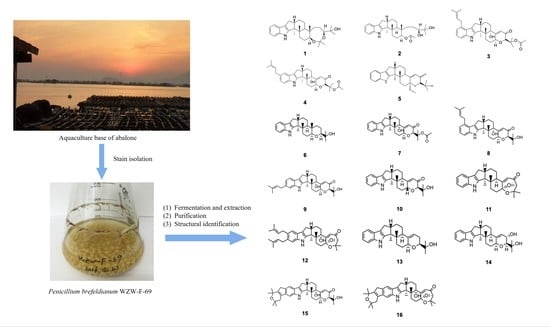
Paspalines C–D and Paxillines B–D: New Indole Diterpenoids from Penicillium brefeldianum WZW-F-69
Abstract
1. Introduction
2. Results and Discussion
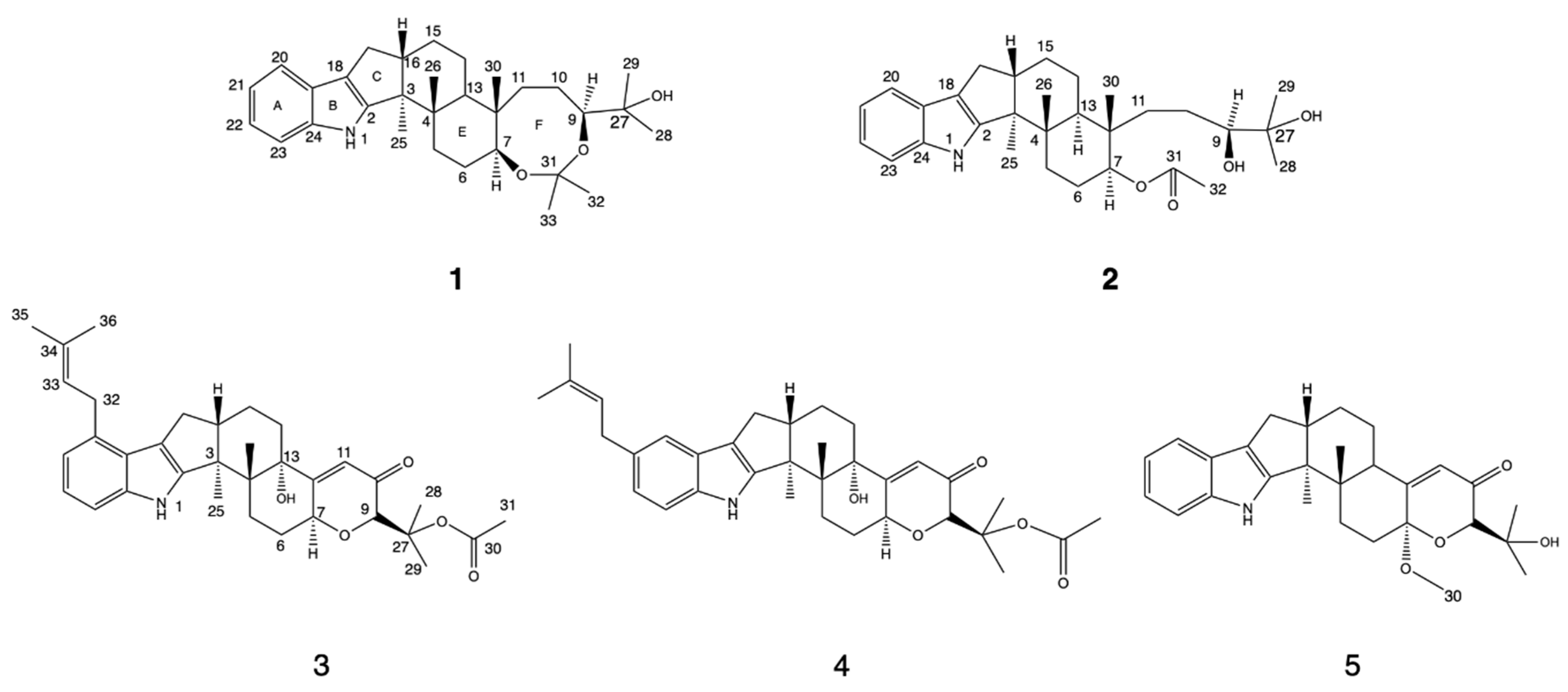
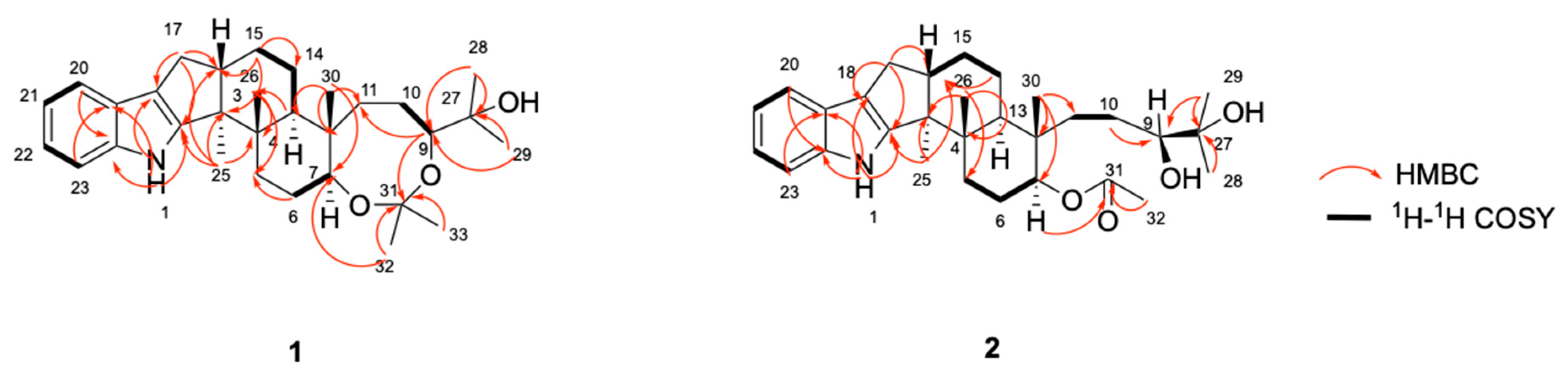
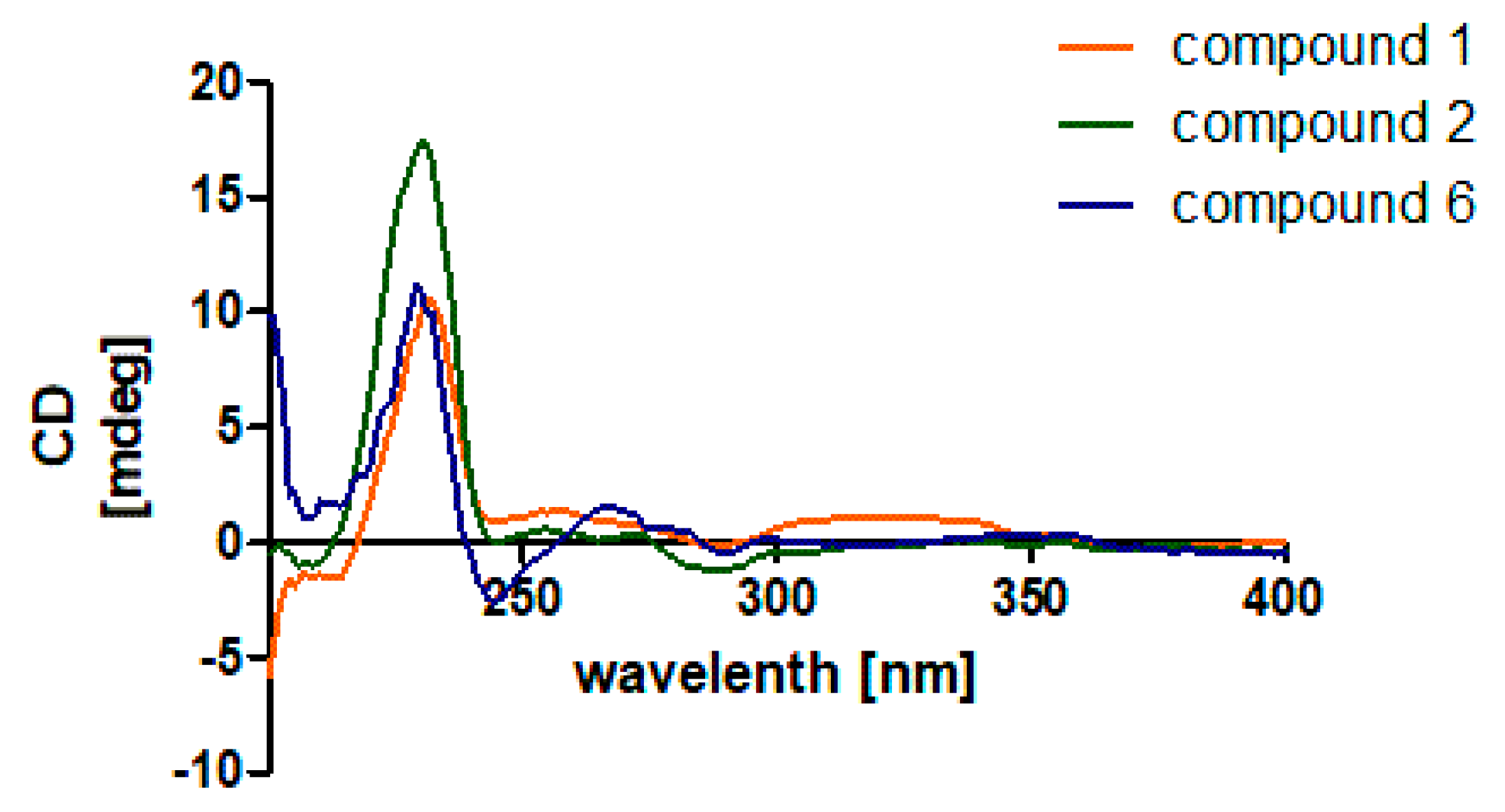
2.1. Structure Elucidation of the New Compounds
2.2. Bioactivities of Compounds
3. Materials and Methods
3.1. General Apparatus
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Isolation and Purification
3.5. Antimicrobial Activity
3.6. Cytotoxic Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ariantari, N.P.; Ancheeva, E.; Wang, C.; Mandi, A.; Knedel, T.-O.; Kurtan, T.; Chaidir, C.; Mueller, W.E.G.; Kassack, M.U.; Janiak, C.; et al. Indole diterpenoids from an endophytic Penicillium sp. J. Nat. Prod. 2019, 82, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Breteche, A.; Duflos, M.; Dassonville, A.; Nourrisson, M.R.; Brelet, J.; Le Baut, G.; Grimaud, N.; Petit, J.Y. New N-pyridinyl(methyl)-indole-2- and 3-(alkyl) carboxamides and derivatives acting as systemic and topical inflammation inhibitors. J. Enzym. Inhib. Med. Chem. 2002, 17, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.Y.; Meng, L.H.; Li, X.; Yang, S.Q.; Li, X.M.; Wang, B.G. Three new indole diterpenoids from the sea-anemone-derived fungus Penicillium sp. AS-79. Mar. Drugs 2017, 15, 137. [Google Scholar] [CrossRef]
