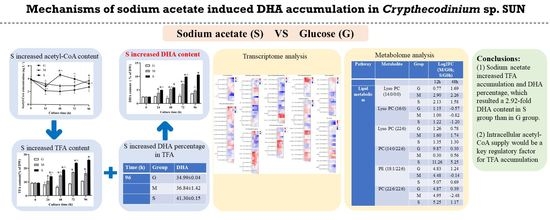
Mechanisms of Sodium-Acetate-Induced DHA Accumulation in a DHA-Producing Microalga, Crypthecodinium sp. SUN
Abstract
:1. Introduction
2. Results and Discussion
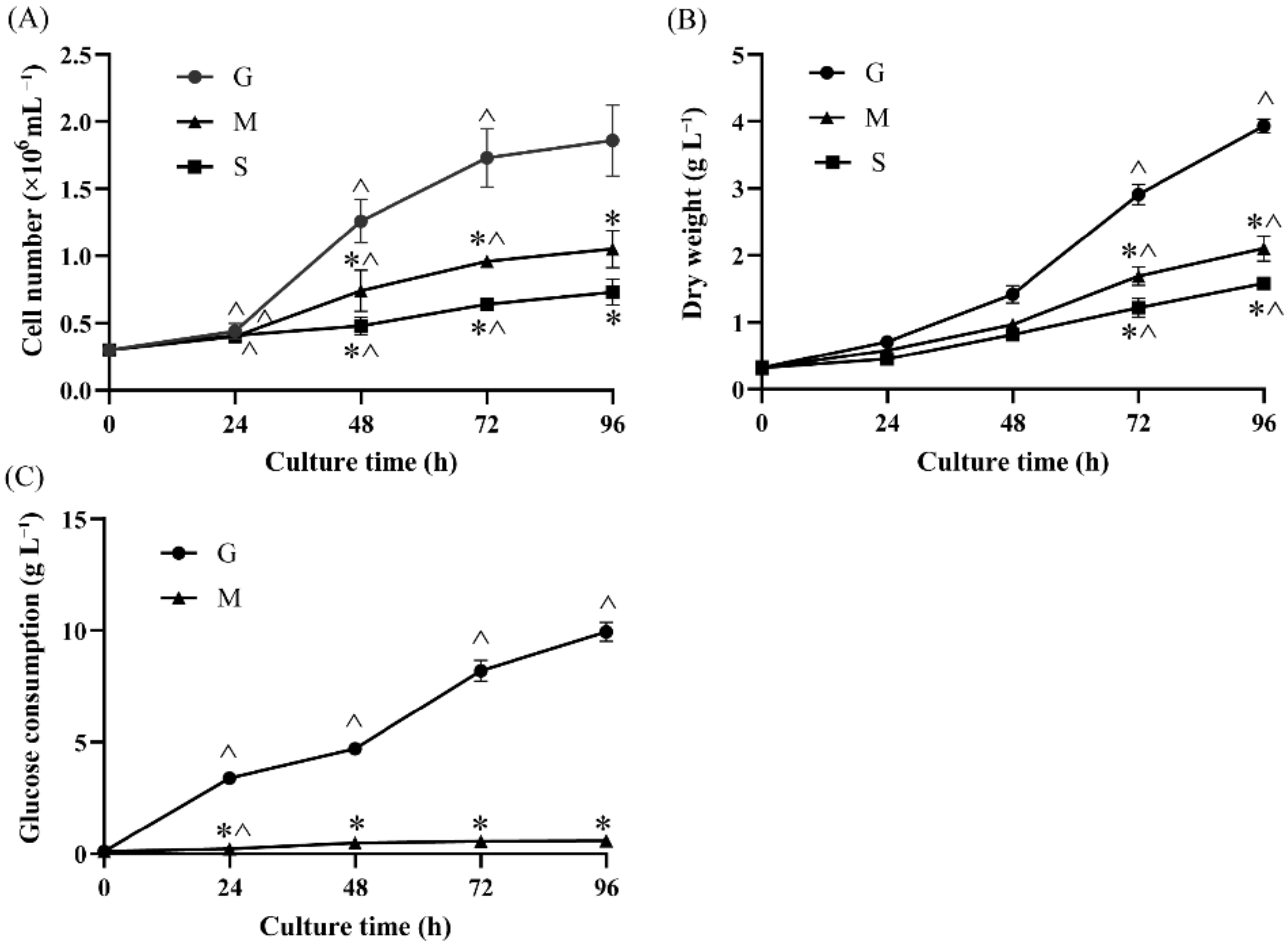
2.1. Effects of Different Carbon Sources on Cell Growth and Glucose Consumption
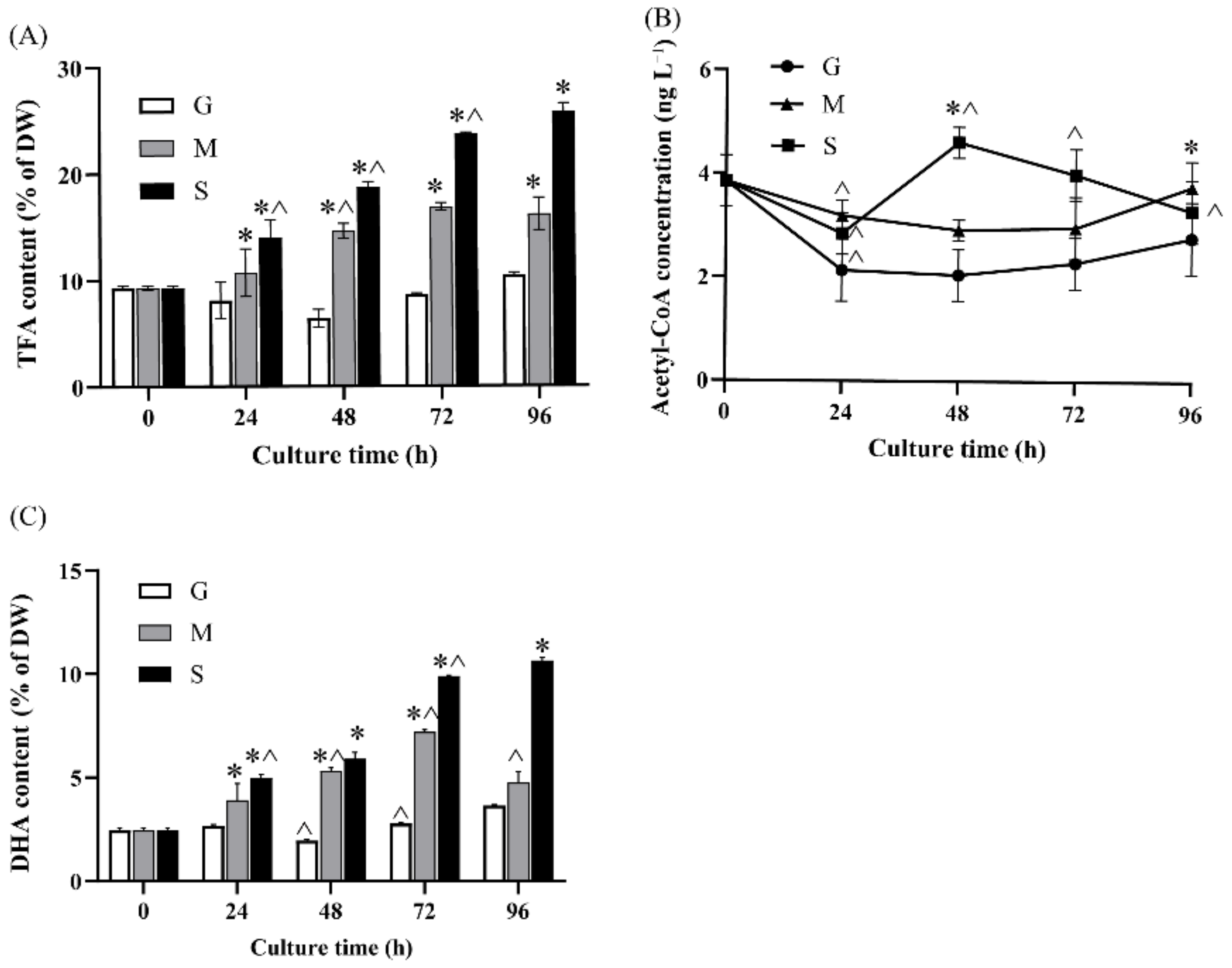
2.2. Effect of Different Carbon Sources on Fatty Acid Biosynthesis and DHA Production
2.3. Effect of Different Carbon Sources on Starch and Protein Contents
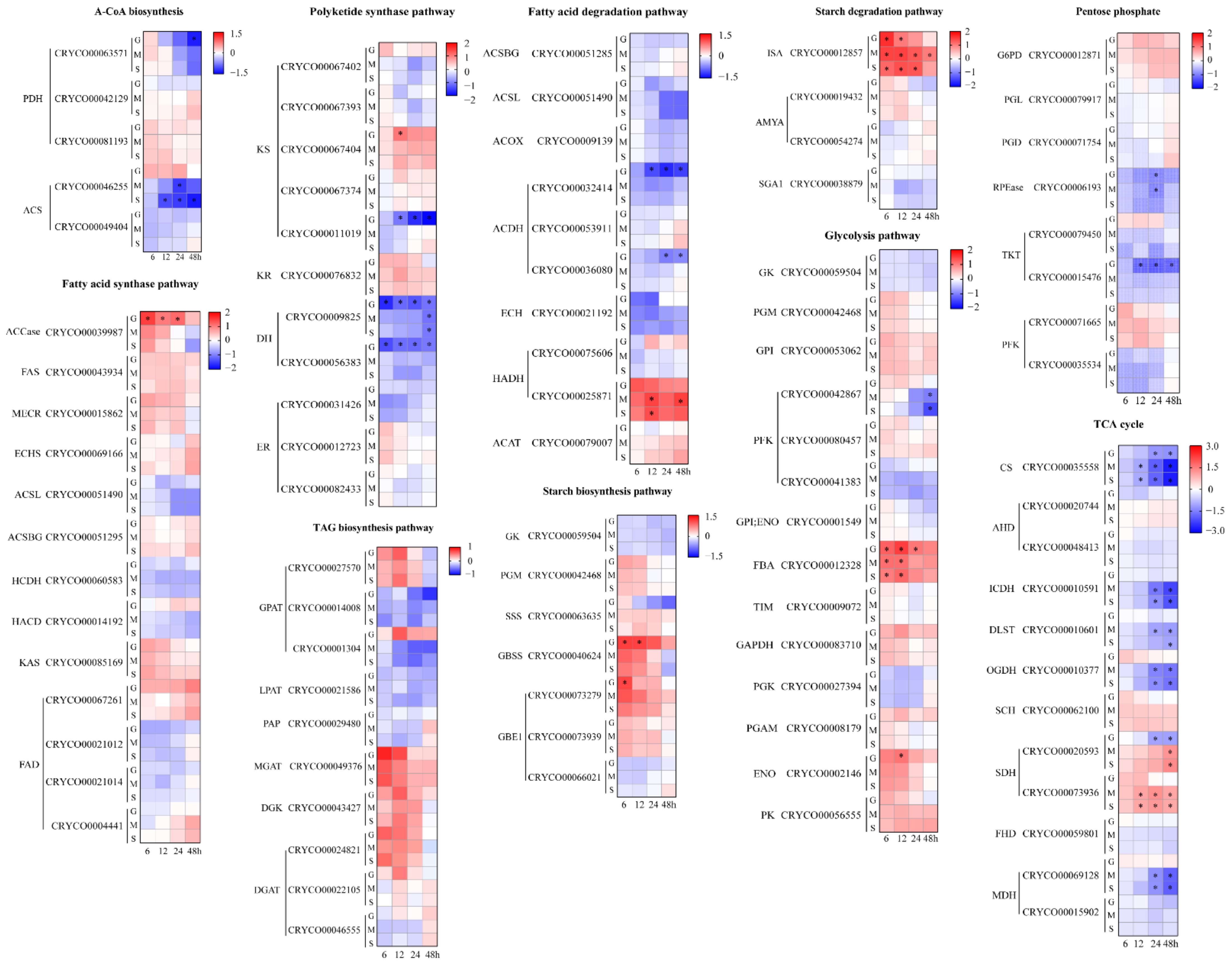
2.4. Comparative Transcriptome Analysis under Different Carbon Source Treatments
2.4.1. Acetyl-CoA Biosynthesis and Lipid Metabolism
2.4.2. Starch Metabolism
2.4.3. Glycolysis, Pentose Phosphate Pathway and the TCA Cycle
2.5. Metabolome Analysis under Different Carbon Source Treatments
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Measurement of Cell Number and Dry Weight
3.3. Quantification of Glucose Consumption
3.4. Fatty Acid Analysis
3.5. Extraction and Quantification of Intracellular Acetyl-CoA
3.6. Quantification of Starch
3.7. Quantification of Protein
3.8. Total RNA Extraction, Quality Control, and cDNA Library Construction
3.9. Read Mapping, Differential Genes Expression Analysis, and Functional Enrichment
3.10. Real-Time Quantitative PCR (qPCR)
3.11. Metabolite Extraction, LC-MS Analysis, and Metabolite Identification
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, J.; Diao, J.; Sun, T.; Shi, M.; Liu, L.; Wang, F.; Chen, L.; Zhang, W. 13C Metabolic Flux Analysis of Enhanced Lipid Accumulation Modulated by Ethanolamine in Crypthecodinium cohnii. Front. Microbiol. 2018, 9, 956. [Google Scholar] [CrossRef] [PubMed]
- Scheibel, S.; de Oliveira, C.A.L.; Boyd, M.D.; Pereira, L.C.; Rodrigues, D.M.; Barion, M.R.L.; Carbonera, F.; Visentainer, J.V.; Huppes, R.R.; Ribeiro, L.B.; et al. DHA from microalgae Schizochytrium spp. (Thraustochytriaceae) modifies the inflammatory response and gonadal lipid profile in domestic cats. Brit. J. Nutr. 2021, 126, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Carrillo, B.E.; Valdes-Ramos, R.; Garcia-Rillo, A.; Gutierrez-Pliego, L.E.; Escoto-Herrera, J.A.; Rosales-Gomez, C. EPA and DHA Consumption Extracted from Microalgae and Its Effect on Anthropometric and Biochemical Parameters. Ann. Nutr. Metab. 2017, 71, 1193. [Google Scholar]
- Schade, S.; Stangl, G.I.; Meier, T. Distinct microalgae species for food-part 2: Comparative life cycle assessment of microalgae and fish for eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), and protein. J. Appl. Phycol. 2020, 32, 2997–3013. [Google Scholar] [CrossRef]
