Ion Mobility-Derived Collision Cross-Sections Add Extra Capability in Distinguishing Isomers and Compounds with Similar Retention Times: The Case of Aphidicolanes
Abstract
:1. Introduction
2. Results and Discussion
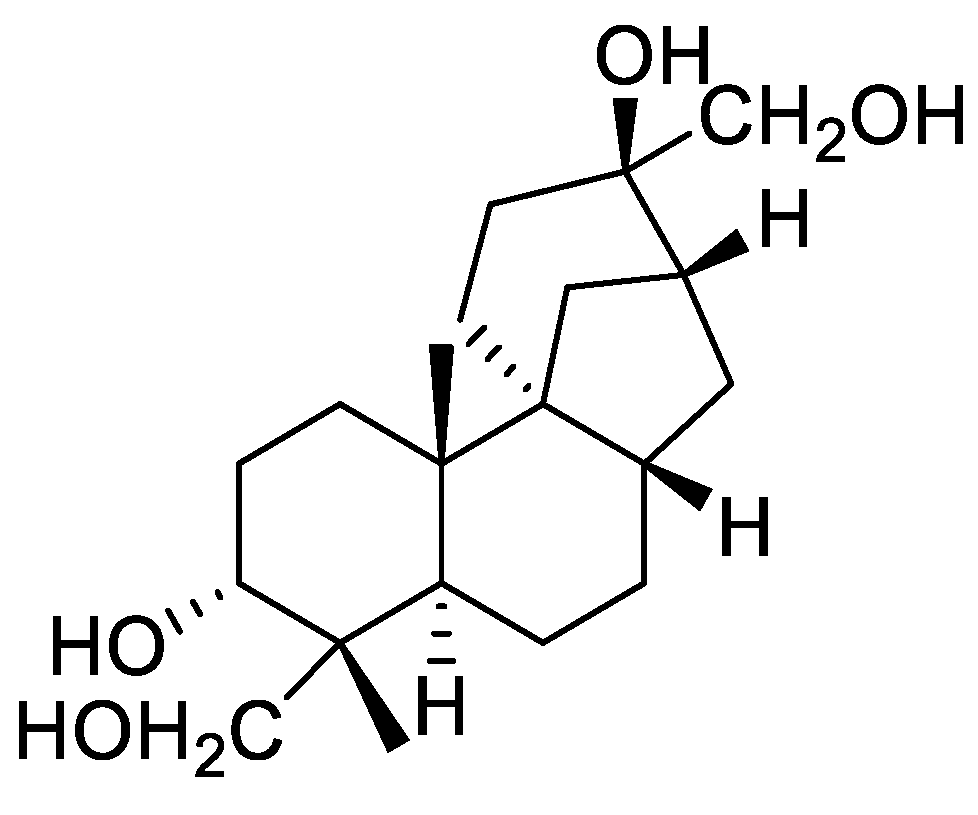
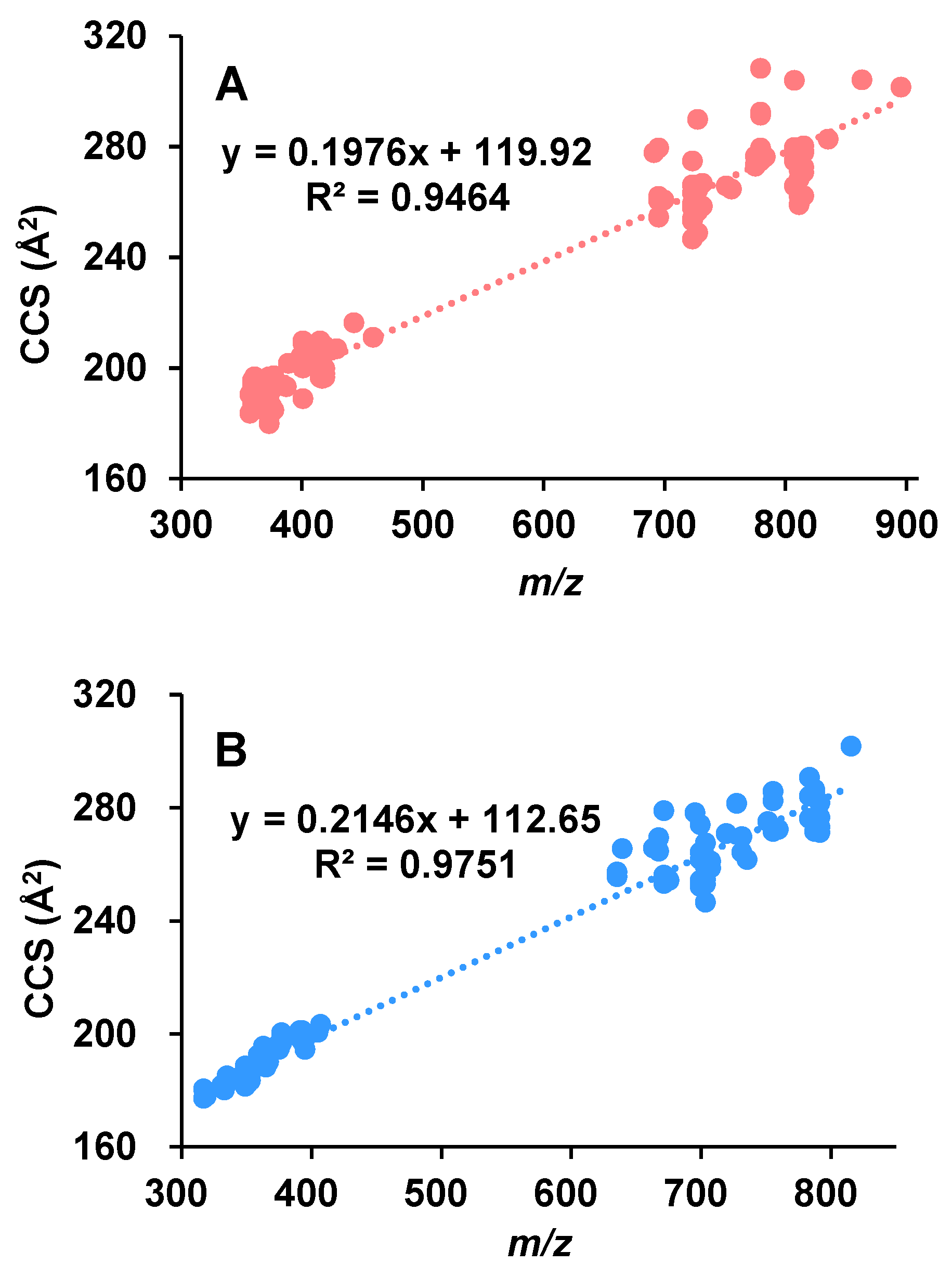
2.1. Ion Mobility-Derived CCS Values for Aphidicolanes
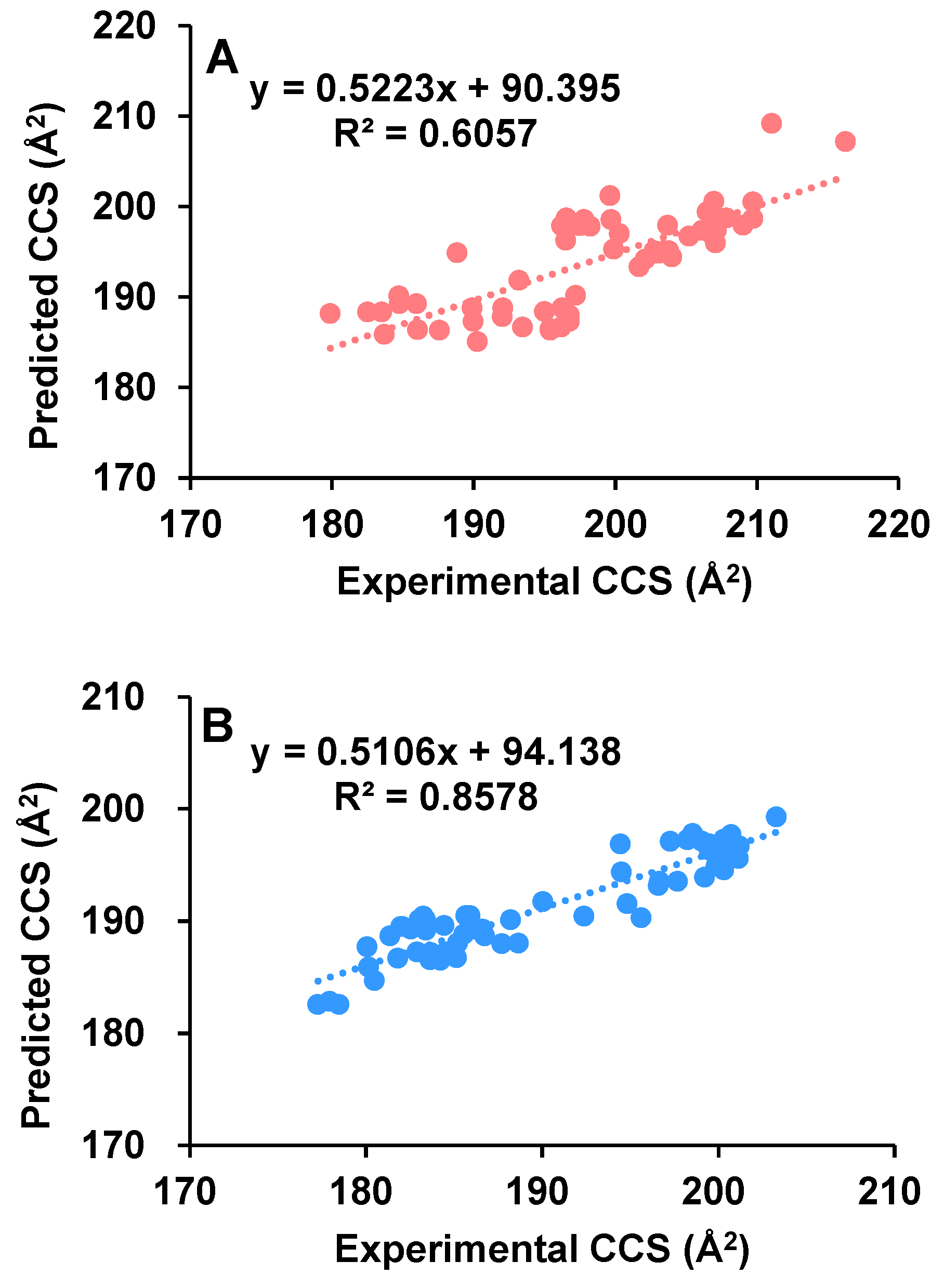
2.2. Comparison of Experimental with Theoretical CCS
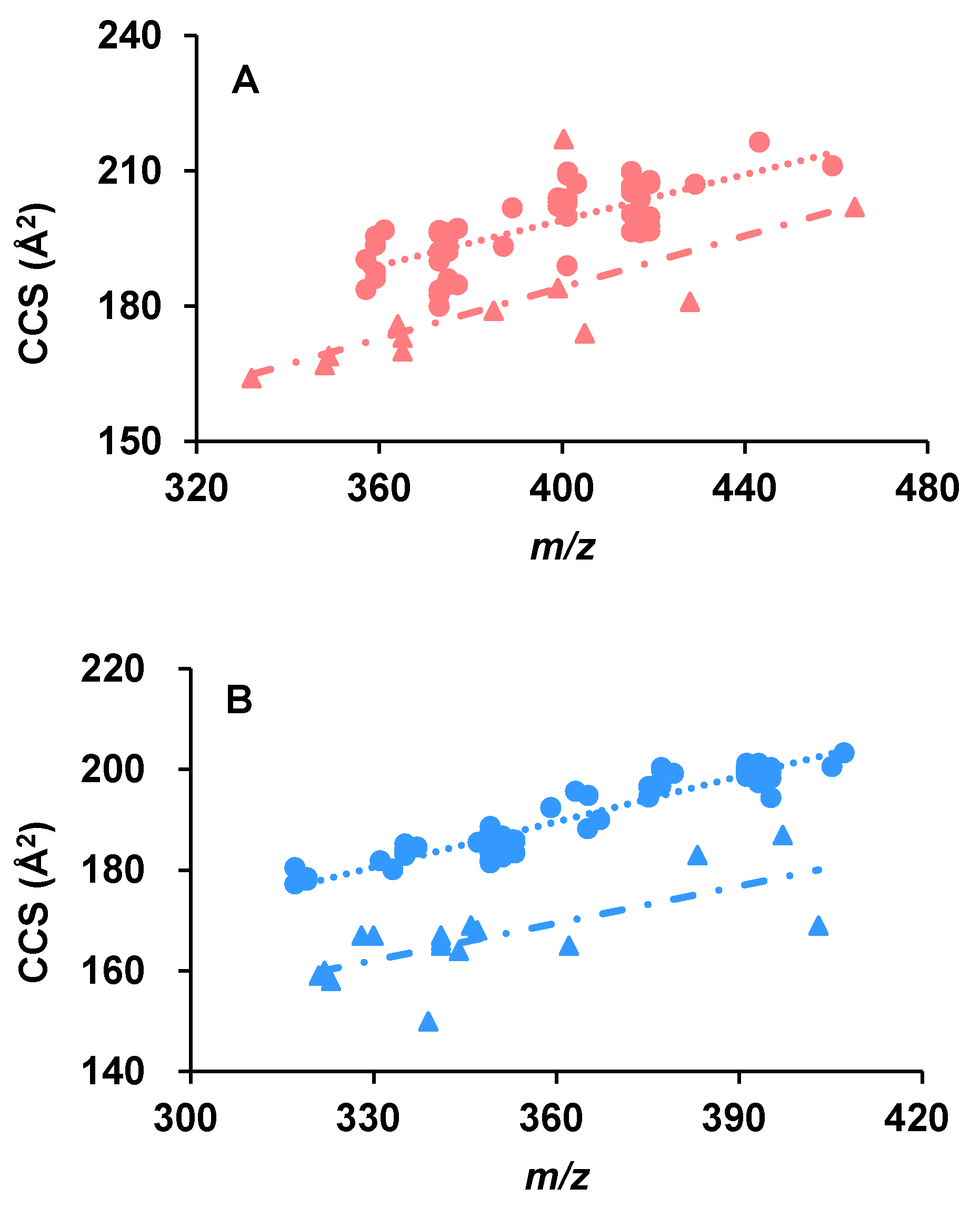
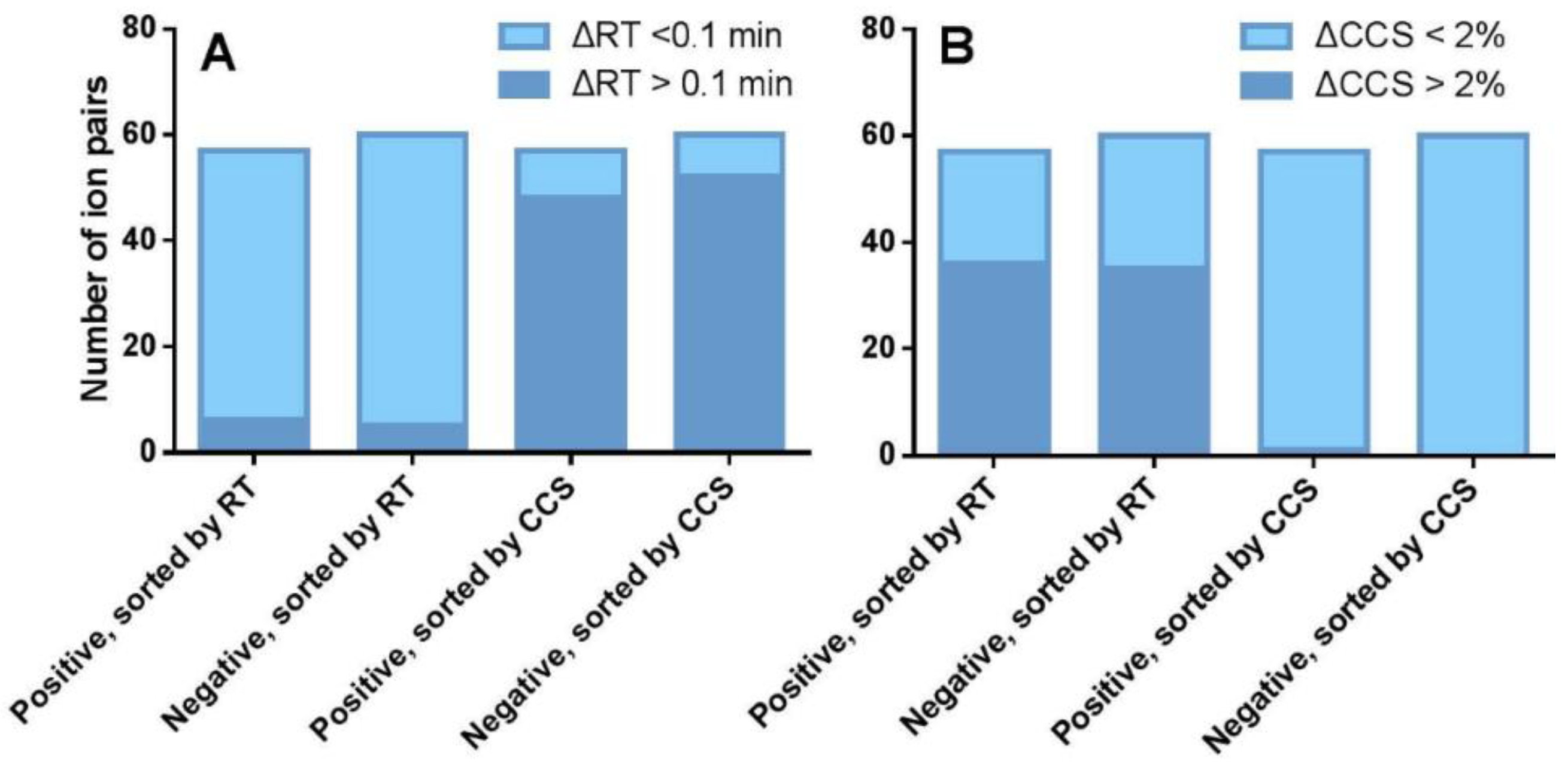
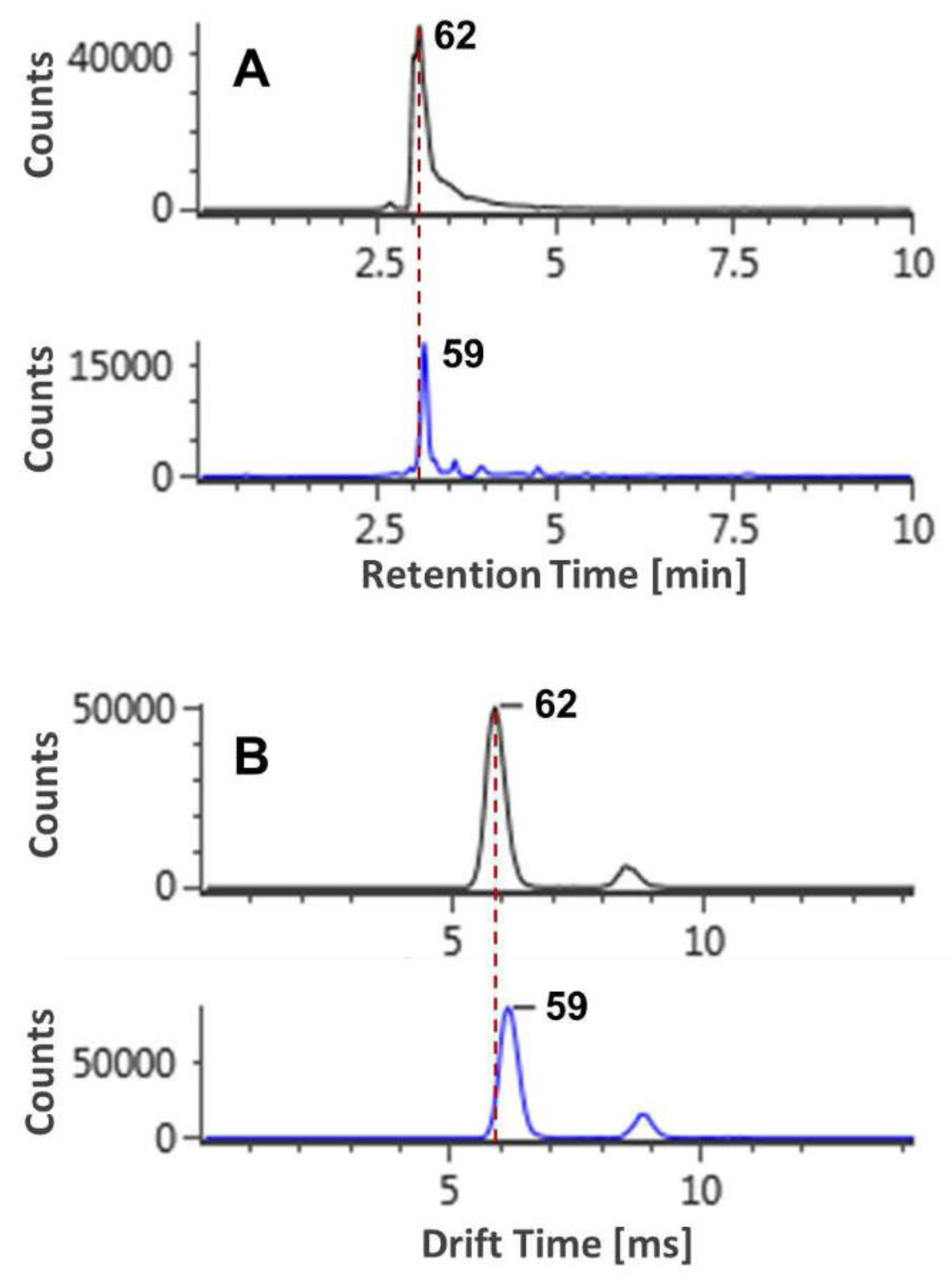
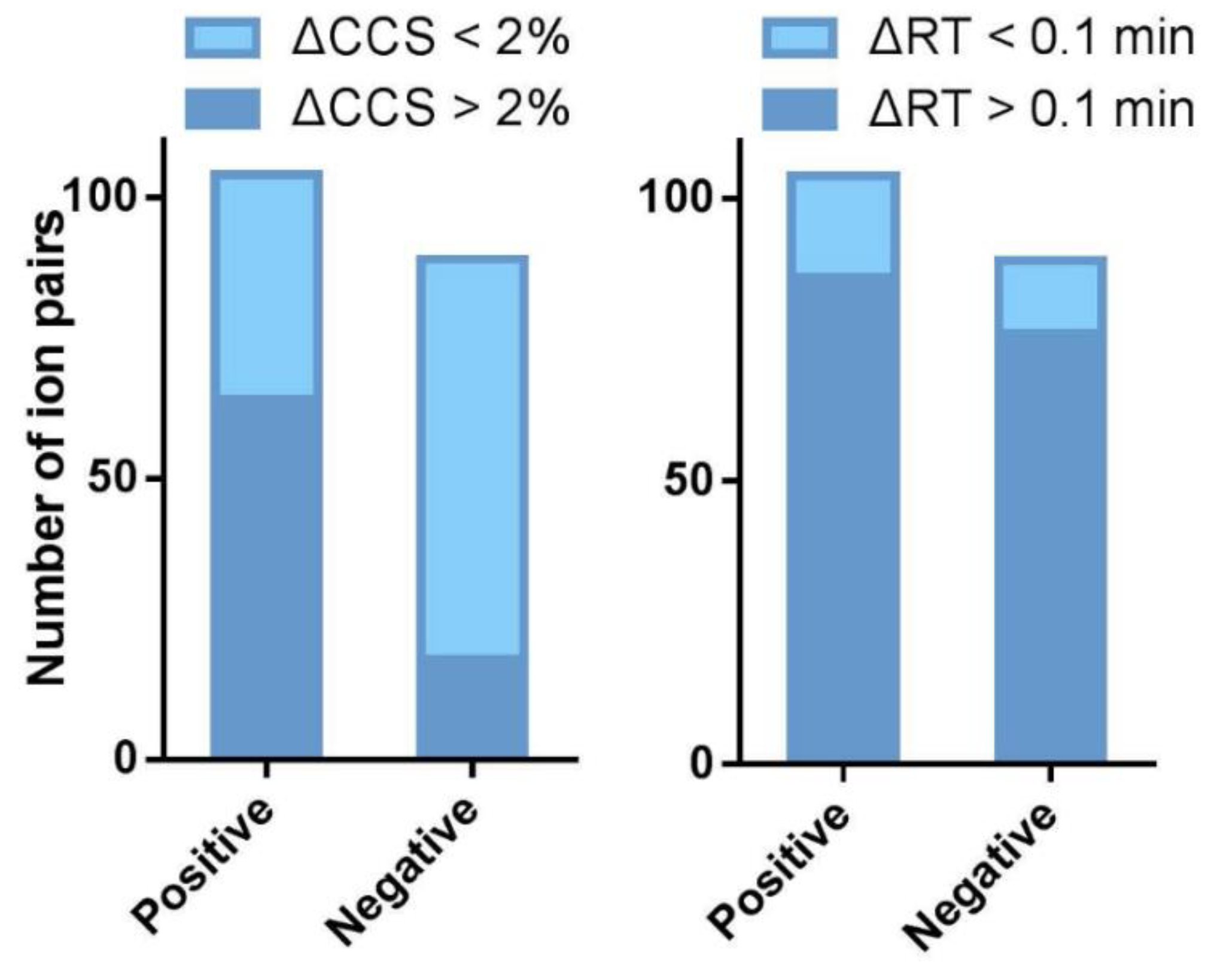
2.3. The CCS Values as an Important Supplementation to the RT Separation
2.4. The Capability of CCS Values in Discriminating Isomers
3. Materials and Methods
3.1. Chemicals, Reagents, and Materials
3.2. Liquid Chromatographic Analysis
3.3. MS Analysis
3.4. CCS Measurements for Aphidicolanes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Aksenov, A.A.; da Silva, R.; Knight, R.; Lopes, N.P.; Dorrestein, P.C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017, 1, 0054. [Google Scholar] [CrossRef]
- Cruesemann, M. Coupling mass spectral and genomic information to improve bacterial natural product discovery workflows. Mar. Drugs 2021, 19, 142. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E.; Zhang, X.; Weitz, K.K.; Monroe, M.E.; Ibrahim, Y.M.; Moore, R.J.; Cha, J.; Sun, X.; Lovelace, E.S.; Wagoner, J.; et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst 2016, 141, 1649–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, J.J.; Rutherford, E. On the passage of electricity through gases exposed to Roentgen rays. Phil. Mag. Ser. 1896, 42, 392–407. [Google Scholar] [CrossRef] [Green Version]
