Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications
Abstract
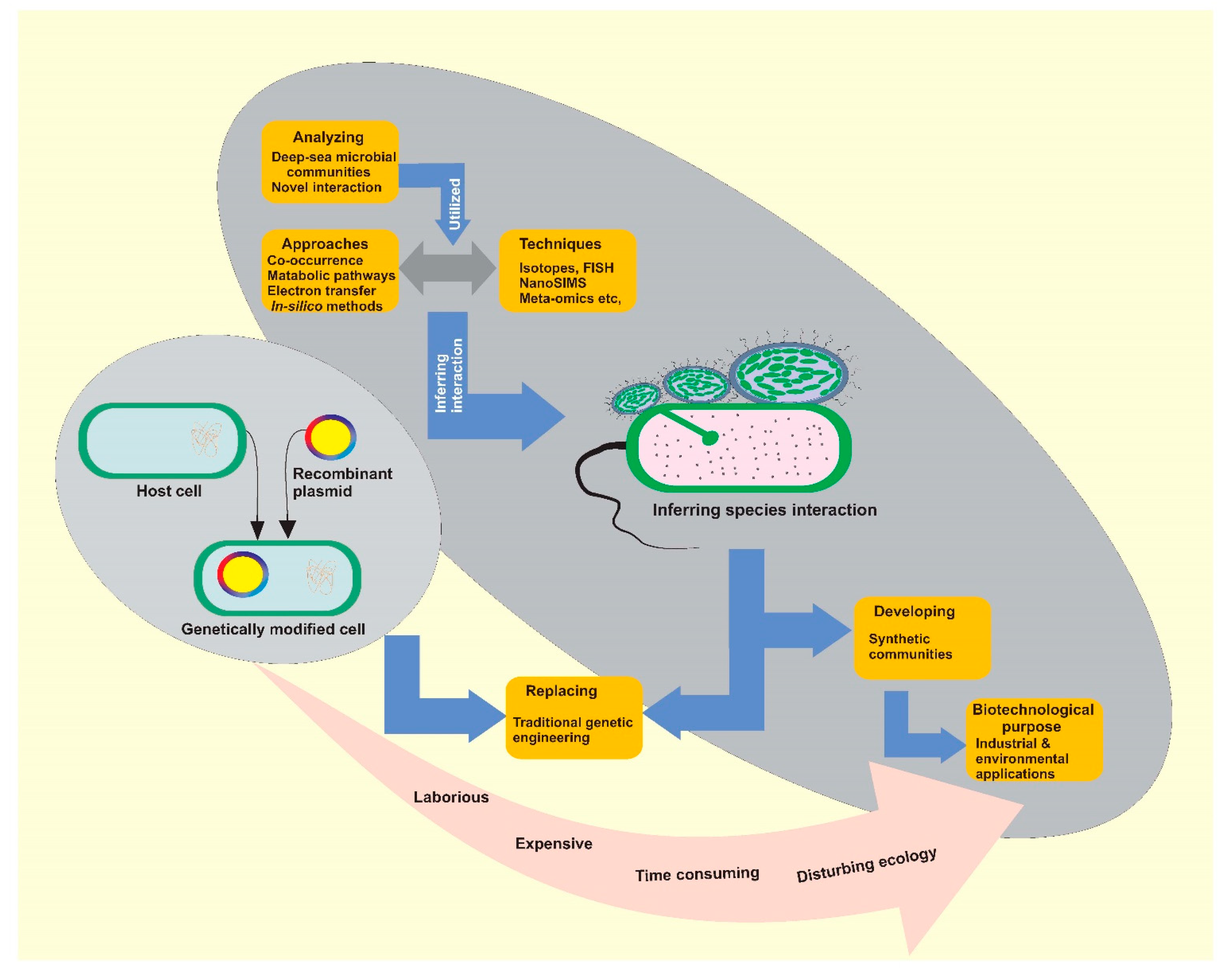
1. Background
2. The Complexity of Microbial Interaction in the Deep-Sea Environment
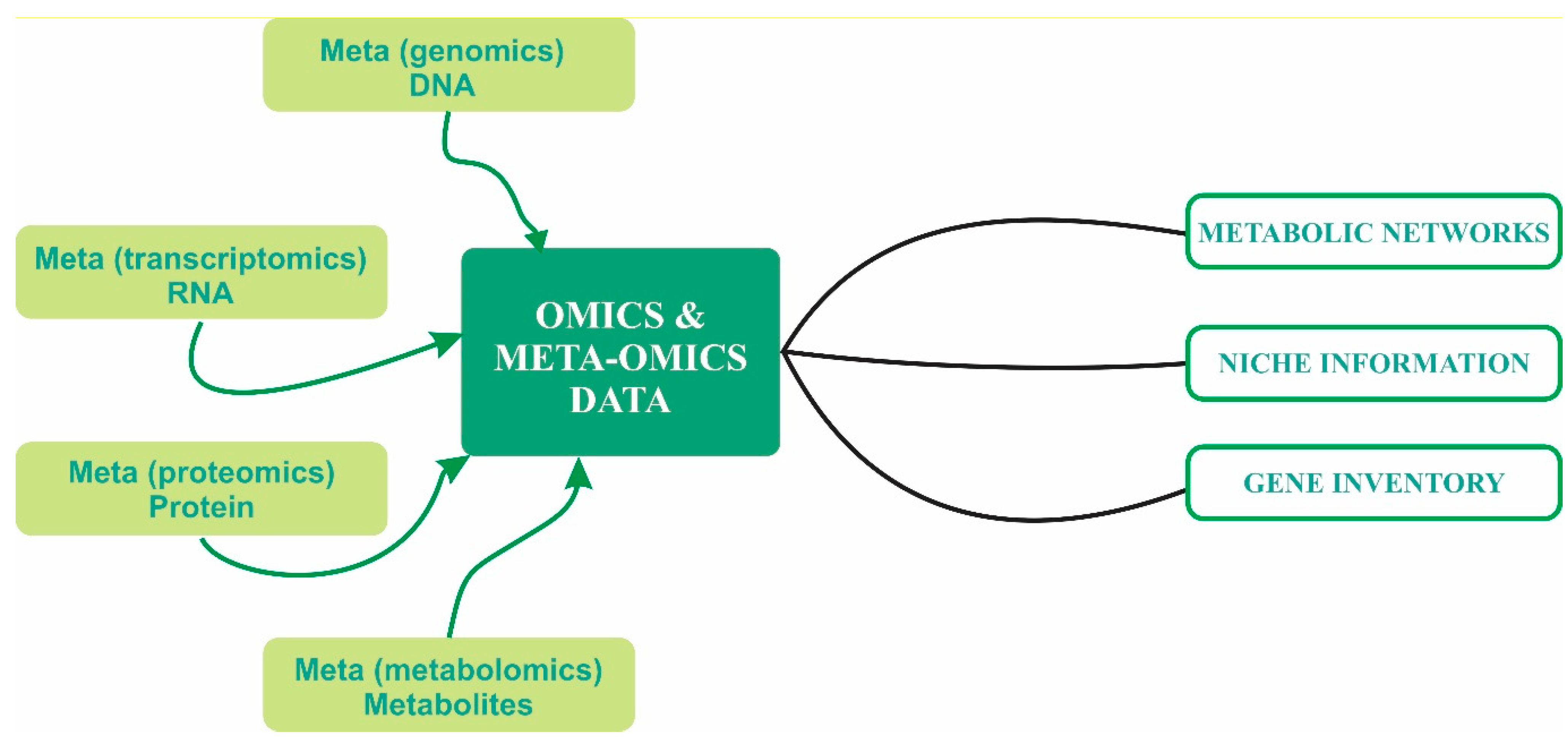
3. Advancements in Molecular Techniques for Exploring Species Interaction
4. Approaches for Exploring Species Interactions
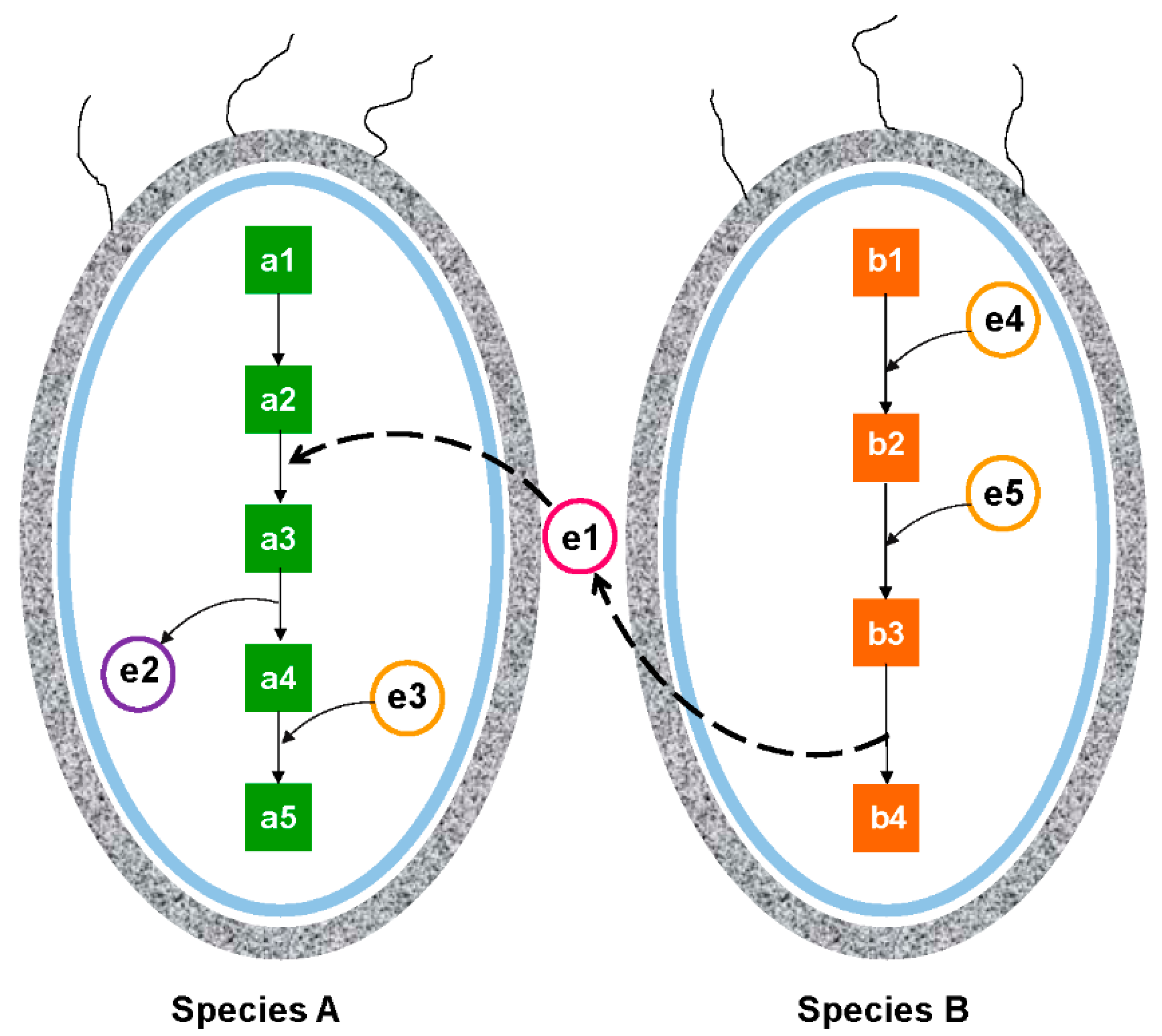
4.1. Inferring Microbial Interactions through Co-Occurrence Pattern Analyses
4.2. Inferring Microbial Interactions through Community Metabolic Pathway Analyses
4.3. Inferring Microbial Interactions through Community Eco-Energetic Modeling
4.4. Synthetic Microbial Communities in Biotechnological and Therapeutic Applications
5. Future Directions
Funding
Conflicts of Interest
References
- Wintermute, E.H.; Silver, P.A. Dynamics in the mixed microbial concourse. Genes Dev. 2010, 24, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.B. Social interaction in synthetic and natural microbial communities. Mol. Syst. Biol. 2011, 7, 483. [Google Scholar] [CrossRef] [PubMed]
- Tanouchi, Y.; Smith, R.P.; You, L. Engineering microbial systems to explore ecological and evolutionary dynamics. Curr. Opin. Biotechnol. 2012, 23, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Mitri, S.; Foster, K.R. The genotypic view of social interactions in microbial communities. Annu. Rev. Genet. 2013, 47, 247–273. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Follows, M.J.; Dutkiewicz, S.; Grant, S.; Chisholm, S.W. Emergent biogeography of microbial communities in a model ocean. Science 2007, 315, 1843–1846. [Google Scholar] [CrossRef]
- Freilich, S.; Kreimer, A.; Meilijson, I.; Gophna, U.; Sharan, R.; Ruppin, E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010, 38, 3857–3868. [Google Scholar] [CrossRef]
- Steele, J.A.; Countway, P.D.; Xia, L.; Vigil, P.D.; Beman, J.M.; Kim, D.Y.; Chow, C.E.; Sachdeva, R.; Jones, A.C.; Schwalbach, M.S.; et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011, 5, 1414–1425. [Google Scholar] [CrossRef]
- Tikariha, H.; Purohit, H.J. Unfolding microbial community intelligence in aerobic and anaerobic biodegradation processes using metagenomics. Arch. Microbiol. 2020, 202, 1269–1274. [Google Scholar] [CrossRef]
- Konopka, A. What is microbial community ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Pande, S.; Kost, C. Bacterial unculturability and the formation of intercellular metabolic networks. Trends Microbiol. 2017, 25, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, O.; Lear, G.; Singhal, N. Metabolic network modeling of microbial interactions in natural and engineered environmental systems. Front. Microbiol. 2016, 7, 673. [Google Scholar] [CrossRef] [PubMed]
- Shou, W.; Ram, S.; Vilar, J.M. Synthetic cooperation in engineered yeast populations. Proc. Natl. Acad. Sci. USA 2007, 104, 1877–1882. [Google Scholar] [CrossRef] [PubMed]
- Hillesland, K.L.; Stahl, D.A. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc. Natl. Acad. Sci. USA 2010, 107, 2124–2129. [Google Scholar] [CrossRef]
- Summers, Z.M.; Fogarty, H.E.; Leang, C.; Franks, A.E.; Malvankar, N.S.; Lovley, D.R. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 2010, 330, 1413–1415. [Google Scholar] [CrossRef]
- Kerner, A.; Park, J.; Williams, A.; Lin, X.N. A programmable Escherichia coli consortium via tunable symbiosis. PLoS ONE 2012, 7, e34032. [Google Scholar] [CrossRef]
- Kerr, B.; Riley, M.A.; Feldman, M.W.; Bohannan, B.J. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 2002, 418, 171–174. [Google Scholar] [CrossRef]
- Balagadde, F.K.; Song, H.; Ozaki, J.; Collins, C.H.; Barnet, M.; Arnold, F.H.; Quake, S.R.; You, L. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 2008, 4, 187. [Google Scholar] [CrossRef]
- Xu, A.; Dolfing, J.; Curtis, T.P.; Montague, G.; Martin, E. Maintenance affects the stability of a two-tiered microbial ‘food chain’? J. Theor. Biol. 2011, 276, 35–41. [Google Scholar] [CrossRef]
