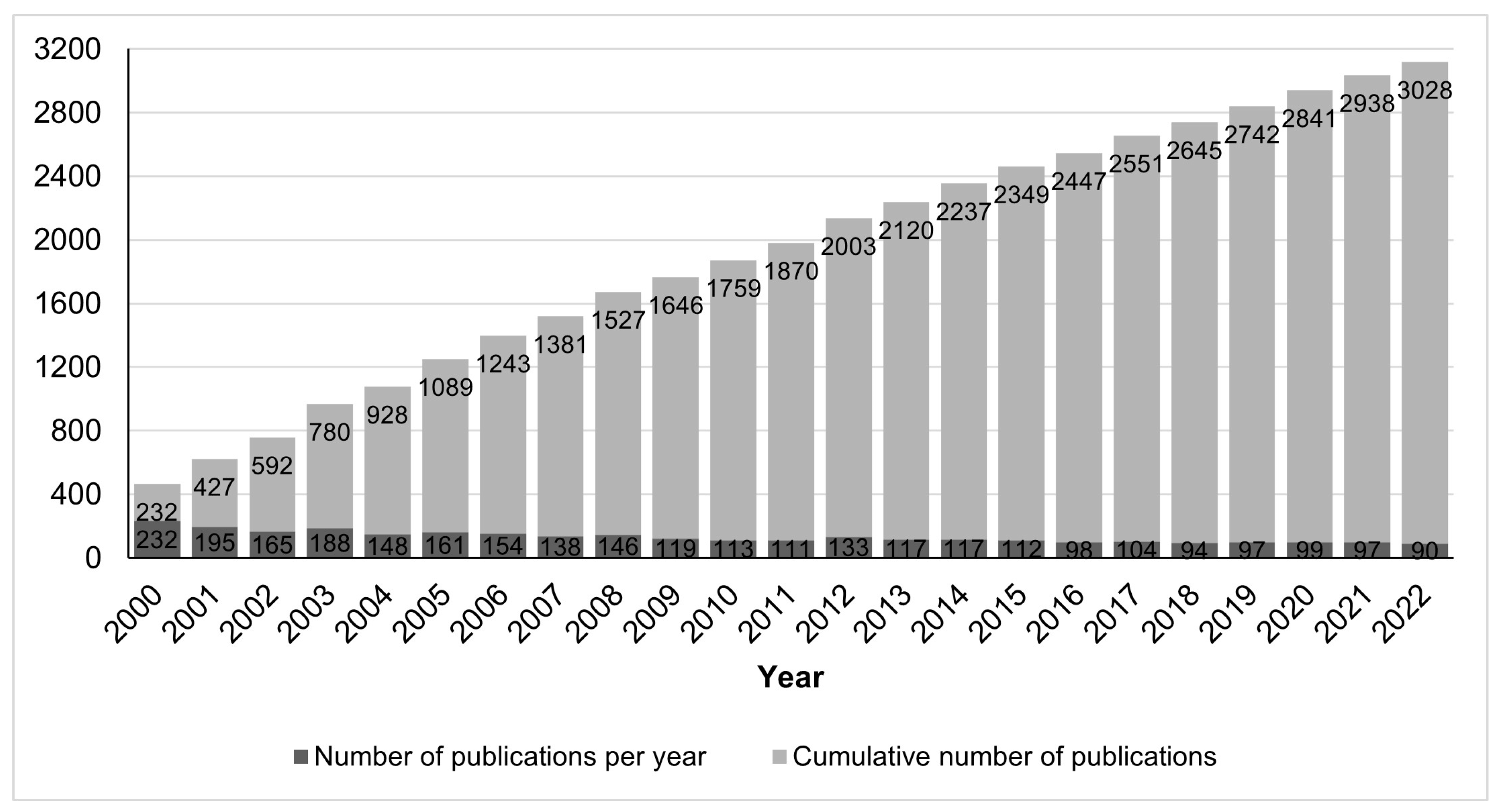
We retrieved information on 3421 peer-reviewed publications in the field of conopeptides in the period from 2000 to 2022, which should be considered as large, and by itself already proves that it is an important research field. Among these articles, 393 are reviews, representing 11% of the research output as well as an average of 18 reviews per year, which suggests that the field is very active and constantly requires an overview of its current trends to catch up with the latest developments. The cumulative number of research articles, shown in Figure 1, indicates a nearly steady growth in the number of articles in the last 10 years, although at a slower rate than the growth in the decade 2000–2009. The average rate in the 2000–2022 period was 132 ± 37 (standard deviation) publications per year. By contrast, toxins from other animal groups attracted less interest than conopeptides during that period, with <80 publications per year for spider or scorpion toxins. Statistics for other animal groups besides cone snails were made using the PubMed database and in the period 2000-2022; for instance, publications on spider and scorpion toxins were retrieved with the search queries “spider AND toxin AND peptide” and “scorpion AND toxin AND peptide”, resulting in 1178 (56/year) and 1563 (74/year) publications, respectively.
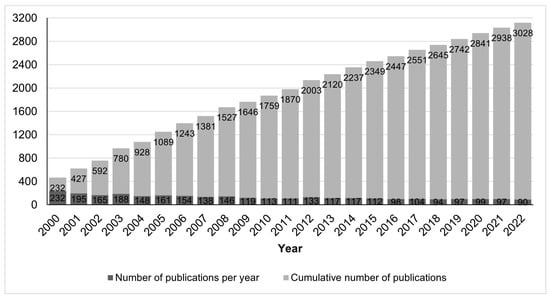
Figure 1.
Number of research publications on conopeptides in the period 2000–2022.
