Spatial Structure of Lectin from the Mussel Mytilus trossulus: In-Sights from Molecular Modelling and Practical Proof
Abstract
1. Introduction
2. Results
2.1. Multiple Alignments of Mytilectin Family Lectins
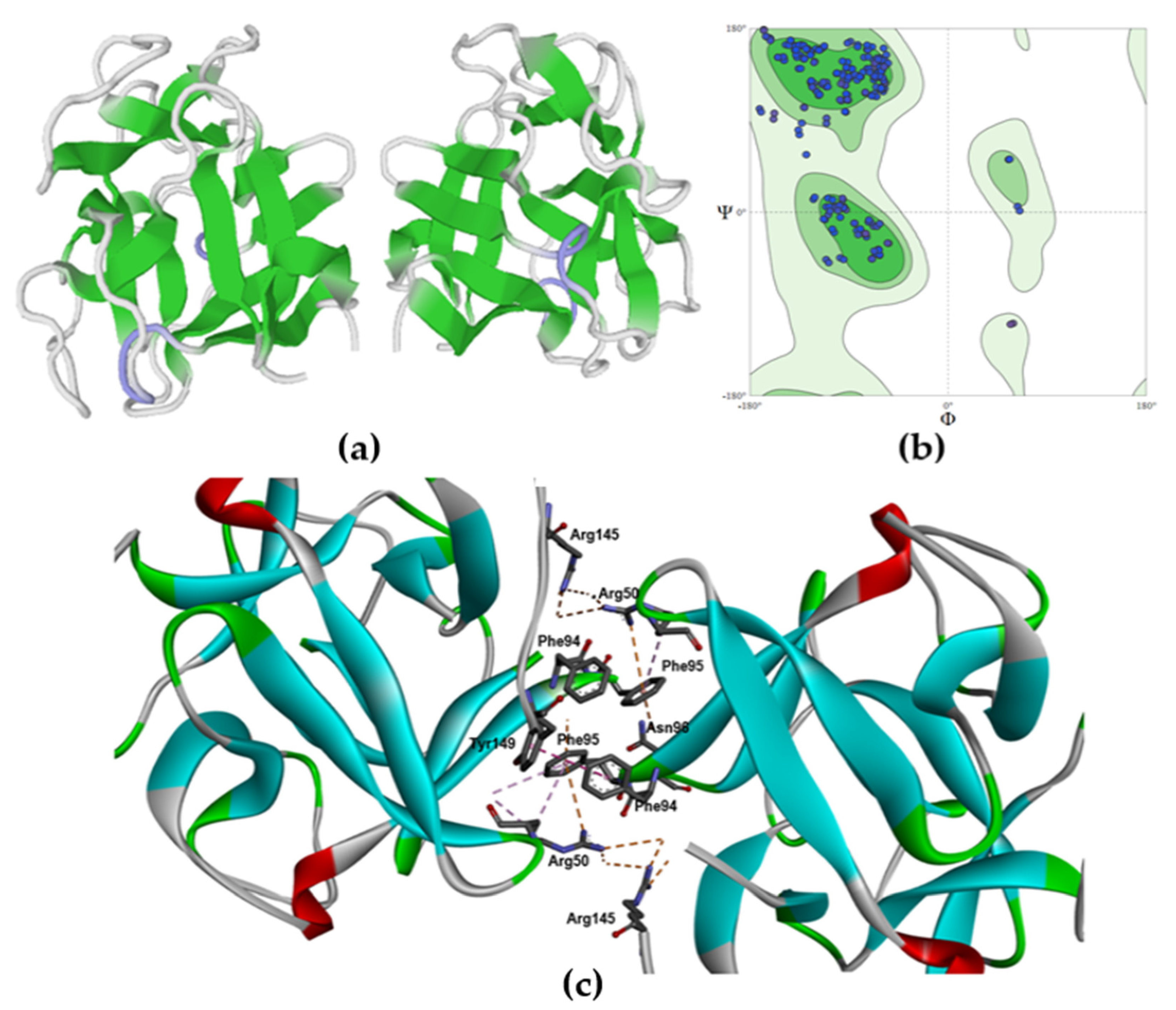
2.2. Molecular Modeling of the MTL Structure
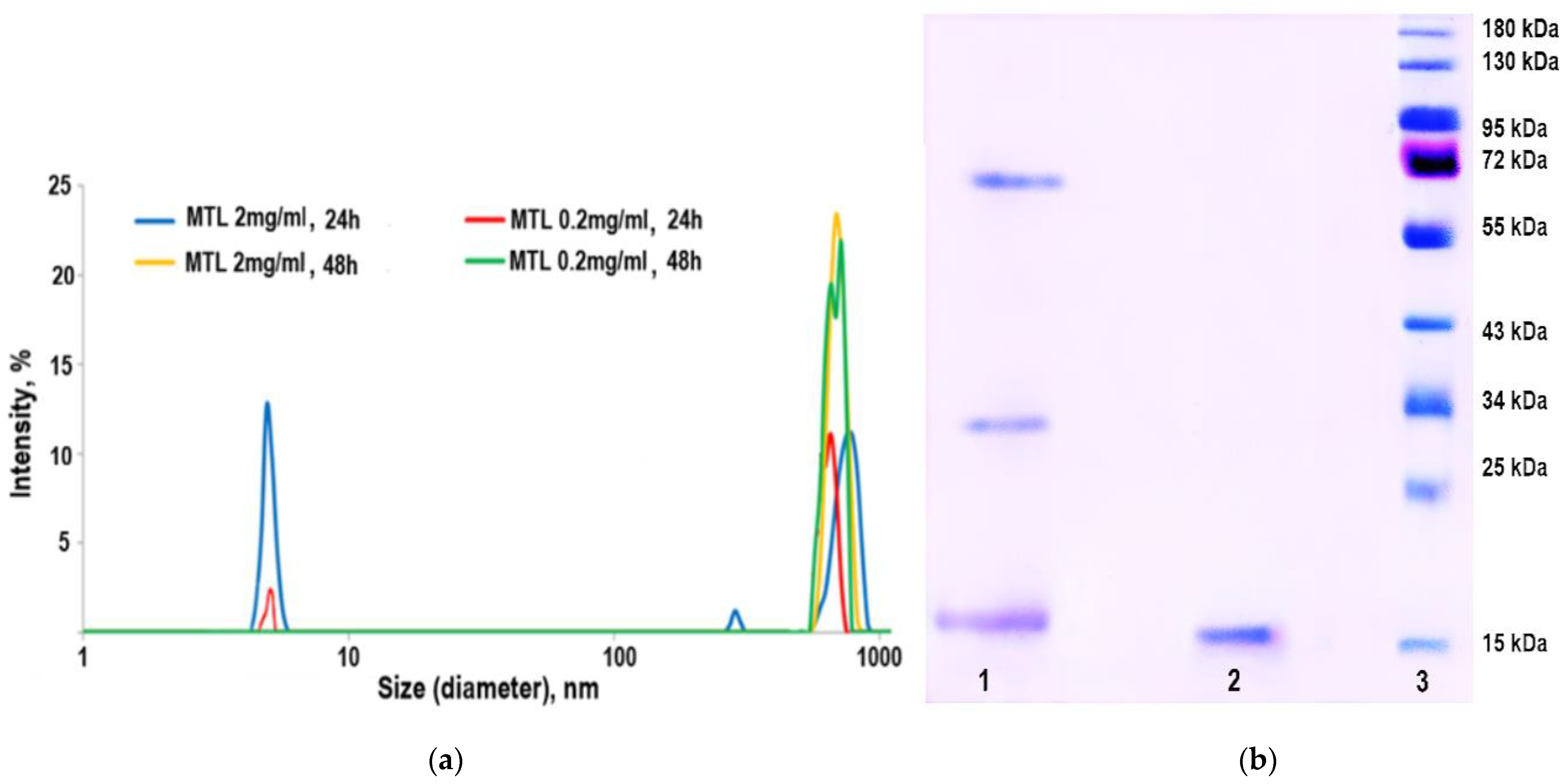
2.3. Practical Proof of MTL Oligomerization
3. Discussion
4. Materials and Methods
4.1. MTL Purification
4.2. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.3. Dynamic Light Scattering Method
4.4. Multiple Alignment Analysis of Amino Acid Sequences of Lectins of the Mytilectin Family
4.5. In Silico Analysis and Quaternary Structure Prediction
4.6. Molecular Modeling of the MTL Structure
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lis, H.; Sharon, N. Lectins: Carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 1998, 98, 637–674. [Google Scholar] [CrossRef] [PubMed]
- Chettri, D.; Boro, M.; Sarkar, L.; Verma, A.K. Lectins: Biological significance to biotechnological application. Carbohydr. Res. 2021, 506, 108367. [Google Scholar] [CrossRef] [PubMed]
- Vasta, G.R.; Ahmed, H.; Feng, C.; Saito, K.; Tasumi, S.; Odom, E.W. Lectin repertoires in invertebrates and ectothermic vertebrates: Structural and functional aspects. In Comprehensive Glycoscience; Elsevier: Amsterdam, The Netherlands, 2021; pp. 74–92. [Google Scholar] [CrossRef]
- Vasta, G.R.; Wang, J.-X. Galectin-mediated immune recognition: Opsonic roles with contrasting outcomes in selected shrimp and bivalve mollusk species. Dev. Comp. Immunol. 2020, 110, 103721. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.; Lu, D.; Ballesteros, A.; Blois, S.M.; Abernathy, K.; Feng, C.; Dimitroff, C.J.; Zmuda, J.; Panico, M.; Dell, A.; et al. Glycan characterization of pregnancy-specific glycoprotein 1 and its identification as a novel galectin-1 ligand. Glycobiology 2020, 30, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Vasta, G.R.; Wang, J.-X. The functional relevance of shrimp C-type lectins in host-pathogen interactions. Dev. Comp. Immunol. 2020, 109, 103708. [Google Scholar] [CrossRef]