- Foster, K.R.; Bell, T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 2012, 22, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Diamond, J.M.; Cody, M.L. Ecology and Evolution of Communities; Belknap Press of Harvard University Press: Cambridge, MA, USA, 1975. [Google Scholar]
- Lidicker, W.Z. A clarification of interactions in ecological systems. BioScience 1979, 29, 475–477. [Google Scholar] [CrossRef]
- Corno, G.; Salka, I.; Pohlmann, K.; Hall, A.R.; Grossart, H.P. Interspecific interactions drive chitin and cellulose degradation by aquatic microorganisms. Aquat. Microb. Ecol. 2015, 76, 27–37. [Google Scholar] [CrossRef]
- Lima-Mendez, G.; Faust, K.; Henry, N.; Decelle, J.; Colin, S.; Carcillo, F.; Chaffron, S.; Ignacio-Espinosa, J.C.; Roux, S.; Vincent, F. Determinants of community structure in the global plankton interactome. Science 2015, 348. [Google Scholar] [CrossRef] [PubMed]
- Gorter, F.A.; Manhart, M.; Ackermann, M. Understanding the evolution of interspecies interactions in microbial communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190256. [Google Scholar] [CrossRef]
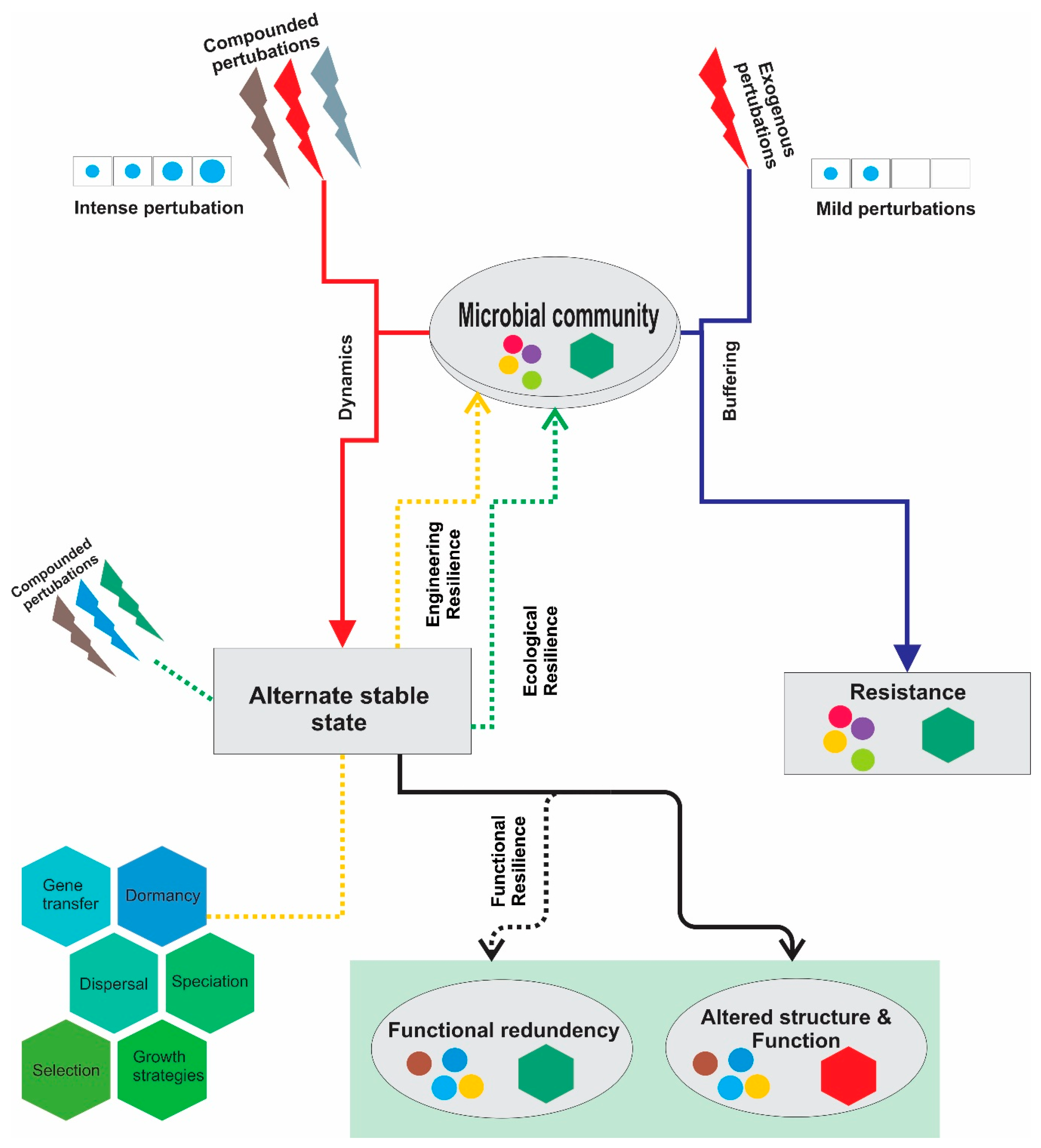
- Angeler, D.G.; Fried-Petersen, H.B.; Allen, C.R.; Garmestani, A.; Twidwell, D.; Chuang, W.-C.; Donovan, V.M.; Eason, T.; Roberts, C.P.; Sundstrom, S.M. Adaptive capacity in ecosystems. In Advances in Ecological Research; Elsevier: Amsterdam, The Netherlands, 2019; Volume 60, pp. 1–24. [Google Scholar]
- Shade, A.; Read, J.S.; Youngblut, N.D.; Fierer, N.; Knight, R.; Kratz, T.K.; Lottig, N.R.; Roden, E.E.; Stanley, E.H.; Stombaugh, J.; et al. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 2012, 6, 2153–2167. [Google Scholar] [CrossRef]
- Philippot, L.; Griffiths, B.S.; Langenheder, S. Microbial community resilience across ecosystems and multiple disturbances. Microbiol. Mol. Biol. Rev. 2021, 85, e00026-20. [Google Scholar] [CrossRef]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.; Relman, D.A. The application of ecological theory toward an understanding of the human microbiome. Science 2012, 336, 1255–1262. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B. Colloquium paper: Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105 (Suppl. 1), 11512–11519. [Google Scholar] [CrossRef]
- Bissett, A.; Burke, C.; Cook, P.L.; Bowman, J.P. Bacterial community shifts in organically perturbed sediments. Environ. Microbiol. 2007, 9, 46–60. [Google Scholar] [CrossRef]
- Yannarell, A.C.; Steppe, T.F.; Paerl, H.W. Disturbance and recovery of microbial community structure and function following Hurricane Frances. Environ. Microbiol. 2007, 9, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Yokokawa, T.; Matsui, K. Biodiversity and multifunctionality in a microbial community: A novel theoretical approach to quantify functional redundancy. Proc. Biol. Sci. 2014, 281, 20132498. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A.; Cram, J.A.; Needham, D.M. Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 2015, 13, 133–146. [Google Scholar] [CrossRef]
- Little, A.E.; Robinson, C.J.; Peterson, S.B.; Raffa, K.F.; Handelsman, J. Rules of engagement: Interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 2008, 62, 375–401. [Google Scholar] [CrossRef]
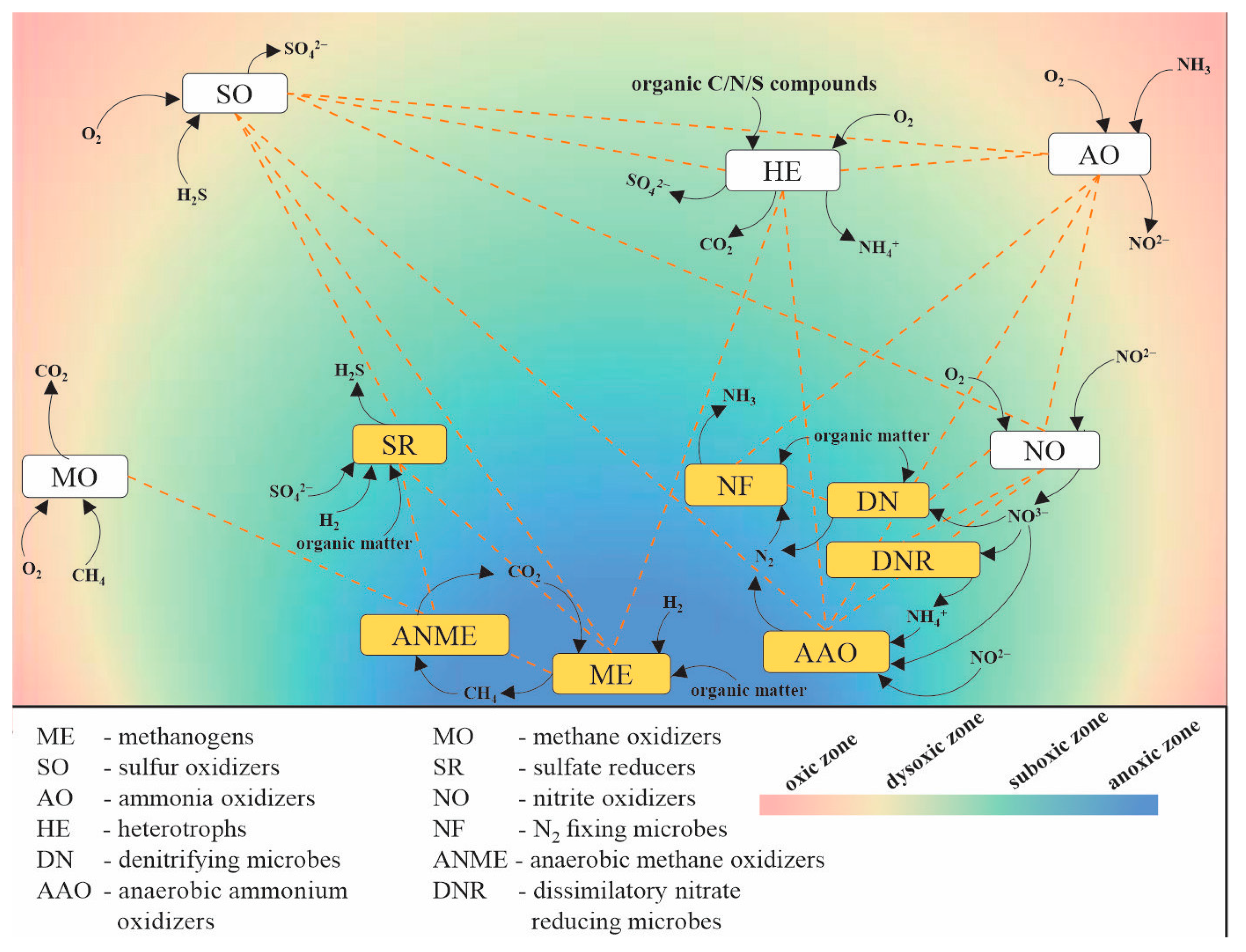
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Zehr, J.P.; Kudela, R.M. Nitrogen cycle of the open ocean: From genes to ecosystems. Ann. Rev. Mar. Sci. 2011, 3, 197–225. [Google Scholar] [CrossRef]
- He, Y.; Li, M.; Perumal, V.; Feng, X.; Fang, J.; Xie, J.; Sievert, S.M.; Wang, F. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 2016, 1, 16035. [Google Scholar] [CrossRef]
- Martin, W.; Baross, J.; Kelley, D.; Russell, M.J. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008, 6, 805–814. [Google Scholar] [CrossRef]
- Jorgensen, B.B.; Boetius, A. Feast and famine--microbial life in the deep-sea bed. Nat. Rev. Microbiol. 2007, 5, 770–781. [Google Scholar] [CrossRef]
- Levin, L.A.; Sibuet, M. Understanding continental margin biodiversity: A new imperative. Ann. Rev. Mar. Sci. 2012, 4, 79–112. [Google Scholar] [CrossRef] [PubMed]
- Bienhold, C.; Pop Ristova, P.; Wenzhofer, F.; Dittmar, T.; Boetius, A. How deep-sea wood falls sustain chemosynthetic life. PLoS ONE 2013, 8, e53590. [Google Scholar] [CrossRef] [PubMed]
- Fulweiler, R.W. Microbiology. Fantastic fixers. Science 2009, 326, 377–378. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Luan, X.; Zhao, J.; Li, J. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Appl. Environ. Microbiol. 2009, 75, 2238–2245. [Google Scholar] [CrossRef]
- Dekas, A.E.; Poretsky, R.S.; Orphan, V.J. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 2009, 326, 422–426. [Google Scholar] [CrossRef]
- Miyazaki, J.; Higa, R.; Toki, T.; Ashi, J.; Tsunogai, U.; Nunoura, T.; Imachi, H.; Takai, K. Molecular characterization of potential nitrogen fixation by anaerobic methane-oxidizing archaea in the methane seep sediments at the number 8 Kumano Knoll in the Kumano Basin, offshore of Japan. Appl. Environ. Microbiol. 2009, 75, 7153–7162. [Google Scholar] [CrossRef]
- Hoehler, T.M.; Jorgensen, B.B. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 2013, 11, 83–94. [Google Scholar] [CrossRef]
- Orsi, W.D. Ecology and evolution of seafloor and subseafloor microbial communities. Nat. Rev. Microbiol. 2018, 16, 671–683. [Google Scholar] [CrossRef]
- Bradley, J.; Arndt, S.; Amend, J.; Burwicz, E.; Dale, A.W.; Egger, M.; LaRowe, D.E. Widespread energy limitation to life in global subseafloor sediments. Sci. Adv. 2020, 6, eaba0697. [Google Scholar] [CrossRef]
- McCollom, T.M. Geochemical constraints on primary productivity in submarine hydrothermal vent plumes. Deep Sea Res. Part I Oceanogr. Res. Pap. 2000, 47, 85–101. [Google Scholar] [CrossRef]
- Bach, W.; Edwards, K.J. Iron and sulfide oxidation within the basaltic ocean crust: Implications for chemolithoautotrophic microbial biomass production. Geochim. Cosmochim. Acta 2003, 67, 3871–3887. [Google Scholar] [CrossRef]
- Graw, M.F.; D’Angelo, G.; Borchers, M.; Thurber, A.R.; Johnson, J.E.; Zhang, C.; Liu, H.; Colwell, F.S. Energy gradients structure microbial communities across sediment horizons in deep marine sediments of the South China sea. Front. Microbiol. 2018, 9, 729. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Chen, C.T.A. Ecological energetic perspectives on responses of nitrogen-transforming chemolithoautotrophic microbiota to changes in the marine environment. Front. Microbiol. 2017, 8, 1246. [Google Scholar] [CrossRef] [PubMed]
- Harcombe, W. Novel cooperation experimentally evolved between species. Evolution 2010, 64, 2166–2172. [Google Scholar] [CrossRef] [PubMed]
- Wintermute, E.H.; Silver, P.A. Emergent cooperation in microbial metabolism. Mol. Syst. Biol. 2010, 6, 407. [Google Scholar] [CrossRef] [PubMed]
- Hom, E.F.; Murray, A.W. Plant-fungal ecology. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science 2014, 345, 94–98. [Google Scholar] [CrossRef]
- Boeuf, D.; Edwards, B.R.; Eppley, J.M.; Hu, S.K.; Poff, K.E.; Romano, A.E.; Caron, D.A.; Karl, D.M.; DeLong, E.F. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc. Natl. Acad. Sci. USA 2019, 116, 11824–11832. [Google Scholar] [CrossRef]
- Baker, B.J.; Appler, K.E.; Gong, X. New microbial biodiversity in marine sediments. Ann. Rev. Mar. Sci. 2021, 13, 161–175. [Google Scholar] [CrossRef]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Mark Welch, D.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef]
- Bartram, A.K.; Lynch, M.D.; Stearns, J.C.; Moreno-Hagelsieb, G.; Neufeld, J.D. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 2011, 77, 3846–3852. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; He, Z.; Wu, L.; Van Nostrand, J.D. Applying GeoChip analysis to disparate microbial communities. Microbe Mag. 2010, 5, 60–65. [Google Scholar] [CrossRef][Green Version]
- Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C.; et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000, 66, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.W.; Chapman, J.; Hugenholtz, P.; Allen, E.E.; Ram, R.J.; Richardson, P.M.; Solovyev, V.V.; Rubin, E.M.; Rokhsar, D.S.; Banfield, J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004, 428, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Tringe, S.G.; von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C.; et al. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Lu, S.; Orcutt, B.N.; Xie, W.; Chen, Y.; Xiao, X.; Edwards, K.J. Discovering the roles of subsurface microorganisms: Progress and future of deep biosphere investigation. Sci. Bull. 2012, 58, 456–467. [Google Scholar] [CrossRef]
- Meyer-Reil, L.-A. Microbial life in sedimentary biofilms—The challenge to microbial ecologists. Mar. Ecol. Prog. Ser. 1994, 112, 303–311. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Berlanga, M.; Guerrero, R. Living together in biofilms: The microbial cell factory and its biotechnological implications. Microb. Cell Fact 2016, 15, 165. [Google Scholar] [CrossRef]
- Williams, P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 2007, 153, 3923–3938. [Google Scholar] [CrossRef]
- Diggle, S.P. Microbial communication and virulence: Lessons from evolutionary theory. Microbiology 2010, 156, 3503–3512. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.M.; Whiteley, M. Special delivery: Vesicle trafficking in prokaryotes. Mol. Microbiol. 2006, 61, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Dubey, G.P.; Ben-Yehuda, S. Intercellular nanotubes mediate bacterial communication. Cell 2011, 144, 590–600. [Google Scholar] [CrossRef]
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Hooshangi, S.; Bentley, W.E. From unicellular properties to multicellular behavior: Bacteria quorum sensing circuitry and applications. Curr. Opin. Biotechnol. 2008, 19, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Bedard, D.L. A case study for microbial biodegradation: Anaerobic bacterial reductive dechlorination of polychlorinated biphenyls—from sediment to defined medium. Annu. Rev. Microbiol. 2008, 62, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, X.; Zhang, C.; Xiao, Z.; Li, Y.; Liang, Y.; Dang, H. Electrostimulated bio-dechlorination of a PCB mixture (Aroclor 1260) in a marine-originated dechlorinating culture. Environ. Pollut. 2021, 291, 118157. [Google Scholar] [CrossRef]
- Cavan, E.L.; Laurenceau-Cornec, E.C.; Bressac, M.; Boyd, P.W. Exploring the ecology of the mesopelagic biological pump. Prog. Oceanogr. 2019, 176, 102125. [Google Scholar] [CrossRef]
- Ambrosino, L.; Tangherlini, M.; Colantuono, C.; Esposito, A.; Sangiovanni, M.; Miralto, M.; Sansone, C.; Chiusano, M.L. Bioinformatics for marine products: An overview of resources, bottlenecks, and perspectives. Mar. Drugs 2019, 17, 576. [Google Scholar] [CrossRef]
- Brenner, K.; You, L.; Arnold, F.H. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef]
- Ben Said, S.; Or, D. Synthetic microbial ecology: Engineering habitats for modular consortia. Front. Microbiol. 2017, 8, 1125. [Google Scholar] [CrossRef] [PubMed]
- Alldredge, A.L.; Silver, M.W. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 1988, 20, 41–82. [Google Scholar] [CrossRef]
- Wright, J.J.; Konwar, K.M.; Hallam, S.J. Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 2012, 10, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Grossart, H.P.; Schweitzer, B.; Ploug, H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 2002, 28, 175–211. [Google Scholar] [CrossRef]
- Bristow, L.A. Anoxia in the snow. Nat. Geosci. 2018, 11, 226–227. [Google Scholar] [CrossRef]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef]