- Barnes, W.S.; Martin, D.W.; Mcdaniel, E.W. Mass spectrographic identification of the ion observed in hydrogen mobility experiments. Phys. Rev. Lett. 1961, 6, 110–111. [Google Scholar] [CrossRef]
- Lee, D.S.; Wu, C.; Hill, H.H. Detection of carbohydrates by electrospray ionization ion mobility spectrometry following microbore high-performance liquid chromatography. J. Chromatogr. A 1998, 822, 1–9. [Google Scholar] [CrossRef]
- Siems, W.; Viehland, L.; Hill, H. Improved momentum-transfer theory for ion mobility. 1. Derivation of the fundamental equation. Anal. Chem. 2012, 84, 9782–9791. [Google Scholar] [CrossRef]
- Bush, M.F.; Campuzano, I.D.G.; Robinson, C.V. Ion mobility mass spectrometry of peptide ions: Effects of drift gas and calibration strategies. Anal. Chem. 2012, 84, 7124–7130. [Google Scholar] [CrossRef]
- Causon, T.J.; Kurulugama, R.T.; Hann, S. Drift-tube ion mobility-mass spectrometry for nontargeted ‘omics. In Ion Mobility-Mass Spectrometry: Methods and Protocols; Paglia, G., Astarita, G., Eds.; Humana: New York, NY, USA, 2020; pp. 79–94. [Google Scholar]
- Reisdorph, R.; Michel, C.; Quinn, K.; Doenges, K.; Reisdorph, N. Untargeted differential metabolomics analysis using drift tube. In Ion Mobility-Mass Spectrometry: Methods and Protocols; Paglia, G., Astarita, G., Eds.; Humana: New York, NY, USA, 2020; pp. 55–94. [Google Scholar]