- Munday-Finch, S.C.; Wilkins, A.L.; Miles, C.O. Isolation of paspaline B, an indole-diterpenoid from Penicillium paxilli. Phytochemistry 1996, 41, 327–332. [Google Scholar] [CrossRef]
- Huang, X.H.; Tomoda, H.; Nishida, H.; Masuma, R.; Omura, S. Terpendoles, novel ACAT inhibitors produced by Albophoma yamanashiensis. I. Production, isolation and biological properties. J. Antibiot. 1995, 48, 1–4. [Google Scholar] [CrossRef]
- Sallam, A.A.; Houssen, W.E.; Gissendanner, C.R.; Orabi, K.Y.; Foudah, A.I.; El Sayed, K.A. Bioguided discovery and pharmacophore modeling of the mycotoxic indole diterpene alkaloids penitrems as breast cancer proliferation, migration, and invasion inhibitors. MedChemComm 2013, 4, 1360–1369. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, Y.; Liu, P.; Fu, P.; Zhu, T.; Wang, W.; Zhu, W. Indole diterpenoids with anti-H1N1 activity from the aciduric fungus Penicillium camemberti-1492. J. Nat. Prod. 2013, 76, 1328–1336. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Noike, M.; Minami, A.; Oikawa, H.; Dairi, T. A fungal prenyltransferase catalyzes the regular di-prenylation at positions 20 and 21 of paxilline. Biosci. Biotechnol. Biochem. 2014, 78, 448–454. [Google Scholar] [CrossRef]