- Ernst, B.; Magnani, J.L. From carbohydrate leads to glycomimetic drugs. Nat. Rev. Drug Discov. 2009, 8, 661–677. [Google Scholar] [CrossRef]
- Kovalchuk, S.N.; Golotin, V.A.; Balabanova, L.A.; Buinovskaya, N.S.; Likhatskaya, G.N.; Rasskazov, V.A. Carbohydrate-binding motifs in a novel type lectin from the sea mussel Crenomytilus grayanus: Homology modeling study and site-specific mutagenesis. Fish Shellfish Immunol. 2015, 47, 565–571. [Google Scholar] [CrossRef]
- Holmskov, U.; Malhotra, R.; Sim, R.B.; Jensenius, J.C. Collectins: Collagenous C-type lectins of the innate immune defense system. Immunol. Today 1994, 15, 67–74. [Google Scholar] [CrossRef]
- Hasan, I.; Gerdol, M.; Fujii, Y.; Rajia, S.; Koide, Y.; Yamamoto, D.; Kawsar, S.; Ozeki, Y. cDNA and gene structure of MytiLec-1, a bacteriostatic R-type lectin from the Mediterranean mussel (Mytilus galloprovincialis). Mar. Drugs 2016, 14, 92. [Google Scholar] [CrossRef]
- García-Maldonado, E.; Cano-Sánchez, P.; Hernández-Santoyo, A. Molecular and functional characterization of a glycosylated galactose-binding lectin from Mytilus californianus. Fish Shellfish Immunol. 2017, 66, 564–574. [Google Scholar] [CrossRef]
- Chikalovets, I.; Filshtein, A.; Molchanova, V.; Mizgina, T.; Lukyanov, P.; Nedashkovskaya, O.; Hua, K.-F.; Chernikov, O. Activity dependence of a novel lectin family on structure and carbohydrate-binding properties. Molecules 2019, 25, 150. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.-H.; Chien, C.-T.H.; Wu, H.-Y.; Huang, K.-F.; Wang, I.; Ho, M.-R.; Tu, I.-F.; Lee, I.-M.; Li, W.; Shih, Y.-L.; et al. A multivalent marine lectin from Crenomytilus grayanus possesses anti-cancer activity through recognizing globotriose Gb3. J. Am. Chem. Soc. 2016, 138, 4787–4795. [Google Scholar] [CrossRef] [PubMed]
- Terada, D.; Voet, A.R.D.; Noguchi, H.; Kamata, K.; Ohki, M.; Addy, C.; Fujii, Y.; Yamamoto, D.; Ozeki, Y.; Tame, J.R.H.; et al. Computational design of a symmetrical β-trefoil lectin with cancer cell binding activity. Sci. Rep. 2017, 7, 5943. [Google Scholar] [CrossRef]