- Mendes, A.; Reis, A.; Vasconcelos, R.; Guerra, P.; Lopes da Silva, T. Crypthecodinium cohnii with emphasis on DHA production: A review. J. Appl. Phycol. 2008, 21, 199–214. [Google Scholar] [CrossRef]
- Taborda, T.; Moniz, P.; Reis, A.; da Silva, T.L. Evaluating low-cost substrates for Crypthecodinium cohnii lipids and DHA production, by flow cytometry. J. Appl. Phycol. 2020, 33, 263–274. [Google Scholar] [CrossRef]
- Sun, D.; Zhang, Z.; Chen, F. Effects of light intensity, light quality, and illumination period on cell growth, TFA accumulation, and DHA production in Crypthecodinium sp. SUN. J. Appl. Phycol. 2018, 30, 1495–1502. [Google Scholar] [CrossRef]
- Ran, W.Y.; Wang, H.T.; Liu, Y.H.; Qi, M.; Xiang, Q.; Yao, C.H.; Zhang, Y.K.; Lan, X.Q. Storage of starch and lipids in microalgae: Biosynthesis and manipulation by nutrients. Bioresour. Technol. 2019, 291, 121894. [Google Scholar] [CrossRef]
- Lv, M.; Wang, F.; Zeng, L.; Bi, Y.; Cui, J.; Liu, L.; Bi, Y.; Chen, L.; Zhang, W. Identification and metabolomic analysis of a starch-deficient Crypthecodinium cohnii mutant reveals multiple mechanisms relevant to enhanced growth and lipid accumulation. Algal Res. 2020, 50, 102001. [Google Scholar] [CrossRef]
- Zhang, L.J.; Pei, H.Y.; Chen, S.Q.; Jiang, L.Q.; Hou, Q.J.; Yang, Z.G.; Yu, Z. Salinity-induced cellular cross-talk in carbon partitioning reveals starch-to-lipid biosynthesis switching in low-starch freshwater algae. Bioresour. Technol. 2018, 250, 449–456. [Google Scholar] [CrossRef]
- Kato, Y.; Ho, S.H.; Vavricka, C.J.; Chang, J.S.; Hasunuma, T.; Kondo, A. Evolutionary engineering of salt-resistant Chlamydomonas sp. strains reveals salinity stress-activated starch-to-lipid biosynthesis switching. Bioresour. Technol. 2017, 245, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Toyoshima, M. Dynamism of Metabolic Carbon Flow of Starch and Lipids in Chlamydomonas debaryana. Front. Plant Sci. 2021, 12, 646498. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.H.; Miao, X.L. Carbon flow conversion induces alkali resistance and lipid accumulation under alkaline conditions based on transcriptome analysis in Chlorella sp. BLD. Chemosphere 2021, 265, 129046. [Google Scholar] [CrossRef] [PubMed]
- Abedini Najafabadi, H.; Malekzadeh, M.; Jalilian, F.; Vossoughi, M.; Pazuki, G. Effect of various carbon sources on biomass and lipid production of Chlorella vulgaris during nutrient sufficient and nitrogen starvation conditions. Bioresour. Technol. 2015, 180, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.Y.; Sun, L.L.; Wu, S.; Liu, C.; Zhao, P.P.; Xie, X.J.; Wang, G.C. Utilization of glucose and acetate by Chlorella and the effect of multiple factors on cell composition. J. Appl. Phycol. 2017, 29, 23–33. [Google Scholar] [CrossRef]