- Reddy, P.; Guthridge, K.; Vassiliadis, S.; Hemsworth, J.; Hettiarachchige, I.; Spangenberg, G.; Rochfort, S. Tremorgenic mycotoxins: Structure diversity and biological activity. Toxins 2019, 11, 302. [Google Scholar] [CrossRef]
- Zhou, L.M.; Kong, F.D.; Fan, P.; Ma, Q.Y.; Xie, Q.Y.; Li, J.H.; Zheng, H.Z.; Zheng, Z.H.; Yuan, J.Z.; Dai, H.F.; et al. Indole-diterpenoids with protein tyrosine phosphatase inhibitory activities from the marine-derived fungus Penicillium sp. KFD28. J. Nat. Prod. 2019, 82, 2638–2644. [Google Scholar] [CrossRef]
- Kim, D.E.; Zweig, J.E.; Newhouse, T.R. Total synthesis of paspaline A and emindole PB enabled by computational augmentation of a transform-guided retrosynthetic strategy. J. Am. Chem. Soc. 2019, 141, 1479–1483. [Google Scholar] [CrossRef]
- Liu, C.; Tagami, K.; Minami, A.; Matsumoto, T.; Frisvad, J.C.; Suzuki, H.; Ishikawa, J.; Gomi, K.; Oikawa, H. Reconstitution of biosynthetic machinery for the synthesis of the highly elaborated indole diterpene penitrem. Angew. Chem. Int. Ed. 2015, 54, 5748–5752. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.I.; Gross, M.; Cramer, B.; Wegner, S.; Hausmann, H.; Hamscher, G.; Usleber, E. Detection of the tremorgenic mycotoxin paxilline and its desoxy analog in ergot of rye and barley: A new class of mycotoxins added to an old problem. Anal. Bioanal. Chem. 2017, 409, 5101–5112. [Google Scholar] [CrossRef] [PubMed]