- Paglia, G.; Astarita, G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat. Protoc. 2017, 12, 797–813. [Google Scholar] [CrossRef]
- Zhang, X.; Quinn, K.; Cruickshank-Quinn, C.; Reisdorph, R.; Reisdorph, N. The application of ion mobility mass spectrometry to metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 60–66. [Google Scholar] [CrossRef]
- Mairinger, T.; Causon, T.J.; Hann, S. The potential of ion mobility–mass spectrometry for non-targeted metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Hamid, Z.; Armirotti, A. Traveling wave ion mobility-mass spectrometry to enhance the detection of low abundance features in untargeted lipidomics. In Ion Mobility-Mass Spectrometry: Methods and Protocols; Paglia, G., Astarita, G., Eds.; Humana: New York, NY, USA, 2020; pp. 103–117. [Google Scholar]
- Hinz, C.; Liggi, S.; Griffin, J.L. The potential of Ion Mobility Mass Spectrometry for high-throughput and high-resolution lipidomics. Curr. Opin. Chem. Biol. 2018, 42, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merciai, F.; Musella, S.; Sommella, E.; Bertamino, A.; D’Ursi, A.M.; Campiglia, P. Development and application of a fast ultra-high performance liquid chromatography-trapped ion mobility mass spectrometry method for untargeted lipidomics. J. Chromatogr. A 2022, 1673, 463124. [Google Scholar] [CrossRef]
- Vissers, J.P.C.; McCullagh, M. An analytical perspective on protein analysis and discovery proteomics by ion mobility-mass spectrometry. In Ion Mobility-Mass Spectrometry: Methods and Protocols; Paglia, G., Astarita, G., Eds.; Humana: New York, NY, USA, 2020; pp. 161–178. [Google Scholar]