3.1. Geographic Regions, Institutions, and Authors
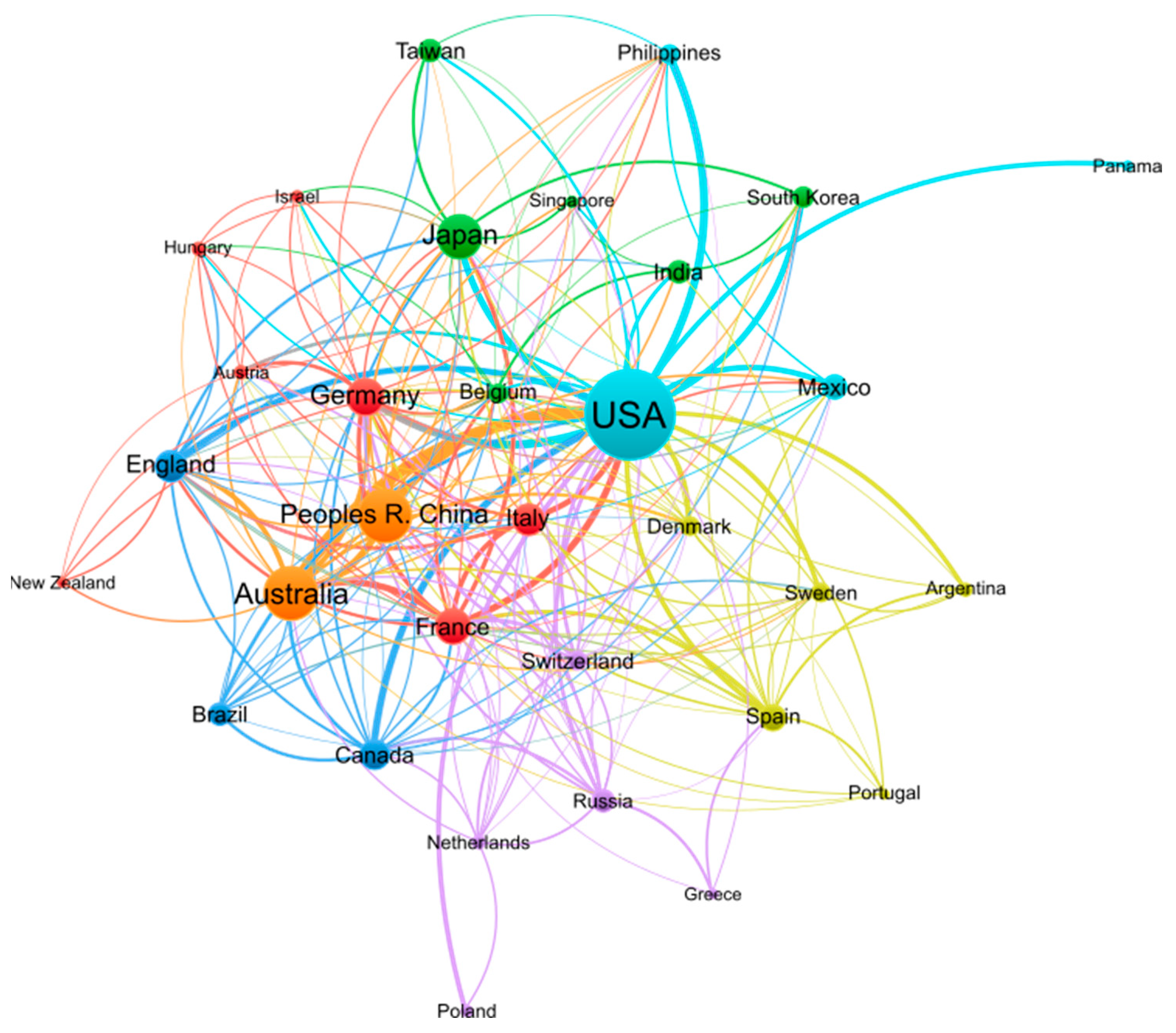
Research on conopeptides is international and is carried out at similar levels in several countries and continents, as shown in Table 1. The USA, where conopeptide research initially sprouted, has the largest publication output from 2000 to 2022, with >1100 publications over that period. The second most productive countries are Australia and China, with ~400 publications. Considering that the pool of scientists in Australia is at least an order-of-magnitude lower than in the USA, Australia’s relative scientific output on conopeptide research is per capita larger than that of the USA. Five European countries, including Germany, France, England, Italy, and Spain, are in the top 10 most productive countries, and together account for >700 publications, and would, therefore place Europe in the second position if its constitutive countries were simultaneously considered. The average number of citations per publication for the top 10 countries is between 23 and 44, therefore, also of a similar level. The impact of the publications made on conopeptides is thus similar in the most productive countries.

Table 1.
Top 10 countries for conopeptide research publications (2000–2022).
Research on conopeptides frequently involves collaborations between laboratories from multiple countries, as illustrated in Figure 2. The 10 countries with the largest publication output are well-connected to each other in this dense network of co-authored publications, suggesting that conopeptide research is the result of international collaborative efforts. International societies related to conopeptide research, such as the International Society on Toxinology (https://www.toxinology.org (accessed on 13 January 2023)) or the International Peptide Society (https://peptidesociety.org (accessed on 13 January 2023)), organize regular symposiums that create opportunities to foster such international collaborations.
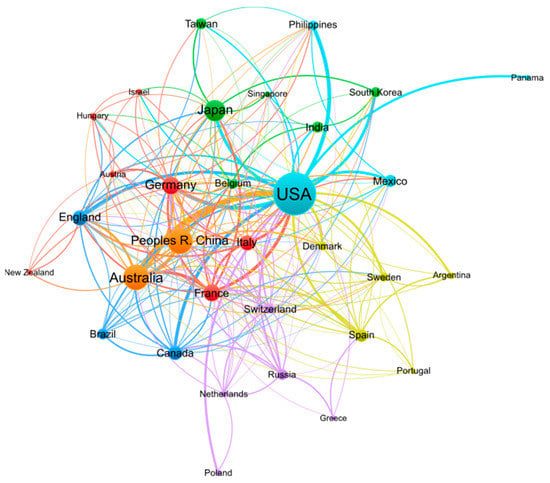
Figure 2.
Co-authorship network of countries engaged in conopeptide research in the period 2000–2022. Figure generated by VOSviewer 1.6.18 software with parameters as follows: Type of analysis—Co-authorship; Unit of analysis—Countries; Counting method—Fractional counting; Minimum number of documents of a country—10; the size of the item label in the visualization was determined by the number of documents; the layout was set with Attraction = −2 and Repulsion = −1. All other parameters were set as default values. A total of 32 out of 75 countries meeting the thresholds are shown in the figure.