- Kozak, L.; Pocsi, I.; Molnar, I.; Kozak, L.; Szilagyi, Z.; Toth, L.; Molnar, I. Tremorgenic and neurotoxic paspaline-derived indole-diterpenes: Biosynthetic diversity, threats and applications. Appl. Microbiol. Biotechnol. 2019, 103, 1599–1616. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.; Parker, E.J.; Koulman, A.; Scott, B. Defining paxilline biosynthesis in Penicillium paxilli: Functional characterization of two cytochrome P450 monooxygenases. J. Biol. Chem. 2007, 282, 16829–16837. [Google Scholar] [CrossRef] [PubMed]
- Fehr, T.; Acklin, W. Isolation of 2 new indole derivatives from the mycelia of Claviceps paspali. Helv. Chim. Acta 1966, 49, 1907–1910. [Google Scholar] [CrossRef]
- Springer, J.P.; Clardy, J. Paspaline and paspalicine, two indole-mevalonate metabolites from Claviceps paspali. Tetrahedron Lett. 1980, 21, 231–234. [Google Scholar] [CrossRef]
- Nozawa, K.; Horie, Y.; Udagawa, S.; Kawai, K.; Yamazaki, M. Studies on fungal products. Part XXIV. Isolation of a new tremorgenic indoloditerpene, 1′-O-acetylpaxilline, from Emericella striata and distribution of paxilline in Emericella spp. Chem. Pharm. Bull. 1989, 37, 1387–1389. [Google Scholar] [CrossRef]
- