Cryptic Diversity of Black Band Disease Cyanobacteria in Siderastrea siderea Corals Revealed by Chemical Ecology and Comparative Genome-Resolved Metagenomics
Abstract
1. Introduction
2. Results
2.1. Collection of S. siderea-Associated Black Band Disease Cyanobacterial Mats
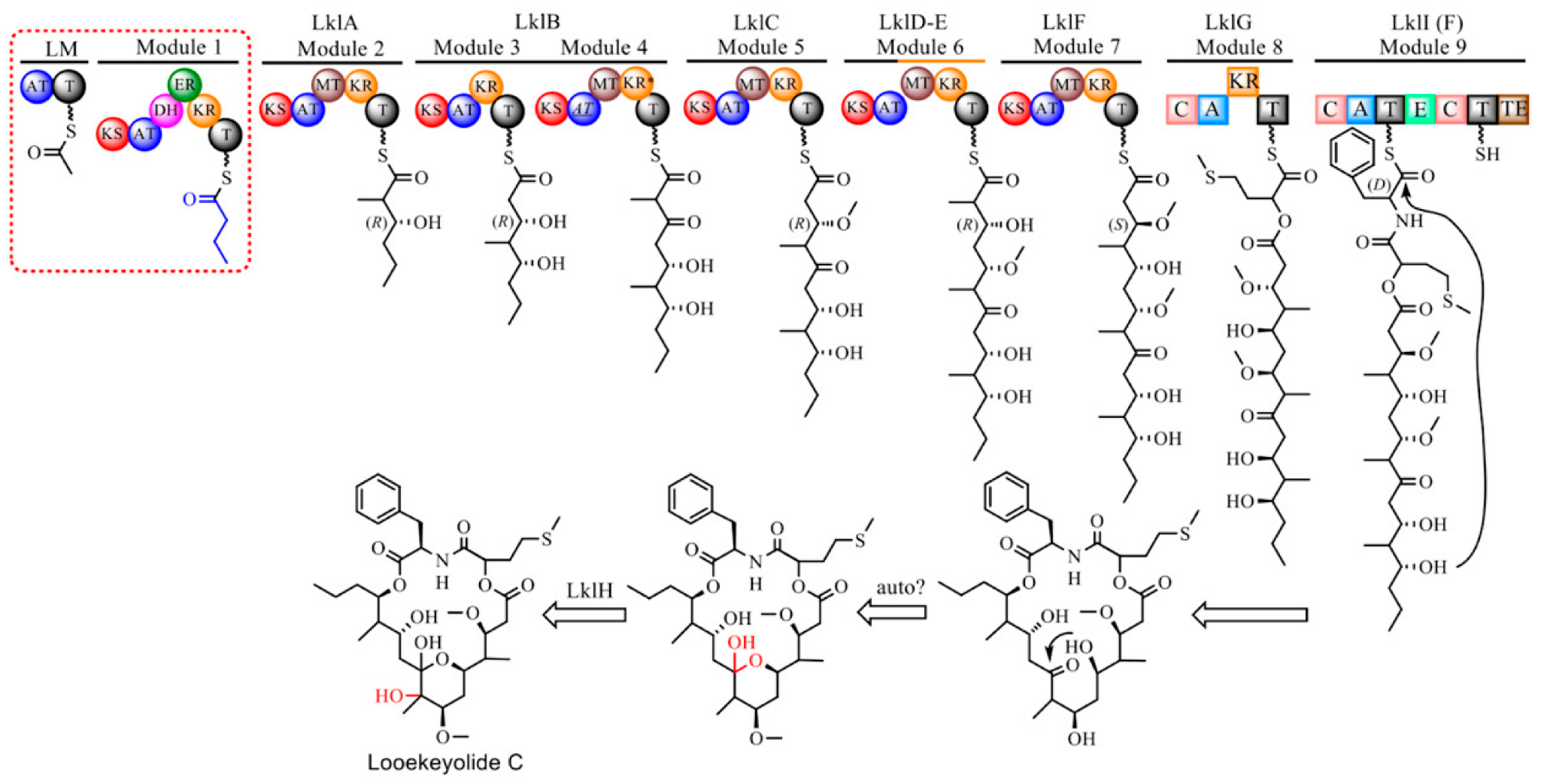
2.2. Isolation and Characterization of Novel Looekeyolides
2.3. Bacterial Composition of Belize Samples
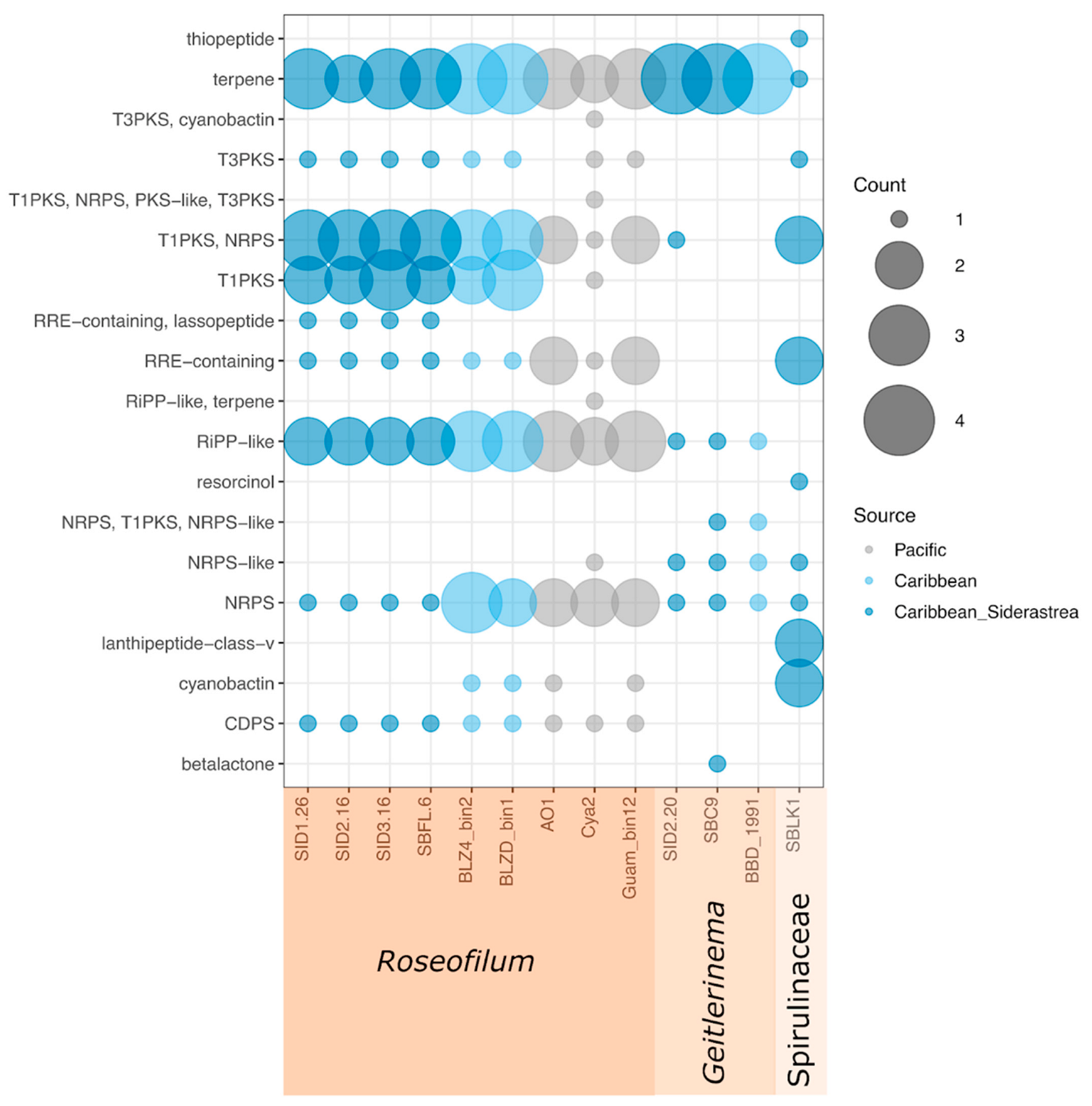
2.4. Metagenome-Assembled Genomes of Cyanobacteria from Belize and Florida
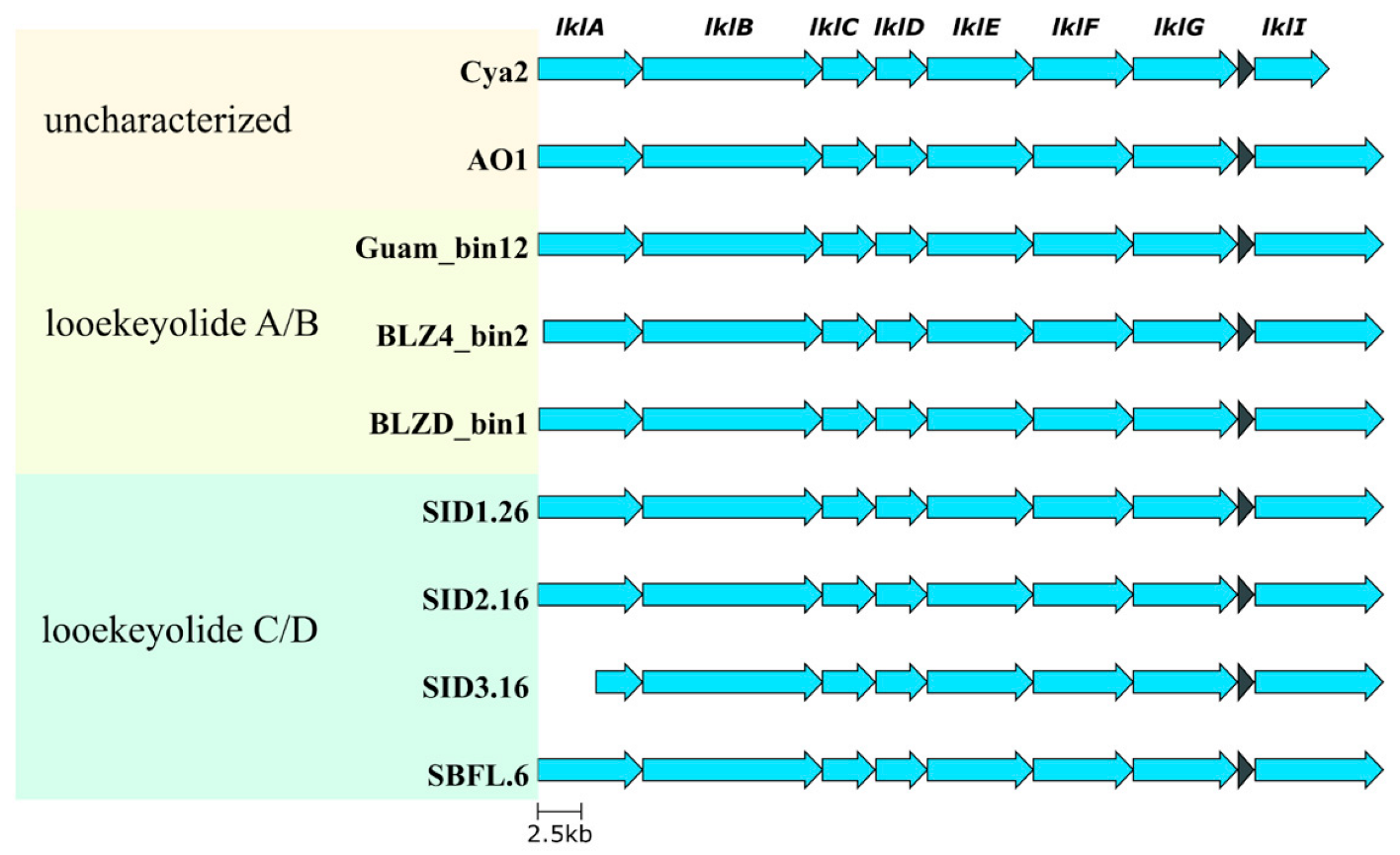
2.5. Comparative Genomics of Black Band Disease-Associated Cyanobacteria
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Enrichment Culturing
4.2. Characterization of Major Secondary Metabolites
4.3. Nucleic Acids Extraction
4.4. V6 Amplicon Libraries of Belize Samples
4.5. Metagenomic Library Preparation
4.6. Metagenomic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leão, T.; Wang, M.; Moss, N.; da Silva, R.; Sanders, J.; Nurk, S.; Gurevich, A.; Humphrey, G.; Reher, R.; Zhu, Q.; et al. A Multi-Omics Characterization of the Natural Product Potential of Tropical Filamentous Marine Cyanobacteria. Mar. Drugs 2021, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The Re-Emergence of Natural Products for Drug Discovery in the Genomics Era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Engene, N.; Gunasekera, S.P.; Gerwick, W.H.; Paul, V.J. Phylogenetic Inferences Reveal a Large Extent of Novel Biodiversity in Chemically Rich Tropical Marine Cyanobacteria. Appl. Environ. Microbiol. 2013, 79, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Yancey, C.E.; Smith, D.J.; Den Uyl, P.A.; Mohamed, O.G.; Yu, F.; Ruberg, S.A.; Chaffin, J.D.; Goodwin, K.D.; Tripathi, A.; Sherman, D.H.; et al. Metagenomic and Metatranscriptomic Insights into Population Diversity of Microcystis Blooms: Spatial and Temporal Dynamics of Mcy Genotypes, Including a Partial Operon That Can Be Abundant and Expressed. Appl. Environ. Microbiol. 2022, 88, e0246421. [Google Scholar] [CrossRef] [PubMed]
- Antonius, A. New Observations on Coral Destruction in Reefs. In Proceedings of the Tenth Meeting of the Association of Island Marine Laboratories of the Caribbean; University of Puerto Rico Mayaguez, Mayagüez, Puerto Rico, 4–7 September 1973; Volume 10. [Google Scholar]
- Roff, G. Earliest Record of a Coral Disease from the Indo-Pacific? Coral Reefs 2016, 35, 457. [Google Scholar] [CrossRef]
- Casamatta, D.; Staníc, D.; Gantar, M.; Richardson, L.L. Characterization of Roseofilum reptotaenium (Oscillatoriales, Cyanobacteria) gen. et sp. nov. isolated from Caribbean black band disease. Phycologia 2012, 51, 489–499. [Google Scholar] [CrossRef]
- Carlton, R.G.; Richardson, L.L. Oxygen and Sulfide Dynamics in a Horizontally Migrating Cyanobacterial Mat: Black Band Disease of Corals. FEMS Microbiol. Ecol. 1995, 18, 155–162. [Google Scholar] [CrossRef]
- Meyer, J.L.; Gunasekera, S.P.; Scott, R.M.; Paul, V.J.; Teplitski, M. Microbiome Shifts and the Inhibition of Quorum Sensing by Black Band Disease Cyanobacteria. ISME J. 2016, 10, 1204–1216. [Google Scholar] [CrossRef]
- Gunasekera, S.P.; Meyer, J.L.; Ding, Y.; Abboud, K.A.; Luo, D.; Campbell, J.E.; Angerhofer, A.; Goodsell, J.L.; Raymundo, L.J.; Liu, J.; et al. Chemical and Metagenomic Studies of the Lethal Black Band Disease of Corals Reveal Two Broadly Distributed, Redox-Sensitive Mixed Polyketide/Peptide Macrocycles. J. Nat. Prod. 2019, 82, 111–121. [Google Scholar] [CrossRef]
- Buerger, P.; Wood-Charlson, E.M.; Weynberg, K.D.; Willis, B.L.; van Oppen, M.J.H. CRISPR-Cas Defense System and Potential Prophages in Cyanobacteria Associated with the Coral Black Band Disease. Front. Microbiol. 2016, 7, 2077. [Google Scholar] [CrossRef]
- Meyer, J.L.; Paul, V.J.; Raymundo, L.J.; Teplitski, M. Comparative Metagenomics of the Polymicrobial Black Band Disease of Corals. Front. Microbiol. 2017, 8, 618. [Google Scholar] [CrossRef] [PubMed]
- Sekar, R.; Kaczmarsky, L.T.; Richardson, L.L. Microbial Community Composition of Black Band Disease on the Coral Host Siderastrea siderea from Three Regions of the Wider Caribbean. Mar. Ecol. Prog. Ser. 2008, 362, 85–98. [Google Scholar] [CrossRef]
- Richardson, L.L.; Sekar, R.; Myers, J.L.; Gantar, M.; Voss, J.D.; Kaczmarsky, L.; Remily, E.R.; Boyer, G.L.; Zimba, P.V. The presence of the cyanobacterial toxin microcystin in black band disease of corals. FEMS Micro. Lett. 2007, 2, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Voss, J.D.; Mills, D.K.; Myers, J.L.; Remily, E.R.; Richardson, L.L. Black Band Disease Microbial Community Variation on Corals in Three Regions of the Wider Caribbean. Microb. Ecol. 2007, 54, 730–739. [Google Scholar] [CrossRef]
- Havermann, S.A.; Foster, J.S. Comparative characterization of the microbial diversities of an artificial microbialite model and a natural stromatolite. Appl. Environ. Micro. 2008, 74, 7410–7421. [Google Scholar] [CrossRef]
- Aeby, G.S.; Work, T.M.; Runyon, C.M.; Shore-Maggio, A.; Ushijima, B.; Videau, P.; Beurmann, S.; Callahan, S.M. First Record of Black Band Disease in the Hawaiian Archipelago: Response, Outbreak Status, Virulence, and a Method of Treatment. PLoS ONE 2015, 10, e0120853. [Google Scholar] [CrossRef]
- Miller, A.W.; Richardson, L.L. A Meta-Analysis of 16S rRNA Gene Clone Libraries from the Polymicrobial Black Band Disease of Corals. FEMS Microbiol. Ecol. 2011, 75, 231–241. [Google Scholar] [CrossRef]
- Sekar, R.; Mills, D.K.; Remily, E.R.; Voss, J.D.; Richardson, L.L. Microbial Communities in the Surface Mucopolysaccharide Layer and the Black Band Microbial Mat of Black Band-Diseased Siderastrea siderea. Appl. Environ. Microbiol. 2006, 72, 5963–5973. [Google Scholar] [CrossRef]
- Den Uyl, P.A.; Richardson, L.L.; Jain, S.; Dick, G.J. Unraveling the Physiological Roles of the Cyanobacterium Geitlerinema sp. BBD and Other Black Band Disease Community Members through Genomic Analysis of a Mixed Culture. PLoS ONE 2016, 11, e0157953. [Google Scholar] [CrossRef]
- Myers, J.L.; Sekar, R.; Richardson, L.L. Molecular Detection and Ecological Significance of the Cyanobacterial Genera Geitlerinema and Leptolyngbya in Black Band Disease of Corals. Appl. Environ. Microbiol. 2007, 73, 5173–5182. [Google Scholar] [CrossRef]
- Sato, Y.; Civiello, M.; Bell, S.C.; Willis, B.L.; Bourne, D.G. Integrated Approach to Understanding the Onset and Pathogenesis of Black Band Disease in Corals: Integrated Approaches to Understand BBD Aetiology. Environ. Microbiol. 2016, 18, 752–765. [Google Scholar] [CrossRef]
- Buerger, P.; Alvarez-Roa, C.; Weynberg, K.D.; Baekelandt, S.; van Oppen, M.J.H. Genetic, Morphological and Growth Characterisation of a New Roseofilum Strain (Oscillatoriales, Cyanobacteria) Associated with Coral Black Band Disease. PeerJ 2016, 4, e2110. [Google Scholar] [CrossRef] [PubMed]
- Gondry, M.; Jacques, I.B.; Thai, R.; Babin, M.; Canu, N.; Seguin, J.; Belin, P.; Pernodet, J.-L.; Moutiez, M. A Comprehensive Overview of the Cyclodipeptide Synthase Family Enriched with the Characterization of 32 New Enzymes. Front. Microbiol. 2018, 9, 46. [Google Scholar] [CrossRef]
- Yao, T.; Liu, J.; Liu, Z.; Li, T.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Gu, Q.; Li, W. Genome Mining of Cyclodipeptide Synthases Unravels Unusual tRNA-Dependent Diketopiperazine-Terpene Biosynthetic Machinery. Nat. Commun. 2018, 9, 4091. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.C.; Teplitski, M.; Gunasekera, S.P.; Paul, V.J.; Luesch, H. Isolation and Biological Evaluation of 8-Epi-Malyngamide C from the Floridian Marine Cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2010, 73, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Morais, J.; Cardoso, A.P.L.R.; Santos, B.A. A Global Synthesis of the Current Knowledge on the Taxonomic and Geographic Distribution of Major Coral Diseases. Environ. Adv. 2022, 8, 100231. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic Insights That Advance the Species Definition for Prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Olm, M.R.; Crits-Christoph, A.; Diamond, S.; Lavy, A.; Matheus Carnevali, P.B.; Banfield, J.F. Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries. MSystems 2020, 5, e00731-19. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a Comprehensive Public Database of Secondary Metabolites from Cyanobacteria. Water Res. 2021, 196, 117017. [Google Scholar] [CrossRef] [PubMed]
- Leão, P.N.; Engene, N.; Antunes, A.; Gerwick, W.H.; Vasconcelos, V. The Chemical Ecology of Cyanobacteria. Nat. Prod. Rep. 2012, 29, 372–391. [Google Scholar] [CrossRef] [PubMed]
- Kar, J.; Ramrao, D.P.; Zomuansangi, R.; Lalbiaktluangi, C.; Singh, S.M.; Joshi, N.C.; Kumar, A.; Kaushalendra; Mehta, S.; Yadav, M.K.; et al. Revisiting the Role of Cyanobacteria-Derived Metabolites as Antimicrobial Agent: A 21st Century Perspective. Front. Microbiol. 2022, 13, 1034471. [Google Scholar] [CrossRef] [PubMed]
- Carpine, R.; Sieber, S. Antibacterial and Antiviral Metabolites from Cyanobacteria: Their Application and Their Impact on Human Health. Curr. Res. Biotechnol. 2021, 3, 65–81. [Google Scholar] [CrossRef]
- Rojas, V.; Rivas, L.; Cárdenas, C.; Guzmán, F. Cyanobacteria and Eukaryotic Microalgae as Emerging Sources of Antibacterial Peptides. Molecules 2020, 25, 5804. [Google Scholar] [CrossRef]
- Curren, E.; Leaw, C.P.; Lim, P.T.; Leong, S.C.Y. The Toxic Cosmopolitan Cyanobacteria Moorena producens: Insights into Distribution, Ecophysiology and Toxicity. Environ. Sci. Pollut. Res. Int. 2022, 29, 78178–78206. [Google Scholar] [CrossRef]
- Dussault, D.; Vu, K.D.; Vansach, T.; Horgen, F.D.; Lacroix, M. Antimicrobial Effects of Marine Algal Extracts and Cyanobacterial Pure Compounds against Five Foodborne Pathogens. Food Chem. 2016, 199, 114–118. [Google Scholar] [CrossRef]
- Gondry, M.; Sauguet, L.; Belin, P.; Thai, R.; Amouroux, R.; Tellier, C.; Tuphile, K.; Jacquet, M.; Braud, S.; Courçon, M.; et al. Cyclodipeptide Synthases Are a Family of tRNA-Dependent Peptide Bond-Forming Enzymes. Nat. Chem. Biol. 2009, 5, 414–420. [Google Scholar] [CrossRef]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A. A New Genome-Mining Tool Redefines the Lasso Peptide Biosynthetic Landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Wang, M.; Fage, C.D.; He, Y.; Mi, J.; Yang, Y.; Li, F.; An, X.; Fan, H.; Song, L.; Zhu, S.; et al. Recent Advances and Perspectives on Expanding the Chemical Diversity of Lasso Peptides. Front. Bioeng. Biotechnol. 2021, 9, 741364. [Google Scholar] [CrossRef]
- Moss, N.A.; Leão, T.; Rankin, M.R.; McCullough, T.M.; Qu, P.; Korobeynikov, A.; Smith, J.L.; Gerwick, L.; Gerwick, W.H. Ketoreductase domain dysfunction expands chemodiversity: Malyngamide biosynthesis in the cyanobacterium Okeania hirsuta. ACS Chem. Biol. 2018, 13, 3385–3395. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Kretsch, A.M.; Daigh, L.M.; Burk, M.J.; Mitchell, D.A. Cell-free biosynthesis to evaluate lasso peptide formation and enzyme-substrate tolerance. J. Am. Chem. Soc. 2021, 15, 5917–5927. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L. A Filtering Method to Generate High Quality Short Reads Using Illumina Paired-End Technology. PLoS ONE 2013, 8, e66643. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Joshi, N. Sickle: A Sliding-Window, Adaptive, Quality—Based Trimming Tool for FastQ Files; UC Davis Bioinformatics Core: Davis, CA, USA, 2011. [Google Scholar]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9783319242774. [Google Scholar]
- Minoche, A.E.; Dohm, J.C.; Himmelbauer, H. Evaluation of Genomic High-Throughput Sequencing Data Generated on Illumina HiSeq and Genome Analyzer Systems. Genome Biol. 2011, 12, R112. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A Fast and Scalable Metagenome Assembler Driven by Advanced Methodologies and Community Practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an Efficient Tool for Accurately Reconstructing Single Genomes from Complex Microbial Communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) Webserver: Taxonomic and Gene Diversity Analysis of Archaea and Bacteria at the Whole Genome Level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The Enveomics Collection: A Toolbox for Specialized Analyses of Microbial Genomes and Metagenomes; PeerJ Preprints: London, UK, 2016. [Google Scholar]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An Interactive Viewer for Bacterial Population Genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive Prediction of Secondary Metabolite Structure and Biological Activity from Microbial Genome Sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Inkscape. Inkscape Project. 2020. Available online: https://inkscape.org (accessed on 22 December 2022).
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
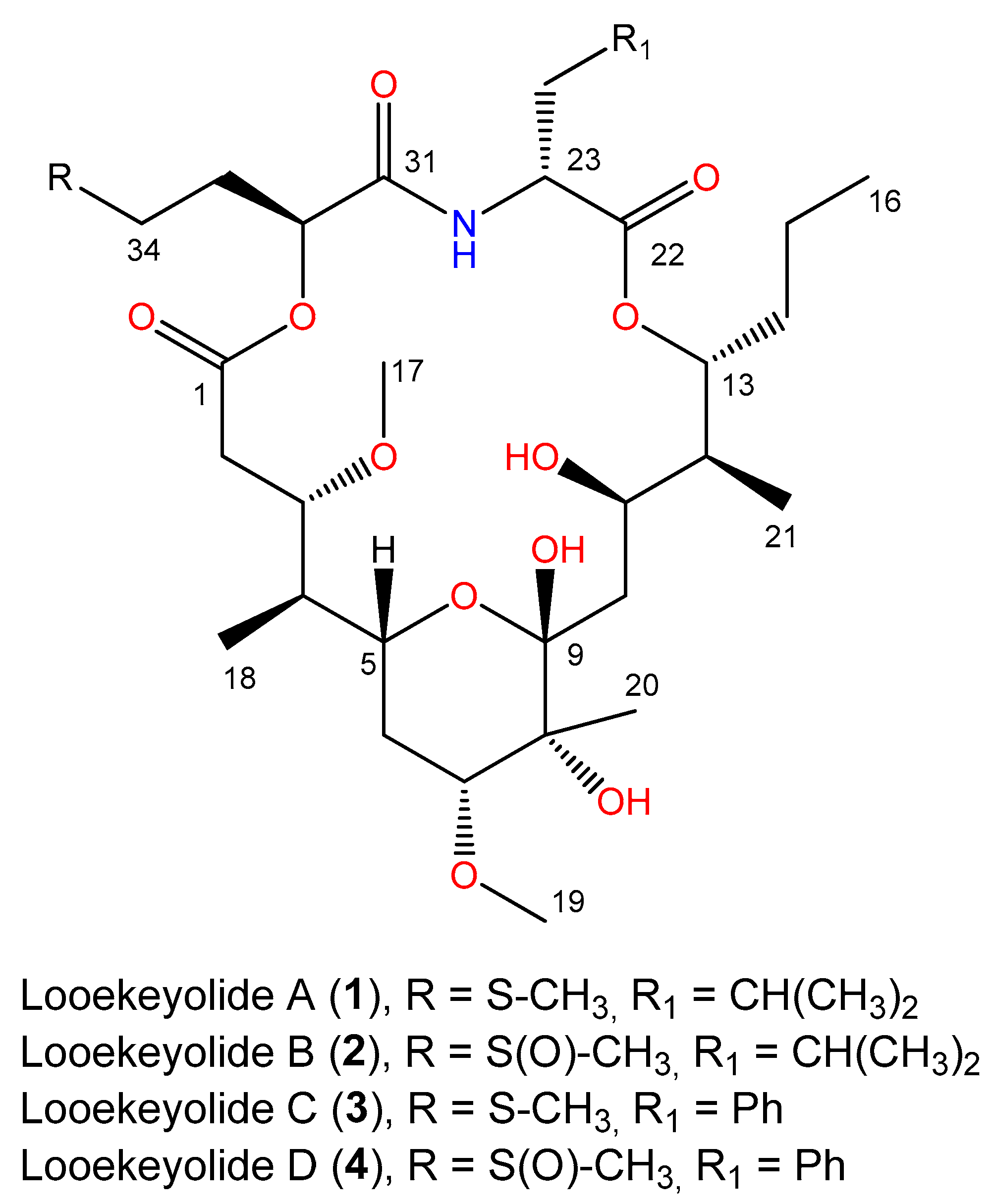



| Position | δC Mult. | δH (J in Hz) | COSY a | HMBC | NOESY b |
|---|---|---|---|---|---|
| 1 | 173.4, C | 2a | |||
| 2a | 33.6, CH2 | 2.52, d (12.0) | 2b, | 2b, 3, 5 | |
| 2b | 2.36, dd (12.0, 10.3) | 2a, 3 | 2a, 3 | ||
| 3 | 77.6, CH | 4.34, dd (10.3, 4.1) | 2b, 4 | 2, 18 | 2a, 2b, 4, 11 |
| 4 | 38.7, CH | 2.21, m | 3, 5, 18 | 6b, 18 | 3, 17, 18 |
| 5 | 70.8, CH | 3.60, ddd (11.0, 11.0, 2.0) | 4, 6a, 6b | 6b, 18 | 2a, 6a, 18 |
| 6a | 31.4, CH2 | 2.08, ddd (12.1, 11.0, 4.8) | 5, 6b, 7 | 5, 6b, 7, 18 | |
| 6b | 1.42, ddd (12.1, 12.0, 1.8) | 5, 6a, | 6a | ||
| 7 | 80.0, CH | 3.44, dd (12.0, 4.8) | 6a, 6b | 6a, 6b, 20 | 6a, 10a, 20 |
| 8 | 74.7, C | 6a, 6b, 20 | |||
| 9 | 102.0, C | 10b, 20 | |||
| 10a | 37.9, CH2 | 1.90, ddd (12.3, 11.0) | 10b, 11 | 7, 10b, 11, 20, 21 | |
| 10b | 1.69, dd (12.3, 2.7) | 10a, 11 | 10a, 11, 12 | ||
| 11 | 66.7, CH | 4.70, ddd (11.0, 1.7, 1.0) | 10a, 10b, 12 | 10a, 12, 21 | 3, 10ab, 14a, 14b |
| 12 | 42.6, CH | 1.52, ddq (7.1, 1.0, 1.0) | 11, 13, 21 | 21 | 10b, 13, 21 |
| 13 | 81.4, CH | 4.99, ddd, (9.6, 6.1, 1.0) | 12, 14a, 14b | 21 | 12, 21 |
| 14a | 35.4, CH2 | 2.15, m | 13, 14b, 15a, 15b | 12, 16 | 14b, 15b |
| 14b | 1.59, m | 13, 14a, 15a, 15b | 14a | ||
| 15a | 20.3, CH2 | 1.36, m | 14a, 14b, 15b, 16 | 16 | 15b, 16 |
| 15b | 1.29, m | 14a, 14b, 15a, 16 | 15a, 16 | ||
| 16 | 14.3, CH3 | 0.93, t (7.5) | 15a, 15b | 15a, 15b | |
| 17 | 57.4, OCH3 | 3.32, s | 4 | ||
| 18 | 9.7, CH3 | 0.81, d (6.9) | 4 | 4, 5, 6a | |
| 19 | 57.7, OCH3 | 3.38, s | 20 | ||
| 20 | 19.6, CH3 | 1.25, s | 7 | 7, 10a, 19 | |
| 21 | 11.9, CH3 | 0.98, d (6.8) | 12 | 12, 13, 10a | |
| 22 | 175.1, C | ||||
| 23 | 53.6, CH | 4.81, dd (10.8, 4.2) | 24a, 24b | 24a, 24b | |
| 24a | 36.0, CH2 | 3.39, m | 23, 24b | 26, 30 | 24b |
| 24b | 2.82, m | 23, 24a | 24a | ||
| 25 | 138.9, C | 27, 29 | |||
| 26 | 130.0, CH | 7.23, d (6.8) | 27 | 28 | 27 |
| 27 | 129.7, CH | 7.27, t (6.8) | 26, 28 | 26, 28 | |
| 28 | 127.9, CH | 7.19, t (6.8) | 27, 29 | 26, 30 | 27, 29 |
| 29 | 129.7, CH | 7.27, t (6.8) | 28, 30 | 28, 30 | |