- Chikalovets, I.V.; Kovalchuk, S.N.; Litovchenko, A.P.; Molchanova, V.I.; Pivkin, M.V.; Chernikov, O.V. A new Gal/GalNAc-specific lectin from the mussel Mytilus trossulus: Structure, tissue specificity, antimicrobial and antifungal activity. Fish Shellfish Immunol. 2016, 50, 27–33. [Google Scholar] [CrossRef]
- Terada, D.; Kawai, F.; Noguchi, H.; Unzai, S.; Hasan, I.; Fujii, Y.; Park, S.-Y.; Ozeki, Y.; Tame, J.R.H. Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types. Sci. Rep. 2016, 6, 28344. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Tauriello, G.; Bienert, S.; Biasini, M.; Johner, N.; Schwede, T. ProMod3—A versatile homology modelling toolbox. PLoS Comput. Biol. 2021, 17, e1008667. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—Distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 2647. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Andreis, C.; Clauwaert, J. Photon correlation spectroscopy and light scattering of eye lens proteins at high concentrations. Biophys. J. 1985, 47, 591–605. [Google Scholar] [CrossRef]
- Chikalovets, I.V.; Kondrashina, A.S.; Chernikov, O.V.; Molchanova, V.I.; Luk’yanov, P.A. Isolation and general characteristics of lectin from the mussel Mytilus trossulus. Chem. Nat. Compd. 2013, 48, 1058–1061. [Google Scholar] [CrossRef]
- Fujii, Y.; Dohmae, N.; Takio, K.; Kawsar, S.M.A.; Matsumoto, R.; Hasan, I.; Koide, Y.; Kanaly, R.A.; Yasumitsu, H.; Ogawa, Y.; et al. A lectin from the mussel Mytilus galloprovincialis has a highly novel primary structure and induces glycan-mediated cytotoxicity of globotriaosylceramide-expressing lymphoma cells. J. Biol. Chem. 2012, 287, 44772–44783. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, S.N.; Chikalovets, I.V.; Chernikov, O.V.; Molchanova, V.I.; Li, W.; Rasskazov, V.A.; Lukyanov, P.A. cDNA cloning and structural characterization of a lectin from the mussel Crenomytilus grayanus with a unique amino acid sequence and antibacterial activity. Fish Shellfish Immunol. 2013, 35, 1320–1324. [Google Scholar] [CrossRef] [PubMed]