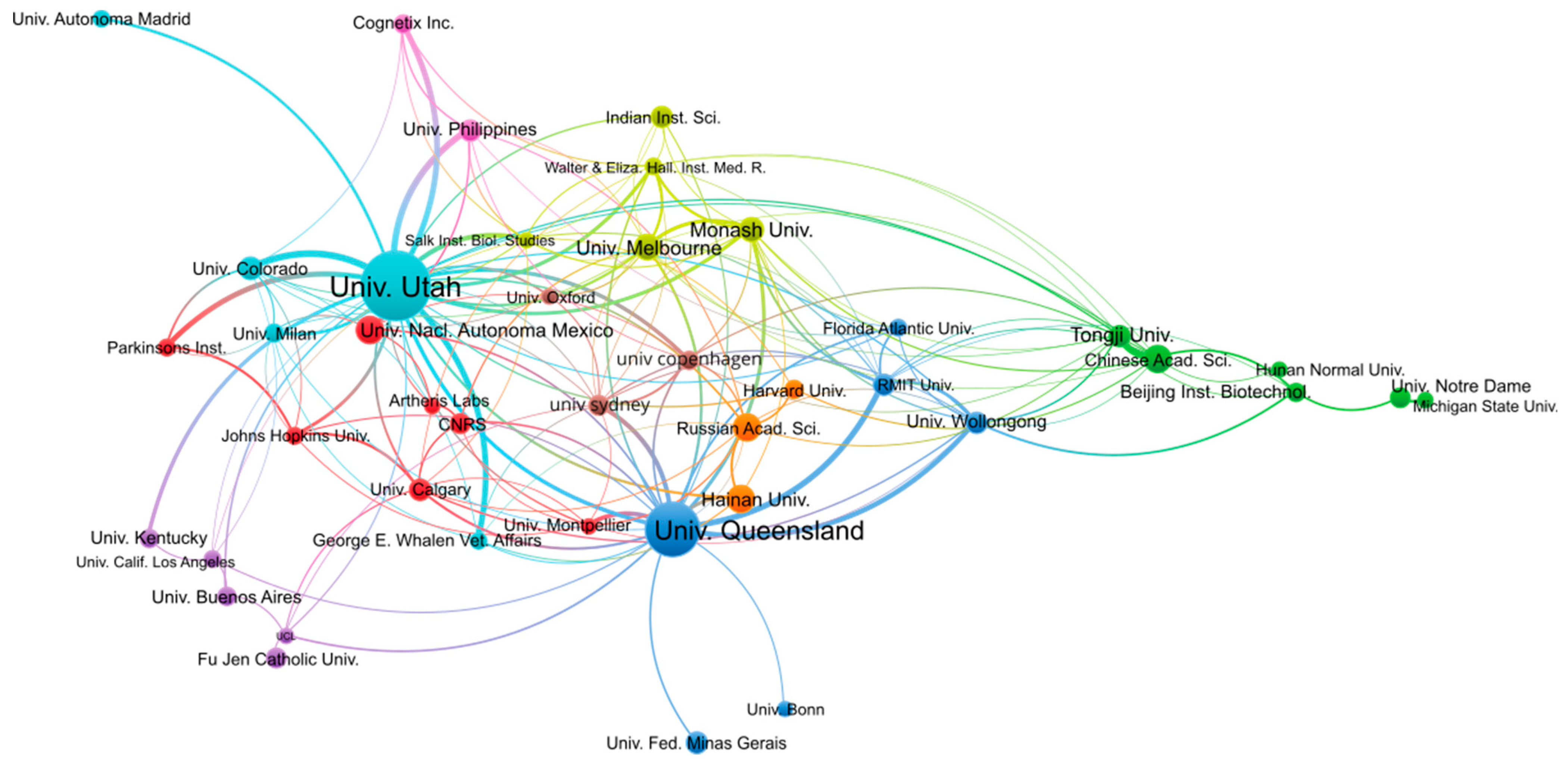
In terms of publication output per institution, the University of Utah (USA) and the University of Queensland (Australia) are the most productive, with 398 and 241 publications, respectively, as shown in Table 2. By contrast, the number of publications from the other institutions from the top 10 most productive institutions reaches only 50–60 publications. As we will see in the analyses per authors, the University of Utah and the University of Queensland host more research groups with interest in conopeptides than other institutions. It is interesting to note that the University of Utah has no geographical access to seas into which cone snails live, in contrast to The University of Queensland, which borders the Coral Sea or Hainan University, which is in the South China Sea. The analysis of co-authored publications between institutions shown in Figure 3 confirms that all institutions are interconnected, whatever their country, but also that strong ties exist within countries. For example, most Australian institutions form a cluster comprising the University of Queensland, RMIT University, and the University of Wollongong. Several Chinese institutions, including Tongji University, the Chinese Academy of Science, and Beijing Institute of Biotechnology, form a cluster that is more isolated than others. By contrast, Hainan University does not belong to that cluster but is co-published with the University of Utah, the University of Queensland, and the Russian Academy of Science. The University of Utah has among the strongest co-publications with other institutions, such as the University of Colorado, the University of the Philippines, or the private company Cognetix, which is a biotech spin-out from the University of Utah.

Table 2.
Top 10 institutions for conopeptide research publications (2000–2022).
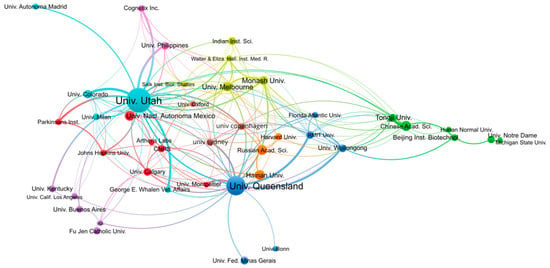
Figure 3.
Co-authorship network of institutions engaged in conopeptide research in the period 2000–2022. Figure was generated using VOSviewer 1.6.18 with the following parameters: Type of analysis = Co-authorship; Unit of analysis = Organizations; Counting method = Fractional; Minimum number of documents of an organization = 20; the size of the item label in the visualization was determined by the number of publications; the layout was set with Attraction = 2 and Repulsion = 0. All other parameters were kept with default values. A total of 43 out of the 1950 organizations met the thresholds. The largest set of connected items of the 42 items is shown in the figure.
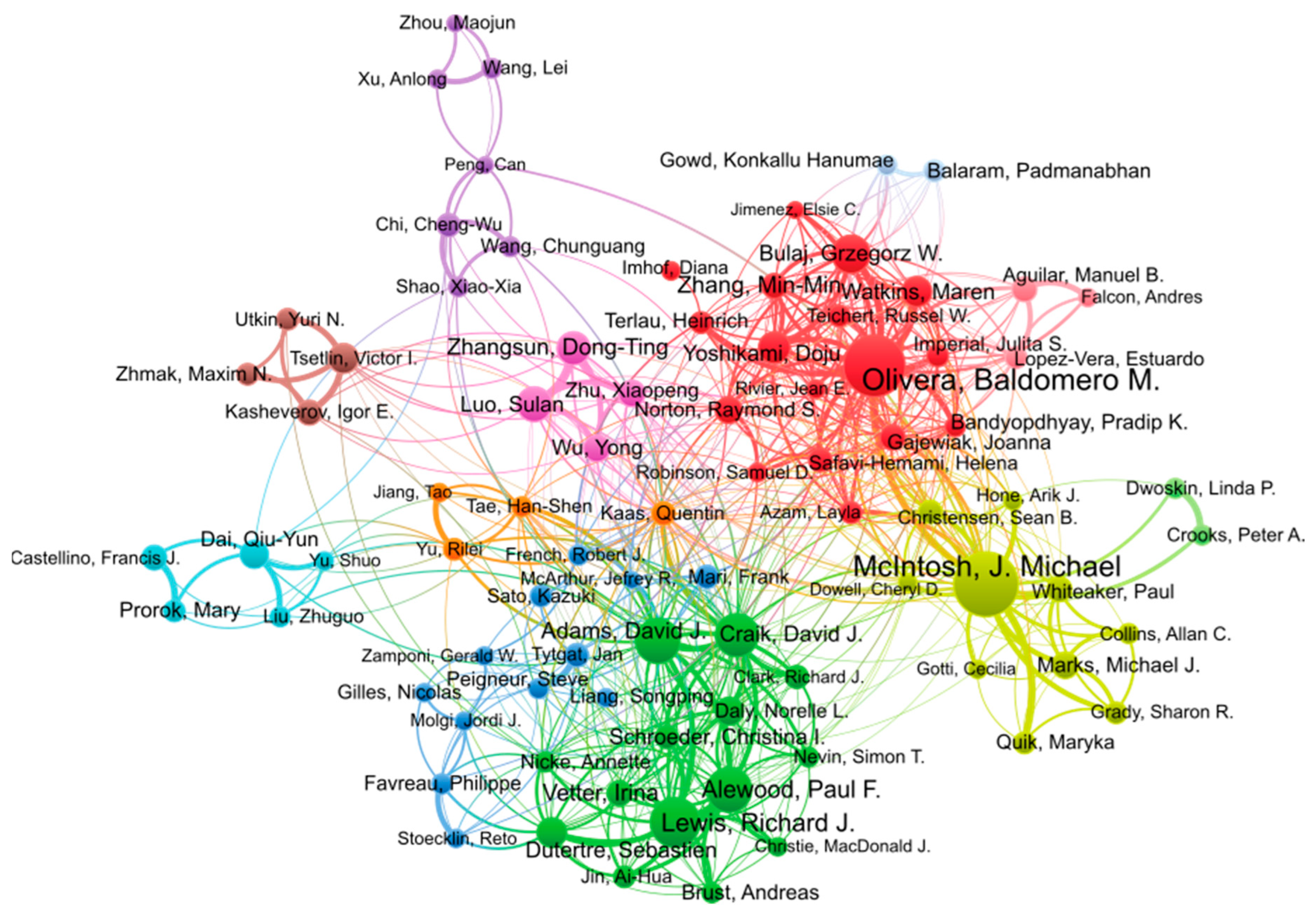
The two most active researchers in conopeptide research in the period 2000–2022 are Michael McIntosh and Baldomero Olivera, with 203 and 182 publications, respectively, as shown in Table 3. Baldomero Olivera founded the field in the 1980s [16] and is one of the two most prolific researchers in this field and in this millennium. His research encompasses the discovery of conopeptides, their pharmacological characterization, and biotechnological applications, especially in medicine. Michael McIntosh is a pharmacologist whose research group is located at the University of Utah, and he has co-authored numerous publications with Olivera, and independently, since 1982 [22]. His focus is on the characterization of ion channels and receptors of the human central nervous system as well as toxins acting on them for physiological studies and drug development. The following four most productive authors in the top 20 list, Richard Lewis, David Craik, David Adams, and Paul Alewood, are all Australian and have, or had, their research group at the University of Queensland. David Adams is currently at the University of Wollongong. As shown in the analysis of co-authorship in Figure 4, these four authors form a tight cluster. They have complementary expertise in natural product discovery (Richard Lewis), peptide chemical synthesis (Paul Alewood, David Craik), structural biology (David Craik), and pharmacology (David Adams), thus enabling productive collaborations for conopeptide discovery and characterization. Interestingly, the clusters of authors (identified by different colors) shown in Figure 4 approximately recapitulate research groups or institutions. For example, the pink cluster is constituted of Sulan Luo (Hainan University) and three other members of her group. The brown cluster also identifies the group of Victor Tsetlin at the Moscow Russian Academy of Science, and the darker blue cluster, albeit more loosely defined, mainly comprises European authors.

Table 3.
Top 20 active authors in conopeptide research (2000–2022).
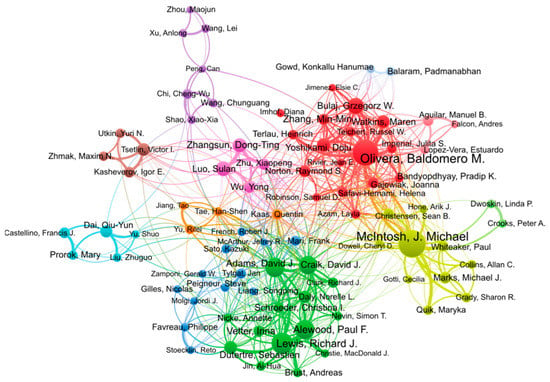
Figure 4.
Co-authorship network of authors contributing to conopeptide research in the period 2000–2022. Figure generated using VOSviewer 1.6.18 with the following parameters: Type of analysis = Co-authorship; Unit of analysis = Authors; Counting method = Fractional; Minimum number of documents of an author = 15; the size of the item label in the visualization was determined by the number of publications; the layout was set with Attraction = 2, Repulsion = −2. All other parameters were kept with default values. Of the 9392 authors, 90 authors met the thresholds. The largest set of connected items of 84 items is shown in the figure.
3.2. Research Trends
The most impactful conopeptide publications in 2000–2022 were investigated both in a global context (Table 4) and within the conopeptide field (Table 5). Six out of the ten most globally influential conopeptide research articles are physiological studies in which conopeptides are used as molecular probes of specific ion channels. In half of these most cited research articles, the α-conotoxin MII was used to identify nicotinic acetylcholine receptors that contain the α6 subunit. Two other articles in that top 10 list focus on the clinical development of the only conopeptide that is approved as a drug, ziconotide (Table 4). Ziconotide, also called ω-conotoxin MVIIA or Prialt, is a conotoxin isolated from Conus magus with potent inhibitory activity of the N-type voltage-gated calcium channel, and is an analgesic used to treat chronic pain and pain experienced by cancer and AIDS patients. This analysis suggests that the practical applications of conopeptides attract the most interest from the scientific community.

Table 4.