| 30 | 130.0, CH | 7.23, d (6.8) | 29 | 28 | 29 |
| 31 | 171.7, C | ||||
| 32 | 73.6, CH | 4.86, dd (8.9, 4.0) | 33 | 33 | 33a, 33b |
| 33 | 25.4, CH2 | 1.83, m | 32, 34a, 34b | 32, 34 | 32, 34 |
| 34a | 48.9, CH2 | 2.58, m | 33, 34b | 33, 35 | 34b |
| 34b | 2.46, m | 33, 34a | 34a | ||
| 35 | 37.3, CH3 | 2.508, 2.501, s | 34 |
| ASV Name | SILVA Classification | Closest BLAST Match (% Similarity for V6 Region) |
|---|---|---|
| Cyano 1 | Roseofilum AO1-A | EF123646 (100%) |
| Cyano 2 | Geitlerinema PCC-7105 | EF110974 (100%), EF372580 (100%) |
| Cyano 3 | Hormoscilla SI04-45 | KY697267 (100%) |
| Cyano 4 | Hormoscilla SI04-45 | KY697265 (100%) |
| Beggiatoa | unclassified genus of Bacteria | KM924160 (96.7%) |
| Ruegeria | Ruegeria | MT484146 (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer, J.L.; Gunasekera, S.P.; Brown, A.L.; Ding, Y.; Miller, S.; Teplitski, M.; Paul, V.J. Cryptic Diversity of Black Band Disease Cyanobacteria in Siderastrea siderea Corals Revealed by Chemical Ecology and Comparative Genome-Resolved Metagenomics. Mar. Drugs 2023, 21, 76. https://doi.org/10.3390/md21020076
Meyer JL, Gunasekera SP, Brown AL, Ding Y, Miller S, Teplitski M, Paul VJ. Cryptic Diversity of Black Band Disease Cyanobacteria in Siderastrea siderea Corals Revealed by Chemical Ecology and Comparative Genome-Resolved Metagenomics. Marine Drugs. 2023; 21(2):76. https://doi.org/10.3390/md21020076
Chicago/Turabian StyleMeyer, Julie L., Sarath P. Gunasekera, Anya L. Brown, Yousong Ding, Stephanie Miller, Max Teplitski, and Valerie J. Paul. 2023. "Cryptic Diversity of Black Band Disease Cyanobacteria in Siderastrea siderea Corals Revealed by Chemical Ecology and Comparative Genome-Resolved Metagenomics" Marine Drugs 21, no. 2: 76. https://doi.org/10.3390/md21020076
APA StyleMeyer, J. L., Gunasekera, S. P., Brown, A. L., Ding, Y., Miller, S., Teplitski, M., & Paul, V. J. (2023). Cryptic Diversity of Black Band Disease Cyanobacteria in Siderastrea siderea Corals Revealed by Chemical Ecology and Comparative Genome-Resolved Metagenomics. Marine Drugs, 21(2), 76. https://doi.org/10.3390/md21020076