- Struwe, W.B.; Harvey, D.J. Ion mobility-mass spectrometry of glycoconjugates. In Ion Mobility-Mass Spectrometry: Methods and Protocols; Paglia, G., Astarita, G., Eds.; Humana: New York, NY, USA, 2020; pp. 203–219. [Google Scholar]
- Schrimpe-Rutledge, A.C.; Sherrod, S.D.; McLean, J.A. Improving the discovery of secondary metabolite natural products using ion mobility–mass spectrometry. Curr. Opin. Chem. Biol. 2018, 42, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Mollerup, C.B.; Mardal, M.; Dalsgaard, P.W.; Linnet, K.; Barron, L.P. Prediction of collision cross section and retention time for broad scope screening in gradient reversed-phase liquid chromatography-ion mobility-high resolution accurate mass spectrometry. J. Chromatogr. A 2018, 1542, 82–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colby, S.M.; Thomas, D.G.; Nunez, J.R.; Baxter, D.J.; Glaesemann, K.R.; Brown, J.M.; Pirrung, M.A.; Govind, N.; Teeguarden, J.G.; Metz, T.O.; et al. ISiCLE: A quantum chemistry pipeline for establishing in silico collision cross section libraries. Anal. Chem. 2019, 91, 4346–4356. [Google Scholar] [CrossRef]
- Plante, P.L.; Francovic-Fontaine, É.; May, J.C.; McLean, J.A.; Baker, E.S.; Laviolette, F.; Marchand, M.; Corbeil, J. Predicting ion mobility collision cross-sections using a deep neural network: DeepCCS. Anal. Chem. 2019, 91, 5191–5199. [Google Scholar] [CrossRef]
- Ross, D.H.; Cho, J.H.; Xu, L. Breaking down structural diversity for comprehensive prediction of ion-neutral collision cross sections. Anal. Chem. 2020, 92, 4548–4557. [Google Scholar] [CrossRef]