- Ramanan, R.; Kim, B.H.; Cho, D.H.; Ko, S.R.; Oh, H.M.; Kim, H.S. Lipid droplet synthesis is limited by acetate availability in starchless mutant of Chlamydomonas reinhardtii. FEBS Lett. 2013, 587, 370–377. [Google Scholar] [CrossRef] [Green Version]
- Lari, Z.; Abrishamchi, P.; Ahmadzadeh, H.; Soltani, N. Differential carbon partitioning and fatty acid composition inmixotrophic and autotrophic cultures of a new marine isolate Tetraselmis sp. KY114885. J. Appl. Phycol. 2019, 31, 201–210. [Google Scholar] [CrossRef]
- Wang, S.K.; Wang, X.; Tian, Y.T.; Tao, H.H.; Sun, X.S. Direct utilization of starch for heterotrophic cultivation of Chlorella pyrenoidosa by co-culture with immobilized Saccharomycopsis fibuligera. Algal Res. 2018, 33, 406–411. [Google Scholar] [CrossRef]
- Rengel, R.; Smith, R.T.; Haslam, R.P.; Sayanova, O.; Vila, M.; Leon, R. Overexpression of acetyl-CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii. Algal Res 2018, 31, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Perez-Garcia, O.; Escalante, F.M.; de-Bashan, L.E.; Bashan, Y. Heterotrophic cultures of microalgae: Metabolism and potential products. Water Res. 2011, 45, 11–36. [Google Scholar] [CrossRef]
- Xing, G.L.; Yuan, H.L.; Yang, J.S.; Li, J.Y.; Gao, Q.X.; Li, W.L.; Wang, E.T. Integrated analyses of transcriptome, proteome and fatty acid profilings of the oleaginous microalga Auxenochlorella protothecoides UTEX 2341 reveal differential reprogramming of fatty acid metabolism in response to low and high temperatures. Algal Res. 2018, 33, 16–27. [Google Scholar] [CrossRef]
- Liu, J.; Huang, J.; Fan, K.W.; Jiang, Y.; Zhong, Y.; Sun, Z.; Chen, F. Production potential of Chlorella zofingienesis as a feedstock for biodiesel. Bioresour. Technol. 2010, 101, 8658–8663. [Google Scholar] [CrossRef] [PubMed]
- Morales-Sanchez, D.; Tinoco-Valencia, R.; Kyndt, J.; Martinez, A. Heterotrophic growth of Neochloris oleoabundans using glucose as a carbon source. Biotechnol. Biofuels 2013, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.B.; Zhao, X.C.; Zhang, Y.L.; Zhou, Y.Y.; Yang, L.B.; Zhang, W.W. Enhanced lipid and biomass production using alcohol wastewater as carbon source for Chlorella pyrenoidosa cultivation in anaerobically digested starch wastewater in outdoors. Bioresour. Technol. 2018, 247, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, Z.; Mao, X.; Wu, T.; Jiang, Y.; Liu, J.; Chen, F. Light enhanced the accumulation of total fatty acids (TFA) and docosahexaenoic acid (DHA) in a newly isolated heterotrophic microalga Crypthecodinium sp. SUN. Bioresour. Technol. 2017, 228, 227–234. [Google Scholar] [CrossRef]