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA 2015, 112, 6449–6454. [Google Scholar] [CrossRef]
- Vergin, K.L.; Jhirad, N.; Dodge, J.; Carlson, C.A.; Giovannoni, S.J. Marine bacterioplankton consortia follow deterministic, non-neutral community assembly rules. Aquat. Microb. Ecol. 2017, 79, 165–175. [Google Scholar] [CrossRef]
- Karl, D.M. Microbial oceanography: Paradigms, processes and promise. Nat. Rev. Microbiol. 2007, 5, 759–769. [Google Scholar] [CrossRef]
- Wong, G.T.F.; Ku, T.-L.; Mulholland, M.; Tseng, C.-M.; Wang, D.-P. The SouthEast Asian Time-series Study (SEATS) and the biogeochemistry of the South China Sea—An overview. Deep Sea Res. Part II Top. Stud. 2007, 54, 1434–1447. [Google Scholar] [CrossRef]
- Faust, K.; Lahti, L.; Gonze, D.; de Vos, W.M.; Raes, J. Metagenomics meets time series analysis: Unraveling microbial community dynamics. Curr. Opin. Microbiol. 2015, 25, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Gause, G.F. Experimental analysis of Vito Volterra’s mathematical theory of the struggle for existence. Science 1934, 79, 16–17. [Google Scholar] [CrossRef]
- Grosskopf, T.; Soyer, O.S. Synthetic microbial communities. Curr. Opin. Microbiol. 2014, 18, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Ding, M.Z.; Jia, X.Q.; Ma, Q.; Yuan, Y.J. Synthetic microbial consortia: From systematic analysis to construction and applications. Chem. Soc. Rev. 2014, 43, 6954–6981. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.J.; Gutierrez-Zamora, M.L.; Manefield, M.; Gieg, L.M. Identification of toluene degraders in a methanogenic enrichment culture. FEMS Microbiol. Ecol. 2014, 89, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Behrens, S.; Losekann, T.; Pett-Ridge, J.; Weber, P.K.; Ng, W.O.; Stevenson, B.S.; Hutcheon, I.D.; Relman, D.A.; Spormann, A.M. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl. Environ. Microbiol. 2008, 74, 3143–3150. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Andreasen, K.; Lee, N.; Wagner, M. Use of microautoradiography and fluorescent hybridization for characterization of microbial activity in activated sludge. Water Sci. Technol. 1999, 39, 1–9. [Google Scholar] [CrossRef]
- Huang, W.E.; Stoecker, K.; Griffiths, R.; Newbold, L.; Daims, H.; Whiteley, A.S.; Wagner, M. Raman-FISH: Combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ. Microbiol. 2007, 9, 1878–1889. [Google Scholar] [CrossRef]
- Guerquin-Kern, J.-L.; Wu, T.-D.; Quintana, C.; Croisy, A. Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry (SIMS microscopy). Biochim. Biophys. Acta 2005, 1724, 228–238. [Google Scholar] [CrossRef]
- Wagner, M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu. Rev. Microbiol. 2009, 63, 411–429. [Google Scholar] [CrossRef] [PubMed]
- Orphan, V.J.; House, C.H.; Hinrichs, K.U.; McKeegan, K.D.; DeLong, E.F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 2002, 99, 7663–7668. [Google Scholar] [CrossRef] [PubMed]
- Hallam, S.J.; Mincer, T.J.; Schleper, C.; Preston, C.M.; Roberts, K.; Richardson, P.M.; DeLong, E.F. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006, 4, e95. [Google Scholar] [CrossRef] [PubMed]
- Raes, J.; Bork, P. Molecular eco-systems biology: Towards an understanding of community function. Nat. Rev. Microbiol. 2008, 6, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Chaffron, S.; Rehrauer, H.; Pernthaler, J.; von Mering, C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010, 20, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A.; Heinrich, F.; Bertilsson, S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012, 6, 330–342. [Google Scholar] [CrossRef]
- Kalenitchenko, D.; Fagervold, S.K.; Pruski, A.M.; Vetion, G.; Yucel, M.; Le Bris, N.; Galand, P.E. Temporal and spatial constraints on community assembly during microbial colonization of wood in seawater. ISME J. 2015, 9, 2657–2670. [Google Scholar] [CrossRef]
- Leinweber, A.; Fredrik Inglis, R.; Kummerli, R. Cheating fosters species co-existence in well-mixed bacterial communities. ISME J. 2017, 11, 1179–1188. [Google Scholar] [CrossRef]
- Williams, R.J.; Howe, A.; Hofmockel, K.S. Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front. Microbiol. 2014, 5, 358. [Google Scholar] [CrossRef]
- Parkes, R.J.; Cragg, B.A.; Bale, S.J.; Getlifff, J.M.; Goodman, K.; Rochelle, P.A.; Fry, J.C.; Weightman, A.J.; Harvey, S.M. Deep bacterial biosphere in Pacific Ocean sediments. Nature 1994, 371, 410–413. [Google Scholar] [CrossRef]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed]
- Ponomarova, O.; Patil, K.R. Metabolic interactions in microbial communities: Untangling the Gordian knot. Curr. Opin. Microbiol. 2015, 27, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Lykidis, A.; Chen, C.L.; Tringe, S.G.; McHardy, A.C.; Copeland, A.; Kyrpides, N.C.; Hugenholtz, P.; Macarie, H.; Olmos, A.; Monroy, O.; et al. Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J. 2011, 5, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Shafquat, A.; Joice, R.; Simmons, S.L.; Huttenhower, C. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol. 2014, 22, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Zhang, T. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J. 2015, 9, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.E.; Wu, S.; Bhattacharjee, A.S.; Hamilton, J.J.; McMahon, K.D.; Goel, R.; Noguera, D.R. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat. Commun. 2017, 8, 15416. [Google Scholar] [CrossRef] [PubMed]
- Ofaim, S.; Ofek-Lalzar, M.; Sela, N.; Jinag, J.; Kashi, Y.; Minz, D.; Freilich, S. Analysis of microbial functions in the rhizosphere using a metabolic-network based framework for metagenomics interpretation. Front. Microbiol. 2017, 8, 1606. [Google Scholar] [CrossRef]
- Embree, M.; Nagarajan, H.; Movahedi, N.; Chitsaz, H.; Zengler, K. Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J. 2014, 8, 757–767. [Google Scholar] [CrossRef]
- Hawley, A.K.; Brewer, H.M.; Norbeck, A.D.; Pasa-Tolic, L.; Hallam, S.J. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc. Natl. Acad. Sci. USA 2014, 111, 11395–11400. [Google Scholar] [CrossRef]
- Verastegui, Y.; Cheng, J.; Engel, K.; Kolczynski, D.; Mortimer, S.; Lavigne, J.; Montalibet, J.; Romantsov, T.; Hall, M.; McConkey, B.J.; et al. Multisubstrate isotope labeling and metagenomic analysis of active soil bacterial communities. mBio 2014, 5, e01157-14. [Google Scholar] [CrossRef]
- Comolli, L.R.; Banfield, J.F. Inter-species interconnections in acid mine drainage microbial communities. Front. Microbiol. 2014, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.-J.; Chen, P.-Y.; Liaw, C.-C.; Lai, Y.-M.; Yang, Y.-L. Bringing microbial interactions to light using imaging mass spectrometry. Nat. Prod. Rep. 2014, 31, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, L.; Qi, Y.; Ma, C. Imaging mass spectrometry of interspecies metabolic exchange revealed the allelopathic interaction between Microcystis aeruginosa and its antagonist. Chemosphere 2020, 259, 127430. [Google Scholar] [CrossRef] [PubMed]
- Klitgord, N.; Segre, D. Environments that induce synthetic microbial ecosystems. PLoS. Comput. Biol. 2010, 6, e1001002. [Google Scholar] [CrossRef]
- Mahadevan, R.; Henson, M.A. Genome-based modeling and design of metabolic interactions in microbial communities. Comput. Struct. Biotechnol. J. 2012, 3, e201210008. [Google Scholar] [CrossRef]
- Zengler, K.; Palsson, B.O. A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 2012, 10, 366–3672. [Google Scholar] [CrossRef]
- Feist, A.M.; Herrgard, M.J.; Thiele, I.; Reed, J.L.; Palsson, B.O. Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 2009, 7, 129–143. [Google Scholar] [CrossRef]
- Cardona, C.; Weisenhorn, P.; Henry, C.; Gilbert, J.A. Network-based metabolic analysis and microbial community modeling. Curr. Opin. Microbiol. 2016, 31, 124–131. [Google Scholar] [CrossRef]