- Heerdt, G.; Zanotto, L.; Souza, P.C.T.; Araujo, G.; Skaf, M.S. Collision cross section calculations using HPCCS. In Ion Mobility-Mass Spectrometry: Methods and Protocols; Paglia, G., Astarita, G., Eds.; Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2084, pp. 297–310. [Google Scholar]
- Zhou, Z.; Shen, X.; Tu, J.; Zhu, Z.J. Large-scale prediction of collision cross-section values for metabolites in ion mobility-mass spectrometry. Anal. Chem. 2016, 88, 11084–11091. [Google Scholar] [CrossRef]
- Zhou, Z.; Tu, J.; Xiong, X.; Shen, X.; Zhu, Z. LipidCCS: Prediction of collision cross-section values for lipids with high precision to support ion mobility-mass spectrometry-based lipidomics. Anal. Chem. 2017, 89, 9559–9566. [Google Scholar] [CrossRef]
- Zhou, Z.; Luo, M.; Chen, X.; Yin, Y.; Xiong, X.; Wang, R.; Zhu, Z.J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat. Commun. 2020, 11, 4334. [Google Scholar] [CrossRef] [PubMed]
- Righetti, L.; Bergmann, A.; Galaverna, G.; Rolfsson, O.; Paglia, G.; Dall’Asta, C. Ion mobility-derived collision cross section database: Application to mycotoxin analysis. Anal. Chim. Acta 2018, 1014, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Regueiro, J.; Negreira, N.; Berntssen, M.H.G. Ion-mobility-derived collision cross section as an additional identification point for multiresidue screening of pesticides in fish feed. Anal. Chem. 2016, 88, 11169–11177. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Williams, J.P.; Menikarachchi, L.; Thompson, J.W.; Tyldesley-Worster, R.; Halldórsson, S.; Rolfsson, O.; Moseley, A.; Grant, D.; Langridge, J.; et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. 2014, 86, 3985–3993. [Google Scholar] [CrossRef] [Green Version]
- Righetti, L.; Fenclova, M.; Dellafiora, L.; Hajslova, J.; Stranska-Zachariasova, M.; Dall’Asta, C. High resolution-ion mobility mass spectrometry as an additional powerful tool for structural characterization of mycotoxin metabolites. Food Chem. 2018, 245, 768–774. [Google Scholar] [CrossRef]
- Goscinny, S.; Joly, L.; De Pauw, E.; Hanot, V.; Eppe, G. Travelling-wave ion mobility time-of-flight mass spectrometry as an alternative strategy for screening of multi-class pesticides in fruits and vegetables. J. Chromatogr. A 2015, 1405, 85–93. [Google Scholar] [CrossRef]
- Groessl, M.; Graf, S.; Knochenmuss, R. High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst 2015, 14, 6904–6911. [Google Scholar] [CrossRef]
- Hines, K.M.; Ross, D.H.; Davidson, K.L.; Bush, M.F.; Xu, L. Large-scale structural characterization of drug and drug-like compounds by high-throughput ion mobility-mass spectrometry. Anal. Chem. 2017, 89, 9023–9030. [Google Scholar] [CrossRef] [Green Version]
- Ahonen, L.; Fasciotti, M.; af Gennas, G.B.; Kotiaho, T.; Daroda, R.J.; Eberlin, M.; Kostiainen, R. Separation of steroid isomers by ion mobility mass spectrometry. J. Chromatogr. A 2013, 1310, 133–137. [Google Scholar] [CrossRef]
- Qian, Y.-X.; Zhao, D.-X.; Wang, H.-D.; Sun, H.; Xiong, Y.; Xu, X.-Y.; Hu, W.-D.; Liu, M.-Y.; Chen, B.-X.; Hu, Y.; et al. An ion mobility-enabled and high-efficiency hybrid scan approach in combination with ultra-high performance liquid chromatography enabling the comprehensive characterization of the multicomponents from Carthamus tinctorius. J. Chromatogr. A 2022, 1667, 462904. [Google Scholar] [CrossRef]
- Niu, S.; Xia, J.M.; Li, Z.; Yang, L.H.; Yi, Z.W.; Xie, C.L.; Peng, G.; Luo, Z.H.; Shao, Z.; Yang, X.W. Aphidicolin Chemistry of the Deep-Sea-Derived Fungus Botryotinia fuckeliana MCCC 3A00494. J. Nat. Prod. 2019, 82, 2307–2331. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Xie, C.L.; Xia, J.M.; Liu, Q.M.; Peng, G.; Liu, G.M.; Yang, X.W. Botryotins A-H, Tetracyclic Diterpenoids Representing Three Carbon Skeletons from a Deep-Sea-Derived Botryotinia fuckeliana. Org. Lett. 2020, 22, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, S.; Taguchi, T.; Ohashi, M.; Oguro, M.; Nagano, H.; Mano, Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 1978, 275, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Sessa, C.; Zucchetti, M.; Davoli, E.; Califano, R.; Cavalli, F.; Frustaci, S.; Gumbrell, L.; Sulkes, A.; Winograd, B.; Dincalci, M. Phase-I and clinical pharmacological evaluation of aphidicolin glycinate. J. Natl. Cancer Inst. 1991, 83, 1160–1164. [Google Scholar] [CrossRef]
- Hanson, J.R. Aphidicolin: Its chemistry and biosynthesis. J. Chem. Res. 2018, 42, 395–401. [Google Scholar] [CrossRef]
- Fenn, L.S.; McLean, J.A. Biomolecular structural separations by ion mobility-mass spectrometry. Anal. Bioanal. Chem. 2008, 391, 905–909. [Google Scholar] [CrossRef]
- Kanu, A.B.; Dwivedi, P.; Tam, M.; Matz, L.; Hill, H.H., Jr. Ion mobility-mass spectrometry. J. Mass Spectrom. 2008, 43, 1–22. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, J.; Xiao, W.; Lin, X.; Zhou, Y.; Qiu, P.; Si, H.; Wu, X.; Niu, S.; Luo, Z.; Yang, X. Ion Mobility-Derived Collision Cross-Sections Add Extra Capability in Distinguishing Isomers and Compounds with Similar Retention Times: The Case of Aphidicolanes. Mar. Drugs 2022, 20, 541. https://doi.org/10.3390/md20090541
Xia J, Xiao W, Lin X, Zhou Y, Qiu P, Si H, Wu X, Niu S, Luo Z, Yang X. Ion Mobility-Derived Collision Cross-Sections Add Extra Capability in Distinguishing Isomers and Compounds with Similar Retention Times: The Case of Aphidicolanes. Marine Drugs. 2022; 20(9):541. https://doi.org/10.3390/md20090541
Chicago/Turabian StyleXia, Jinmei, Wenhai Xiao, Xihuang Lin, Yiduo Zhou, Peng Qiu, Hongkun Si, Xiaorong Wu, Siwen Niu, Zhuhua Luo, and Xianwen Yang. 2022. "Ion Mobility-Derived Collision Cross-Sections Add Extra Capability in Distinguishing Isomers and Compounds with Similar Retention Times: The Case of Aphidicolanes" Marine Drugs 20, no. 9: 541. https://doi.org/10.3390/md20090541
APA StyleXia, J., Xiao, W., Lin, X., Zhou, Y., Qiu, P., Si, H., Wu, X., Niu, S., Luo, Z., & Yang, X. (2022). Ion Mobility-Derived Collision Cross-Sections Add Extra Capability in Distinguishing Isomers and Compounds with Similar Retention Times: The Case of Aphidicolanes. Marine Drugs, 20(9), 541. https://doi.org/10.3390/md20090541