- Notova, S.; Bonnardel, F.; Lisacek, F.; Varrot, A.; Imberty, A. Structure and engineering of tandem repeat lectins. Curr. Opin. Struct. Biol. 2020, 62, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Tosatto, S.C.E.; Schomburg, D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins Struct. Funct. Bioinforma. 2008, 71, 261–277. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]

| Template (PDB ID) | Seq Identity | Oligo-state | QMEANDisCo | Description |
|---|---|---|---|---|
| 6bfm.1.A | 84.00 | homo-dimer | 0.89 ± 0.05 | M. californianus lectin |
| 6bfm.1.B | 84.00 | homo-dimer | 0.89 ± 0.05 | M. californianus lectin |
| 3wmu.1.A | 83.78 | homo-dimer | 0.90 ± 0.05 | M. galloprovincialis lectin |
| 3wmu.1.B | 83.78 | homo-dimer | 0.90 ± 0.05 | M. galloprovincialis lectin |
| 5f8s.1.B | 83.33 | homo-dimer | 0.90 ± 0.05 | C. grayanus lectin |
| 5duy.1.A | 83.33 | monomer | 0.89 ± 0.07 | C. grayanus lectin |
| 5vbk.1.A | 84.00 | monomer | 0.88 ± 0.07 | M. californianus lectin |
| 5xg5.1.A | 56.62 | monomer | 0.88 ± 0.07 | MITSUBA-1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filshtein, A.P.; Chikalovets, I.V.; Mizgina, T.O.; Lukyanov, P.A.; Hua, K.-F.; Chernikov, O.V. Spatial Structure of Lectin from the Mussel Mytilus trossulus: In-Sights from Molecular Modelling and Practical Proof. Mar. Drugs 2023, 21, 10. https://doi.org/10.3390/md21010010
Filshtein AP, Chikalovets IV, Mizgina TO, Lukyanov PA, Hua K-F, Chernikov OV. Spatial Structure of Lectin from the Mussel Mytilus trossulus: In-Sights from Molecular Modelling and Practical Proof. Marine Drugs. 2023; 21(1):10. https://doi.org/10.3390/md21010010
Chicago/Turabian StyleFilshtein, Alina P., Irina V. Chikalovets, Tatyana O. Mizgina, Pavel A. Lukyanov, Kuo-Feng Hua, and Oleg V. Chernikov. 2023. "Spatial Structure of Lectin from the Mussel Mytilus trossulus: In-Sights from Molecular Modelling and Practical Proof" Marine Drugs 21, no. 1: 10. https://doi.org/10.3390/md21010010
APA StyleFilshtein, A. P., Chikalovets, I. V., Mizgina, T. O., Lukyanov, P. A., Hua, K.-F., & Chernikov, O. V. (2023). Spatial Structure of Lectin from the Mussel Mytilus trossulus: In-Sights from Molecular Modelling and Practical Proof. Marine Drugs, 21(1), 10. https://doi.org/10.3390/md21010010