- Borenstein, E.; Kupiec, M.; Feldman, M.W.; Ruppin, E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc. Natl. Acad. Sci. USA 2008, 105, 14482–14487. [Google Scholar] [CrossRef]
- Handorf, T.; Christian, N.; Ebenhoh, O.; Kahn, D. An environmental perspective on metabolism. J. Theor. Biol. 2008, 252, 530–537. [Google Scholar] [CrossRef]
- Borenstein, E.; Feldman, M.W. Topological signatures of species interactions in metabolic networks. J. Comput. Biol. 2009, 16, 191–200. [Google Scholar] [CrossRef]
- Roling, W.F.; Ferrer, M.; Golyshin, P.N. Systems approaches to microbial communities and their functioning. Curr. Opin. Biotechnol. 2010, 21, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Morine, M.J.; Gu, H.; Myers, R.A.; Bielawski, J.P. Trade-offs between efficiency and robustness in bacterial metabolic networks are associated with niche breadth. J. Mol. Evol. 2009, 68, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Freilich, S.; Kreimer, A.; Borenstein, E.; Yosef, N.; Sharan, R.; Gophna, U.; Ruppin, E. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 2009, 10, R61. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef]
- Le Bris, N.; Yücel, M.; Das, A.; Sievert, S.M.; LokaBharathi, P.; Girguis, P.R. Hydrothermal energy transfer and organic carbon production at the deep seafloor. Front. Mar. Sci. 2019, 5, 531. [Google Scholar] [CrossRef]
- Wu, W.F.; Wang, F.P.; Li, J.H.; Yang, X.W.; Xiao, X.; Pan, Y.X. Iron reduction and mineralization of deep-sea iron reducing bacterium Shewanella piezotolerans WP3 at elevated hydrostatic pressures. Geobiology 2013, 11, 593–601. [Google Scholar]
- Melton, E.D.; Swanner, E.D.; Behrens, S.; Schmidt, C.; Kappler, A. The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle. Nat. Rev. Microbiol. 2014, 12, 797–808. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Liu, J.; Levar, C.; Edwards, M.J.; Babauta, J.T.; Kennedy, D.W.; Shi, Z.; Beyenal, H.; Bond, D.R.; et al. A trans-outer membrane porin-cytochrome protein complex for extracellular electron transfer by Geobacter sulfurreducens PCA. Environ. Microbiol. Rep. 2014, 6, 776–785. [Google Scholar] [CrossRef]
- Bird, L.J.; Bonnefoy, V.; Newman, D.K. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 2011, 19, 330–340. [Google Scholar] [CrossRef]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Hashimoto, K.; Watanabe, K. Microbial interspecies electron transfer via electric currents through conductive minerals. Proc. Natl. Acad. Sci. USA 2012, 109, 10042–10046. [Google Scholar] [CrossRef] [PubMed]
- Knittel, K.; Boetius, A. Anaerobic oxidation of methane: Progress with an unknown process. Annu. Rev. Microbiol. 2009, 63, 311–334. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K. Anaerobic oxidation of methane with sulfate: On the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 2011, 14, 292–299. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, S.E.; Chadwick, G.L.; Kempes, C.P.; Orphan, V.J. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 2015, 526, 531–535. [Google Scholar] [CrossRef]
- Wegener, G.; Krukenberg, V.; Riedel, D.; Tegetmeyer, H.E.; Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 2015, 526, 587–590. [Google Scholar] [CrossRef]
- Dodsworth, J.A.; McDonald, A.I.; Hedlund, B.P. Calculation of total free energy yield as an alternative approach for predicting the importance of potential chemolithotrophic reactions in geothermal springs. FEMS Microbiol. Ecol. 2012, 81, 446–454. [Google Scholar] [CrossRef][Green Version]
- Bradley, J.A.; Amend, J.P.; LaRowe, D.E. Bioenergetic controls on microbial ecophysiology in marine sediments. Front. Microbiol. 2018, 9, 180. [Google Scholar] [CrossRef]
- Sebastian, M.; Forn, I.; Auladell, A.; Gomez-Letona, M.; Sala, M.M.; Gasol, J.M.; Marrase, C. Differential recruitment of opportunistic taxa leads to contrasting abilities in carbon processing by bathypelagic and surface microbial communities. Environ. Microbiol. 2021, 23, 190–206. [Google Scholar] [CrossRef]
- Farag, I.F.; Biddle, J.F.; Zhao, R.; Martino, A.J.; House, C.H.; Leon-Zayas, R.I. Metabolic potentials of archaeal lineages resolved from metagenomes of deep Costa Rica sediments. ISME J. 2020, 14, 1345–1358. [Google Scholar] [CrossRef]
- Dombrowski, N.; Seitz, K.W.; Teske, A.P.; Baker, B.J. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 2017, 5, 106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kim, S.K. Research and application of marine microbial enzymes: Status and prospects. Mar. Drugs 2010, 8, 1920–1934. [Google Scholar] [CrossRef] [PubMed]
- Raddadi, N.; Cherif, A.; Daffonchio, D.; Neifar, M.; Fava, F. Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 2015, 99, 7907–7913. [Google Scholar] [CrossRef]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and hot extremozymes: Industrial relevance and current trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Tshikantwa, T.S.; Ullah, M.W.; He, F.; Yang, G. Current trends and potential applications of microbial interactions for human welfare. Front. Microbiol. 2018, 9, 1156. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Rehman, Z.U.; Leiknes, T. Quorum-quenching bacteria isolated from Red Sea sediments reduce biofilm formation by Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 1354. [Google Scholar] [CrossRef]
- Packiavathy, I.; Kannappan, A.; Thiyagarajan, S.; Srinivasan, R.; Jeyapragash, D.; Paul, J.B.J.; Velmurugan, P.; Ravi, A.V. AHL-lactonase producing Psychrobacter sp. from Palk Bay sediment mitigates quorum sensing-mediated virulence production in Gram negative bacterial pathogens. Front. Microbiol. 2021, 12, 634593. [Google Scholar] [CrossRef]
- Hays, S.G.; Patrick, W.G.; Ziesack, M.; Oxman, N.; Silver, P.A. Better together: Engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 2015, 36, 40–49. [Google Scholar] [CrossRef]
- Anderson, J.C.; Clarke, E.J.; Arkin, A.P.; Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006, 355, 619–627. [Google Scholar] [CrossRef]
- Chifiriuc, M.C.; Grumezescu, A.M.; Lazar, V. Quorum sensing inhibitors from the sea: Lessons from marine symbiotic relationships. Curr. Org. Chem. 2014, 18, 823–839. [Google Scholar] [CrossRef]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation--a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef] [PubMed]
- Gilman, S.E.; Urban, M.C.; Tewksbury, J.; Gilchrist, G.W.; Holt, R.D. A framework for community interactions under climate change. Trends Ecol. Evol. 2010, 25, 325–331. [Google Scholar] [CrossRef]
- Pacheco, A.R.; Segre, D. A multidimensional perspective on microbial interactions. FEMS Microbiol. Lett. 2019, 366, fnz125. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Acinas, S.G.; Bork, P.; Bowler, C.; Tara Oceans, C.; Eveillard, D.; Gorsky, G.; Guidi, L.; Iudicone, D.; Karsenti, E.; et al. Tara Oceans: Towards global ocean ecosystems biology. Nat. Rev. Microbiol. 2020, 18, 428–445. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawaz, M.Z.; Subin Sasidharan, R.; Alghamdi, H.A.; Dang, H. Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications. Mar. Drugs 2022, 20, 108. https://doi.org/10.3390/md20020108
Nawaz MZ, Subin Sasidharan R, Alghamdi HA, Dang H. Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications. Marine Drugs. 2022; 20(2):108. https://doi.org/10.3390/md20020108
Chicago/Turabian StyleNawaz, Muhammad Zohaib, Raghul Subin Sasidharan, Huda Ahmed Alghamdi, and Hongyue Dang. 2022. "Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications" Marine Drugs 20, no. 2: 108. https://doi.org/10.3390/md20020108
APA StyleNawaz, M. Z., Subin Sasidharan, R., Alghamdi, H. A., & Dang, H. (2022). Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications. Marine Drugs, 20(2), 108. https://doi.org/10.3390/md20020108