- Shen, X.-F.; Liu, J.-J.; Chauhan, A.S.; Hu, H.; Ma, L.-L.; Lam, P.K.S.; Zeng, R.J. Combining nitrogen starvation with sufficient phosphorus supply for enhanced biodiesel productivity of Chlorella vulgaris fed on acetate. Algal Res. 2016, 17, 261–267. [Google Scholar] [CrossRef]
- Li, Y.; Huang, A.; Gu, W.; Wu, S.; Xie, X.; Wang, G. Effects of inorganic carbon and light on acetate assimilation by Nannochloropsis oceanica (Eustigmatophyceae) in mixotrophic cultivation. Eur. J. Phycol. 2019, 55, 64–75. [Google Scholar] [CrossRef]
- Pereira, L.T.; Lisboa, C.R.; Costa, J.A.V.; da Rosa, L.M.; de Carvalho, L.F. Evaluation of protein content and antimicrobial activity of biomass from Spirulina cultivated with residues from the brewing process. J. Chem. Technol. Biot. 2022, 97, 160–166. [Google Scholar] [CrossRef]
- Higgins, B.T.; Wang, Q.C.; Du, S.D.; Hennebelle, M.; Taha, A.Y.; Fiehn, O.; VanderGheynst, J.S. Impact of thiamine metabolites and spent medium from Chlorella sorokiniana on metabolism in the green algae Auxenochlorella prototheciodes. Algal Res. 2018, 33, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Han, D.X.; Gerken, H.G.; Li, Y.T.; Sommerfeld, M.; Hu, Q.; Xu, J. Molecular mechanisms for photosynthetic carbon partitioning into storage neutral lipids in Nannochloropsis oceanica under nitrogen-depletion conditions. Algal Res. 2015, 7, 66–77. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, F.; Wang, Z.; Zhou, R.; Tang, Y.; Li, Y. The potential growth and lipid accumulation in Coccomyxa subellipsoidea triggered by glucose combining with sodium acetate. World J. Microbiol. Biotechnol. 2019, 35, 110. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Zheng, H.Q.; Chen, C.N.N. Comparative transcriptome analysis reveals a potential photosynthate partitioning mechanism between lipid and starch biosynthetic pathways in green microalgae. Algal Res. 2016, 16, 54–62. [Google Scholar] [CrossRef]
- Chen, D.; Yuan, X.; Liang, L.M.; Liu, K.; Ye, H.Y.; Liu, Z.Y.; Liu, Y.F.; Huang, L.Q.; He, W.J.; Chen, Y.Q.; et al. Overexpression of acetyl-CoA carboxylase increases fatty acid production in the green alga Chlamydomonas reinhardtii. Biotechnol. Lett. 2019, 41, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Ren, L.J.; Zhao, Q.Y.; Ji, X.J.; Huang, H. Enhancement of lipid accumulation in microalgae by metabolic engineering. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Jenke-Kodama, H.; Sandmann, A.; Muller, R.; Dittmann, E. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 2005, 22, 2027–2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, T.; Chauhan, R.S. Comparative transcriptomics reveals molecular components associated with differential lipid accumulation between Microalgal sp., Scenedesmus dimorphus and Scenedesmus quadricauda. Algal Res. 2016, 19, 109–122. [Google Scholar] [CrossRef]
- Chu, F.F.; Cheng, J.; Zhang, X.D.; Ye, Q.; Chen, S.T.; Zhou, J.H.; Cen, K.F. Transcriptome and key gene expression related to carbon metabolism and fatty acid synthesis of Chlorella vulgaris under a nitrogen starvation and phosphorus repletion regime. J. Appl. Phycol. 2019, 31, 2881–2893. [Google Scholar] [CrossRef]
- Lau, C.C.; Loh, S.H.; Aziz, A.; Cha, T.S. Effects of disrupted omega-3 desaturase gene construct on fatty acid composition and expression of four fatty acid biosynthetic genes in transgenic Chlorella vulgaris. Algal Res. 2017, 26, 143–152. [Google Scholar] [CrossRef]
- Kaye, Y.; Grundman, O.; Leu, S.; Zarka, A.; Zorin, B.; Didi-Cohen, S.; Khozin-Goldberg, I.; Boussiba, S. Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: Overexpression of endogenous Δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res. 2015, 11, 387–398. [Google Scholar] [CrossRef]
- Pei, G.; Li, X.; Liu, L.; Liu, J.; Wang, F.; Chen, L.; Zhang, W. De novo transcriptomic and metabolomic analysis of docosahexaenoic acid (DHA)-producing Crypthecodinium cohnii during fed-batch fermentation. Algal Res. 2017, 26, 380–391. [Google Scholar] [CrossRef]
- Takeuchi, T.; Benning, C. Nitrogen-dependent coordination of cell cycle, quiescence and TAG accumulation in Chlamydomonas. Biotechnol. Biofuels 2019, 12, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Q.W.; Chen, C.; He, T.W.; Fan, J.H. Circadian rhythm promotes the biomass and amylose hyperaccumulation by mixotrophic cultivation of marine microalga Platymonas helgolandica. Biotechnol. Biofuels Bioprod. 2022, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Gargouri, M.; Feng, J.; Park, J.J.; Gao, D.F.; Miao, C.; Dong, T.; Gang, D.R.; Chen, S.L. Regulation of starch and lipid accumulation in a microalga Chlorella sorokiniana. Bioresour. Technol. 2015, 180, 250–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Meng, T.; Hang, W.; Cao, X.; Ni, H.; Shi, Y.; Li, Q.; Xiong, Y.; He, N. Regulation of glucose and glycerol for production of docosahexaenoic acid in Schizochytrium limacinum SR21 with metabolomics analysis. Algal Res. 2021, 58, 102415. [Google Scholar] [CrossRef]
- Osada, K.; Maeda, Y.; Yoshino, T.; Nojima, D.; Bowler, C.; Tanaka, T. Enhanced NADPH production in the pentose phosphate pathway accelerates lipid accumulation in the oleaginous diatom Fistulifera solaris. Algal Res. 2017, 23, 126–134. [Google Scholar] [CrossRef]
- Riaz, S.; Sui, Z.; Niaz, Z.; Khan, S.; Liu, Y.; Liu, H. Distinctive Nuclear Features of Dinoflagellates with A Particular Focus on Histone and Histone-Replacement Proteins. Microorganisms 2018, 6, 128. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.M.; Mi, Y.W.; Zhao, C.; Wei, Q. A comprehensive review on carbon source effect of microalgae lipid accumulation for biofuel production. Sci. Total Environ. 2022, 806, 151387. [Google Scholar] [CrossRef]
- Han, M.A.; Hong, S.J.; Kim, Z.H.; Cho, B.K.; Lee, H.; Choi, H.K.; Lee, C.G. Enhanced Production of Fatty Acids via Redirection of Carbon Flux in Marine Microalga Tetraselmis sp. J. Microbiol. Biotechnol. 2018, 28, 267–274. [Google Scholar] [CrossRef] [Green Version]
- Kugler, A.; Zorin, B.; Didi-Cohen, S.; Sibiryak, M.; Gorelova, O.; Ismagulova, T.; Kokabi, K.; Kumari, P.; Lukyanov, A.; Boussiba, S.; et al. Long-Chain Polyunsaturated Fatty Acids in the Green Microalga Lobosphaera incisa Contribute to Tolerance to Abiotic Stresses. Plant Cell Physiol. 2019, 60, 1205–1223. [Google Scholar] [CrossRef]
- Saqib, A.A.N.; Whitney, P.J. Differential behaviour of the dinitrosalicylic acid (DNS) reagent towards mono- and di-saccharide sugars. Biomass Bioenergy 2011, 35, 4748–4750. [Google Scholar] [CrossRef]
- Avidan, O.; Brandis, A.; Rogachev, I.; Pick, U. Enhanced acetyl-CoA production is associated with increased triglyceride accumulation in the green alga Chlorella desiccata. J. Exp. Bot. 2015, 66, 3725–3735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Sun, D.Z.; Wu, T.; Li, Y.L.; Lee, Y.K.; Liu, J.; Chen, F. The synergistic energy and carbon metabolism under mixotrophic cultivation reveals the coordination between photosynthesis and aerobic respiration in Chlorella zofingiensis. Algal Res. 2017, 25, 109–116. [Google Scholar] [CrossRef]
- Bainor, A.; Chang, L.; McQuade, T.J.; Webb, B.; Gestwicki, J.E. Bicinchoninic acid (BCA) assay in low volume. Anal. Biochem. 2011, 410, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Sirikhachornkit, A.; Suttangkakul, A.; Vuttipongchaikij, S.; Juntawong, P. De novo transcriptome analysis and gene expression profiling of an oleaginous microalga Scenedesmus acutus TISTR8540 during nitrogen deprivation-induced lipid accumulation. Sci. Rep. 2018, 8, 3668. [Google Scholar] [CrossRef] [Green Version]

| Time (h) | Group ^ | C14:0 | C16:0 | C16:1 | C18:0 | C18:1 | DHA | SFAs a | MUFAs b | PUFAs c |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | G | 2.63 ± 0.22 | 13.94 ± 0.62 | 34.24 ± 1.19 | 14.82 ± 1.19 | 6.86 ± 0.27 | 26.40 ± 0.03 | 32.14 ± 0.39 | 41.10 ± 0.93 | 26.76 ± 1.21 |
| 24 | G | 2.84 ± 0.02 | 11.74 ± 0.02 | 30.22 ± 0.99 | 13.19 ± 0.73 | 5.76 ± 0.01 | 34.56 ± 0.05 | 28.57 ± 3.34 | 35.98 ± 4.39 | 35.45 ± 6.63 |
| M | 3.42 ± 0.07 * | 16.83 ± 0.38 * | 25.50 ± 0.59 * | 8.46 ± 0.23 * | 7.78 ± 0.02 | 36.32 ± 0.81 | 29.63 ± 0.22 | 34.48 ± 1.82 | 36.62 ± 3.75 | |
| S | 3.04 ± 0.10 * | 18.13 ± 0.17 * | 25.62 ± 0.79 * | 6.78 ± 0.59 * | 8.86 ± 0.01 * | 36.04 ± 0.15 | 28.91 ± 2.18 | 33.28 ± 0.50 | 37.09 ± 0.71 | |
| 48 | G | 3.22 ± 0.05 | 13.77 ± 0.01 | 32.06 ± 0.48 | 12.57 ± 0.37 | 5.95 ± 0.00 | 31.04 ± 0.07 | 30.15 ± 2.54 | 38.02 ± 2.70 | 31.83 ± 5.20 |
| M | 3.91 ± 0.02 | 19.50 ± 0.04 * | 22.28 ± 0.49 * | 5.31 ± 0.37 * | 7.36 ± 0.02 | 39.71 ± 0.15 * | 30.13 ± 0.99 | 32.13 ± 1.18 * | 35.86 ± 1.99 * | |
| S | 3.56 ± 0.02 | 18.11 ± 0.12 * | 25.24 ± 0.32 * | 9.13 ± 0.28 * | 6.89 ± 0.02 | 35.40 ± 0.30 | 32.01 ± 0.81 | 29.64 ± 1.72 * | 40.22 ± 2.72 * | |
| 72 | G | 3.21 ± 0.01 | 17.95 ± 0.02 | 29.34 ± 0.05 | 8.44 ± 0.02 | 7.40 ± 0.01 | 32.18 ± 0.05 | 30.50 ± 0.09 | 36.73 ± 0.30 | 32.77 ± 0.32 |
| M | 2.95 ± 0.01 | 18.52 ± 0.02 | 22.11 ± 0.23 * | 4.83 ± 0.18 * | 6.76 ± 0.01 | 42.83 ± 0.09 * | 27.72 ± 0.51 | 28.95 ± 2.40 * | 39.66 ± 3.53 * | |
| S | 4.00 ± 0.06 * | 18.98 ± 0.05 | 22.49 ± 1.36 * | 7.15 ± 1.00 | 6.46 ± 0.00 | 39.23 ± 0.04 * | 31.38 ± 1.13 | 28.87 ± 0.75 * | 43.41 ± 1.26 * | |
| 96 | G | 3.47 ± 0.01 | 19.15 ± 0.01 | 27.06 ± 0.11 | 6.28 ± 0.08 | 7.75 ± 0.01 | 34.99 ± 0.04 | 29.69 ± 0.42 | 34.81 ± 0.35 | 35.50 ± 0.76 |
| M | 1.84 ± 0.08 * | 15.87 ± 0.62 * | 26.95 ± 1.49 | 8.52 ± 0.80 | 8.00 ± 0.02 | 36.84 ± 1.42 | 27.69 ± 1.72 | 27.58 ± 0.77 * | 41.70 ± 1.07 * | |
| S | 3.92 ± 0.02 * | 19.64 ± 0.09 | 21.00 ± 0.39 * | 5.77 ± 0.27 | 6.58 ± 0.01 | 41.30 ± 0.15 * | 30.72 ± 0.31 | 34.95 ± 2.28 | 37.36 ± 4.00 |
| Pathway | Metabolite | Group | Log2FC # | VIP | p Value | |||
|---|---|---|---|---|---|---|---|---|
| 12 h | 48 h | 12 h | 48 h | 12 h | 48 h | |||
| Lipid metabolism | Lyso PC (14:0/0:0) | G | 0.77 | 1.69 | 0.51 | 1.51 | 0.0002 | 0.0000 |
| M | 2.90 | 2.26 | 1.65 | 1.34 | 0.0000 | 0.0000 | ||
| S | 2.13 | 1.58 | 1.20 | 0.93 | 0.0000 | 0.0018 | ||
| Lyso PC (16:0) | G | 1.15 | −0.57 | 5.09 | 3.43 | 0.0000 | 0.0250 | |
| M | 1.00 | −0.82 | 4.62 | 2.98 | 0.0001 | 0.0068 | ||
| S | 1.22 | −1.20 | 5.46 | 3.66 | 0.0000 | 0.0027 | ||
| Lyso PC (22:6) | G | 1.26 | 0.78 | 9.93 | 11.10 | 0.0000 | 0.0000 | |
| M | 1.60 | 1.74 | 12.17 | 13.97 | 0.0000 | 0.0000 | ||
| S | 1.35 | 1.30 | 10.80 | 9.70 | 0.0000 | 0.0113 | ||
| PC (14:0/22:6) | G | 9.87 | 0.30 | 1.25 | 0.02 | 0.0001 | 0.5223 | |
| M | 0.30 | 0.56 | 0.02 | 0.01 | 0.5223 | 0.5691 | ||
| S | 11.26 | 5.25 | 2.11 | 0.18 | 0.0001 | 0.2076 | ||
| PE (18:1/22:6) | G | 4.83 | 1.24 | 2.29 | 0.78 | 0.0181 | 0.0346 | |
| M | 4.48 | −0.14 | 2.19 | 0.05 | 0.0035 | 0.8663 | ||
| S | 5.07 | 0.69 | 2.83 | 0.28 | 0.0025 | 0.2742 | ||
| PC (22:6/22:6) | G | 4.87 | 0.39 | 2.05 | 0.16 | 0.0000 | 0.5500 | |
| M | 4.95 | −2.48 | 2.14 | 0.32 | 0.0000 | 0.0245 | ||
| S | 5.25 | 1.17 | 2.41 | 0.33 | 0.0000 | 0.1300 | ||
| Energy metabolism | Citric acid | G | −2.08 | −0.50 | 1.66 | 1.62 | 0.0000 | 0.0000 |
| M | 0.47 | 0.96 | 1.18 | 1.99 | 0.0000 | 0.0000 | ||
| S | 0.64 | 1.38 | 1.44 | 2.72 | 0.0000 | 0.0000 | ||
| Pyruvate metabolism | S- Lactoylglutathione | G | −6.02 | 0.33 | 5.47 | 3.64 | 0.0000 | 0.0149 |
| M | −4.09 | −4.57 | 5.48 | 5.82 | 0.0000 | 0.0000 | ||
| S | −4.06 | −5.03 | 5.53 | 6.15 | 0.0000 | 0.0000 | ||
| Pentose phosphate pathway | Gluconolactone | G | −1.42 | −0.80 | 1.71 | 2.27 | 0.0000 | 0.0000 |
| M | −1.18 | −1.34 | 1.66 | 1.77 | 0.0000 | 0.0001 | ||
| S | −1.78 | −2.51 | 1.92 | 2.28 | 0.0000 | 0.0000 | ||
| Gluconic acid | G | 0.75 | 1.29 | 1.10 | 2.60 | 0.0000 | 0.0000 | |
| M | −1.38 | 0.35 | 1.09 | 0.37 | 0.0000 | 0.4299 | ||
| S | −2.36 | −0.56 | 1.27 | 0.70 | 0.0000 | 0.0286 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Tian, W.; Fu, Z.; Ye, W.; Zhang, X.; Zhang, Z.; Sun, D. Mechanisms of Sodium-Acetate-Induced DHA Accumulation in a DHA-Producing Microalga, Crypthecodinium sp. SUN. Mar. Drugs 2022, 20, 508. https://doi.org/10.3390/md20080508
Li Y, Tian W, Fu Z, Ye W, Zhang X, Zhang Z, Sun D. Mechanisms of Sodium-Acetate-Induced DHA Accumulation in a DHA-Producing Microalga, Crypthecodinium sp. SUN. Marine Drugs. 2022; 20(8):508. https://doi.org/10.3390/md20080508
Chicago/Turabian StyleLi, Yiming, Weina Tian, Zhongxiang Fu, Wenqi Ye, Xinwei Zhang, Zhao Zhang, and Dongzhe Sun. 2022. "Mechanisms of Sodium-Acetate-Induced DHA Accumulation in a DHA-Producing Microalga, Crypthecodinium sp. SUN" Marine Drugs 20, no. 8: 508. https://doi.org/10.3390/md20080508
APA StyleLi, Y., Tian, W., Fu, Z., Ye, W., Zhang, X., Zhang, Z., & Sun, D. (2022). Mechanisms of Sodium-Acetate-Induced DHA Accumulation in a DHA-Producing Microalga, Crypthecodinium sp. SUN. Marine Drugs, 20(8), 508. https://doi.org/10.3390/md20080508