Ten most cited research articles in the conopeptide field published in 2000–2022.

Table 5.
The ten 2000–2022 conopeptide publications that were the most cited by the 3421 conopeptide publications in 2000–2022.
Six of the ten most cited publications within the conopeptide field, which are listed in Table 5, are reviews (two being the most cited publications), which were either published by the group of Baldomero Olivera or by research groups from the University of Queensland. Three out of the four most field-cited conopeptide research articles are not in the list of the most globally cited publications from Table 4, suggesting that the main interests within the field only partly overlap the interest from researchers out of the field. These differing points of interests are in the discovery of novel conopeptides or in the design of conopeptide variants with altered selectivity. One of these most field-cited articles indeed describes ConoServer [8], which is an expert database on conopeptides and provides essential knowledge and bioinformatic resources for the conopeptide discovery. Another highly cited article focuses on the design of conotoxin MII variants with increased selectivity for α3- or α6-containing nAChR subtypes, with the latter being involved in addiction. The third most field-cited research article reports on the discovery of native peptide CVID, which has a similar molecular target to ziconotide but acts with greater selectivity. Unsurprisingly, the only research article that is both in the most globally cited and field-cited publication lists focuses on the clinical development of ziconotide [23]. This article could, therefore, be described as the article that had the most impact. Ziconotide was discovered by Michael McIntosh as he was working in the laboratory of Baldomero Olivera. After successful clinical development, this conopeptide was the first peptide toxin to be approved by the FDA. After that milestone was reached in 2004, the focus of conopeptide research shifted, as highlighted in Figure 5 by a change in keyword bursts.
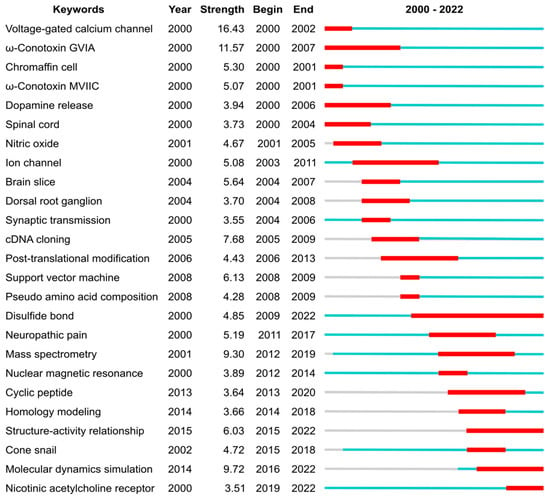
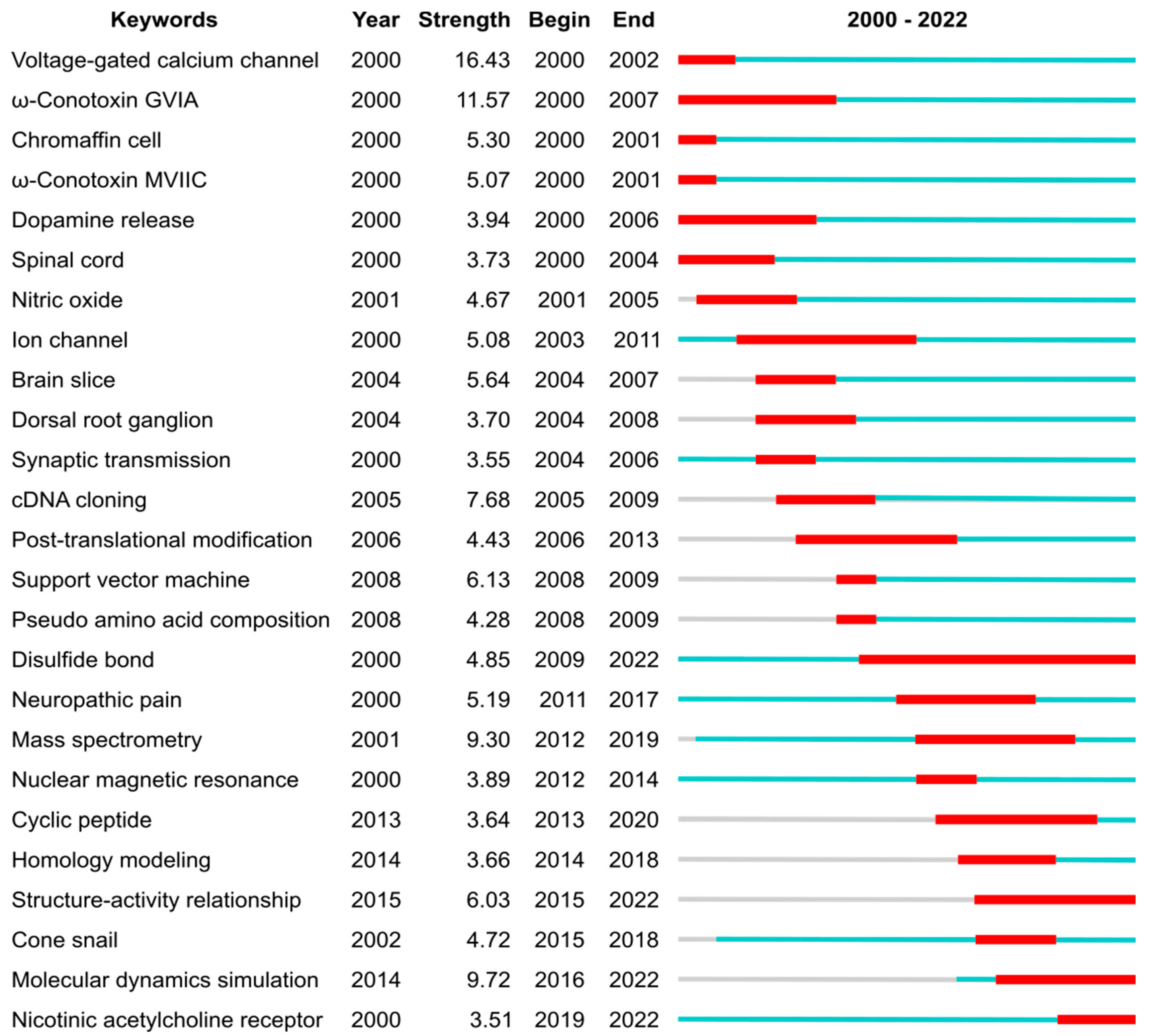
Figure 5.
Top 25 keywords with the strongest citation bursts, which represent a frequency surge in a keyword in conopeptide publications during the period 2000–2022. The blue lines depict the periods during when each keyword was used, and the red segments indicate the burst period. The “Year” column is the earliest year when the corresponding keyword had been published. The “Strength” column is the frequency of the corresponding keyword that had appeared over time. The “Begin” and “End” columns are the year of the beginning and end of the burst period, respectively. The analysis was carried out using CiteSpace 6.1.R6 (64-bit) Basic, with parameters as follows: Time Slicing: from 2000 JAN to 2022 DEC, Years Per Slice: 1; Term Source: Title, Abstract and Author Keywords (DE); Node Types: Keyword; Burstness with Minimum Duration: 1.
Before 2004, the conopeptide literature frequently mentioned voltage-gated calcium channels and spinal cords (both keywords), underpinning the efforts that would ultimately lead to ziconotide approval. Several conotoxins that block voltage-gated calcium channels were heavily mentioned, including GVIA (keyword with highest strength) and MVIIC (keyword), which are used as pharmacological tools due to their very selective and potent (~1 nM IC50) activity [1,24]. As discussed above, the approval of ziconotide by the FDA in 2004 was a game changer for peptide toxin research. It also created a drive to find better peptide-based alternatives because (i) ziconotide has poor bioavailability and it is required to be delivered intrathecally, and (ii) this drug is linked to a number of undesirable side effects.
After 2004 and until ca. 2010, the most cited keywords in conopeptide publications relate to physiological and pharmacological studies (dorsal root ganglion, brain slice, ion channel, cDNA cloning), indicating that the broader characterization of conopeptide pharmacological activities was taking place. Although voltage-gated calcium channels are the most cited keyword by research articles from the 2000—2022 period (369 articles), three other ion channels targets were also the focus of numerous articles: the nicotinic acetylcholine receptors (keyword in 265 articles), the voltage-gated sodium channel (121 articles), and the voltage-gated potassium channels (77 articles). During the 2004—2010 period, two conopeptides entered clinical development but were dropped: conantokin-G (CGX-1007), which targets the NMDA receptor and was developed to reduce ischaemic damage in stroke [25], and contulakin-G (CGX-1160), which targets the neurotensin receptor and has analgesic properties [26,27]. Conotoxin MrIA (Xen2174), which inhibits the norepinephrine transporter, went into clinical trial for severe cancer pain in 2008 [28] but it was subsequently dropped [29].
From ca. 2010, efforts were made to improve the pharmaceutical properties of conopeptides through peptide engineering, such as backbone cyclization (keyword) [30]. For example, the backbone cyclization of conotoxin Vc1.1 engendered oral activity in an animal model of neuropathic pain [31]. Rather than screening natural peptides, a range of studies focused on rationally designing conopeptide variants by establishing structure–activity relationships (keyword) and studying the three-dimensional structures of the conopeptides and in complex with their target. Most of the three-dimensional structures of conopeptides were determined by nuclear magnetic resonance (keyword) [5,6], and the complexes between conopeptides and molecular targets were principally modelled using homology modelling (keyword) and molecular dynamic simulations (keyword) [32,33,34]. More recently, several experimental structures of complexes between conopeptides and their targets were determined [34]. For example, the complex between ziconotide and voltage-gated calcium channels was recently determined by cryo-electron microscopy [35], and several structures of complexes between conopeptides and the acetylcholine-binding protein, which is a structural surrogate of the nicotinic acetylcholine receptor, were studied by X-ray crystallography [5]. After 2010, neuronal nicotinic acetylcholine receptors (keyword) gained an increase in focus as a pharmaceutical target for conopeptides [7,36]. These receptors are associated with several diseases and conditions, such as addictions, epilepsy, pain, and Alzheimer’s and Parkinson’s diseases [36]. The exquisite selectivity and potency of some conopeptides for certain subtypes of these receptors made them valuable tools in physiological studies that aim to tease out their involvement in these pathologies [7,37]. Through structure–activity relationship (keyword) studies, a range of conopeptide variants that showed greater selectivity and potency were created, generating valuable new probes and potential new drug leads [38]. For example, RgIA4 is a promising analgesic drug candidate to treat chemotherapy-induced neuropathic pain. It was designed by the substitution of 5 out of the 13 positions (38%) of Conus regius conotoxin RgIA [39,40].
Simultaneously as the medicinal chemistry developments took place, the discovery of native conopeptides was accelerated by advances in complementary transcriptomics and proteomics techniques (“mass spectrometry” burst from 2012), rapidly increasing our knowledge in conopeptide natural diversity. These omics studies provided clues on how cone snails generate a massive diversity of toxins, in the range of 1000–2000 per species, from a limited number of genes [41,42,43]. This rapidly growing pool of sequence information was also used to suggest variants for pharmaceutically interesting toxins, therefore supporting conopeptide engineering studies.
Several classification schemes have been developed to describe conopeptides, variously classifying them in terms of their evolutionary relationships (gene superfamilies), activity (pharmaceutical families), disulfide bond numbers and sequence similarity (conopeptides classes), and the pattern of cysteines in their primary sequence (cysteine frameworks are a proxy to describe disulfide bond patterns) [44]. The large amount of sequence and structure data on conopeptides, as well as their evolving nomenclature and classification schemes, prompted the creation of the database ConoServer, the 2012 publication of which is among the top ten field-cited conopeptide publications [8].
Disulfide bonds (keyword) are important because they stabilize peptide three-dimensional structures and are determinants for the overall peptide fold. The importance of the disulfide bonds for conopeptide research is evidenced by the longest citation burst from our analysis, from 2009 until 2022. The structure of proteins and peptides being determinant of their function means that there is in turn a relationship between the disulfide bond pattern and the biological activity of conopeptides [1], albeit conopeptides displaying certain disulfide bond patterns, such as the one forming a cystine knot, is associated with a multiple class of pharmacological targets [44]. There are still large gaps in our knowledge of conopeptides as most of the cysteine frameworks have not yet been structurally studied (17 out of 31 cysteine frameworks) or pharmacologically characterized (19 out of 31 cysteine frameworks).
4. Conclusions
The first description of cone snail envenomation was made in 1848 in the scientific report of the HMS Samarang expedition in the East Indies and Southern China [45]. The venom of cone snails, therefore, has generated curiosity and attracted attention for more than 170 years. Our analysis from the years 2000 to 2022 shows that the field of conopeptides is highly active and is carried out worldwide in a spirit of collaboration. Conopeptides indeed do attract “considerable attention”. Since the approval of ziconotide, a major focus of conopeptide research has been on pharmaceutical applications [46] and the use of conopeptides as molecular probes in neuroscience. With the regain in interest from pharmaceutical companies for peptides and the attractive combination of selectivity and potency, newly discovered or designed conopeptides are promising candidates to undergo clinical development and perhaps will join the >80 peptides that have been approved by the FDA and EMA [47,48]. In terms of peptide discovery, <10% of the total pool of conopeptide sequences have been discovered, and only ~300 (<1% of the total) conopeptides have been pharmacologically characterized. We expect that the recent developments in machine learning, which have already revolutionized structural biology [49], could be applied to characterize conopeptide structures and similar algorithms developed to predict their activity, thus potentially helping to discover new “pharmaceutical treasures”.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21030154/s1, Code S1: “01-retype.py” Reclassifies the publications as “Article” or “Review” according to PubMed; Code S2: “02-rekeywords.py” Modify keywords to increase consistency. The list of keyword changes is provided in Table S1; Code S3: Code S3: “03-authors_replace.py” Modify authors first names to increase consistency. The list of first name changes is provided in Table S2; Table S1: List of keyword changes; Table S2: The list of the authors modifications.
Author Contributions
Conceptualization L.T.T.N., D.J.C. and Q.K.; methodology L.T.T.N. and Q.K.; data curation L.T.T.N. and Q.K., writing L.T.T.N., D.J.C. and Q.K. All authors have read and agreed to the published version of the manuscript.
Funding
Work in DJC’s laboratory on conopeptides is supported by a grant from the Australian NHMRC (GNT2009564) and by the facilities of the Australian Research Council Centre of Excellence for Innovations in Peptide and Protein Science (CE200100012).
Data Availability Statement
The code and data used in this study are provided in the Supplementary Materials.
Acknowledgments
We would like to thank David Adams for providing comments and suggestions on our manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Terlau, H.; Olivera, B.M. Conus Venoms: A Rich Source of Novel Ion Channel-Targeted Peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Olivera, B. Conus Venom Peptides: Reflections from the Biology of Clades and Species. Annu. Rev. Ecol. Syst. 2002, 33, 25–47. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of Separate Predation- and Defence-Evoked Venoms in Carnivorous Cone Snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef]
- Olivera, B.M.; Seger, J.; Horvath, M.P.; Fedosov, A.E. Prey-Capture Strategies of Fish-Hunting Cone Snails: Behavior, Neurobiology and Evolution. Brain. Behav. Evol. 2015, 86, 58–74. [Google Scholar] [CrossRef]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, Synthesis, and Structure-Activity Relationships of Conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.-H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.W.A.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and Biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef] [PubMed]
- Azam, L.; McIntosh, J.M. α-Conotoxins as Pharmacological Probes of Nicotinic Acetylcholine Receptors. Acta Pharmacol. Sin. 2009, 30, 771–783. [Google Scholar] [CrossRef]
- Kaas, Q.; Yu, R.; Jin, A.-H.; Dutertre, S.; Craik, D.J. ConoServer: Updated Content, Knowledge, and Discovery Tools in the Conopeptide Database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef]
- Wang, C.-Z.; Chi, C.-W. Conus Peptides—A Rich Pharmaceutical Treasure. Acta Biochim. Biophys. Sin. 2004, 36, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Bowling, J.J.; Kochanowska, A.J.; Kasanah, N.; Hamann, M.T. Nature’s Bounty—Drug Discovery from the Sea. Expert Opin. Drug Discov. 2007, 2, 1505–1522. [Google Scholar] [CrossRef]
- Wang, F.; Chang, S.; Wei, D. Prediction of Conotoxin Type Based on Long Short-Term Memory Network. Math. Biosci. Eng. MBE 2021, 18, 6700–6708. [Google Scholar] [CrossRef] [PubMed]
- Kohn, A.J.; Saunders, P.R.; Wiener, S. Preliminary Studies on the Venom of the Marine Snail Conus. Ann. New York Acad. Sci. 1960, 90, 706–725. [Google Scholar] [CrossRef] [PubMed]
- Whyte, J.M.; Endean, R. Pharmacological Investigation of the Venoms of the Marine Snails Conus Textile and Conus Geographus. Toxicon 1962, 1, 25–31. [Google Scholar] [CrossRef]
- Olivera, B.M.; Cruz, L.J. Conotoxins, in Retrospect. Toxicon 2001, 39, 7–14. [Google Scholar] [CrossRef]
- Gray, W.R.; Luque, A.; Olivera, B.M.; Barrett, J.; Cruz, L.J. Peptide Toxins from Conus Geographus Venom. J. Biol. Chem. 1981, 256, 4734–4740. [Google Scholar] [CrossRef]
- Olivera, B.M.; Gray, W.R.; Zeikus, R.; McIntosh, J.M.; Varga, J.; Rivier, J.; de Santos, V.; Cruz, L.J. Peptide Neurotoxins from Fish-Hunting Cone Snails. Science 1985, 230, 1338–1343. [Google Scholar] [CrossRef]
- Olivera, B.M.; Rivier, J.; Clark, C.; Ramilo, C.A.; Corpuz, G.P.; Abogadie, F.C.; Mena, E.E.; Woodward, S.R.; Hillyard, D.R.; Cruz, L.J. Diversity of Conus Neuropeptides. Science 1990, 249, 257–263. [Google Scholar] [CrossRef]
- Zhu, X.; Zhou, Y.; Yuan, G.; Shi, J.; Shi, S.; Zhang, L.; Chai, R.; Du, Y.; Duan, C.; Hu, Y. Bibliometric Analysis of Nicotinic Acetylcholine Receptors Channel Research (2000–2020). Channels 2021, 15, 298–309. [Google Scholar] [CrossRef]
- van Eck, N.J.; Waltman, L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef]
- van Eck, N.J.; Waltman, L. Visualizing Bibliometric Networks. In Measuring Scholarly Impact: Methods and Practice; Ding, Y., Rousseau, R., Wolfram, D., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 285–320. ISBN 978-3-319-10377-8. [Google Scholar]
- Chen, C. CiteSpace II: Detecting and Visualizing Emerging Trends and Transient Patterns in Scientific Literature. J. Am. Soc. Inf. Sci. Technol. 2006, 57, 359–377. [Google Scholar] [CrossRef]
- McIntosh, M.; Cruz, L.J.; Hunkapiller, M.W.; Gray, W.R.; Olivera, B.M. Isolation and Structure of a Peptide Toxin from the Marine Snail Conus Magus. Arch. Biochem. Biophys. 1982, 218, 329–334. [Google Scholar] [CrossRef]
- Miljanich, G.P. Ziconotide: Neuronal Calcium Channel Blocker for Treating Severe Chronic Pain. Curr. Med. Chem. 2004, 11, 3029–3040. [Google Scholar] [CrossRef]
- Hillyard, D.R.; Monje, V.D.; Mintz, I.M.; Bean, B.P.; Nadasdi, L.; Ramachandran, J.; Miljanich, G.; Azimi-Zoonooz, A.; McIntosh, J.M.; Cruz, L.J. A New Conus Peptide Ligand for Mammalian Presynaptic Ca2+ Channels. Neuron 1992, 9, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.J.; Ling, G.; McCabe, R.T.; Tortella, F.C. Intrathecal CGX-1007 Is Neuroprotective in a Rat Model of Focal Cerebral Ischemia. Neuroreport 2002, 13, 821–824. [Google Scholar] [CrossRef]
- Han, T.S.; Teichert, R.W.; Olivera, B.M.; Bulaj, G. Conus Venoms—A Rich Source of Peptide-Based Therapeutics. Curr. Pharm. Des. 2008, 14, 2462–2479. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.; Kaas, Q.; Craik, D.J. Hormone-like Conopeptides—New Tools for Pharmaceutical Design. RSC Med. Chem. 2020, 11, 1235–1251. [Google Scholar] [CrossRef]
- Brust, A.; Palant, E.; Croker, D.E.; Colless, B.; Drinkwater, R.; Patterson, B.; Schroeder, C.I.; Wilson, D.; Nielsen, C.K.; Smith, M.T.; et al. χ-Conopeptide Pharmacophore Development: Toward a Novel Class of Norepinephrine Transporter Inhibitor (Xen2174) for Pain. J. Med. Chem. 2009, 52, 6991–7002. [Google Scholar] [CrossRef]
- Okkerse, P.; Hay, J.L.; Sitsen, E.; Dahan, A.; Klaassen, E.; Houghton, W.; Groeneveld, G.J. Pharmacokinetics and Pharmacodynamics of Intrathecally Administered Xen2174, a Synthetic Conopeptide with Norepinephrine Reuptake Inhibitor and Analgesic Properties. Br. J. Clin. Pharmacol. 2017, 83, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.J.; Craik, D.J. Engineering Cyclic Peptide Toxins. Methods Enzymol. 2012, 503, 57–74. [Google Scholar] [CrossRef]
- Clark, R.J.; Jensen, J.; Nevin, S.T.; Callaghan, B.P.; Adams, D.J.; Craik, D.J. The Engineering of an Orally Active Conotoxin for the Treatment of Neuropathic Pain. Angew. Chem. 2010, 49, 6545–6548. [Google Scholar] [CrossRef]
- Mansbach, R.A.; Travers, T.; McMahon, B.H.; Fair, J.M.; Gnanakaran, S. Snails In Silico: A Review of Computational Studies on the Conopeptides. Mar. Drugs 2019, 17, 145. [Google Scholar] [CrossRef]
- Kaas, Q.; Craik, D.J. Bioinformatics-Aided Venomics. Toxins 2015, 7, 2159–2187. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.; Kuyucak, S.; Schroeder, C.I.; Kaas, Q. Modelling the Interactions between Animal Venom Peptides and Membrane Proteins. Neuropharmacology 2017, 127, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yao, X.; Yan, N. Structure of Human Cav2.2 Channel Blocked by the Painkiller Ziconotide. Nature 2021, 596, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Abraham, N.; Lewis, R.J. Neuronal Nicotinic Acetylcholine Receptor Modulators from Cone Snails. Mar. Drugs 2018, 16, 208. [Google Scholar] [CrossRef] [PubMed]
- Giribaldi, J.; Dutertre, S. α-Conotoxins to Explore the Molecular, Physiological and Pathophysiological Functions of Neuronal Nicotinic Acetylcholine Receptors. Neurosci. Lett. 2018, 679, 24–34. [Google Scholar] [CrossRef]
- Turner, M.W.; Marquart, L.A.; Phillips, P.D.; McDougal, O.M. Mutagenesis of α-Conotoxins for Enhancing Activity and Selectivity for Nicotinic Acetylcholine Receptors. Toxins 2019, 11, 113. [Google Scholar] [CrossRef]
- Romero, H.K.; Christensen, S.B.; Mannelli, L.D.C.; Gajewiak, J.; Ramachandra, R.; Elmslie, K.S.; Vetter, D.E.; Ghelardini, C.; Iadonato, S.P.; Mercado, J.L.; et al. Inhibition of α9α10 Nicotinic Acetylcholine Receptors Prevents Chemotherapy-Induced Neuropathic Pain. Proc. Natl. Acad. Sci. USA 2017, 114, E1825–E1832. [Google Scholar] [CrossRef]
- Huynh, P.N.; Christensen, S.B.; McIntosh, J.M. RgIA4 Prevention of Acute Oxaliplatin-Induced Cold Allodynia Requires α9-Containing Nicotinic Acetylcholine Receptors and CD3+ T-Cells. Cells 2022, 11, 3561. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.; Dutertre, S.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic Messiness in the Venom Duct of Conus Miles Contributes to Conotoxin Diversity. Mol. Cell. Proteom. 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.; Kaas, Q.; Jones, A.; Alewood, P.; Lewis, R. Deep Venomics Reveals the Mechanism for Expanded Peptide Diversity in Cone Snail Venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef]
- Davis, J.; Jones, A.; Lewis, R.J. Remarkable Inter- and Intra-Species Complexity of Conotoxins Revealed by LC/MS. Peptides 2009, 30, 1222–1227. [Google Scholar] [CrossRef]
- Kaas, Q.; Westermann, J.-C.; Craik, D.J. Conopeptide Characterization and Classifications: An Analysis Using ConoServer. Toxicon 2010, 55, 1491–1509. [Google Scholar] [CrossRef]
- Adams, A.; Reeve, L.; Gray, J.E.; Richardson, J.; White, A.; Wing, W.; Marryat, F.; Hawkins, B.W.; Benjamin, W.; Sowerby, G.B.; et al. The Zoology of the Voyage of H.M.S. Samarang, under the Command of Captain Sir Edward Belcher, C.B., F.R.A.S., F.G.S., during the Years 1843–1846; Reeve and Benham: London, UK, 1848. [Google Scholar]
- Safavi-Hemami, H.; Brogan, S.E.; Olivera, B.M. Pain Therapeutics from Cone Snail Venoms: From Ziconotide to Novel Non-Opioid Pathways. J. Proteom. 2019, 190, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, B.G.; Albericio, F. Peptide Therapeutics 2.0. Mol. Basel Switz. 2020, 25, 2293. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).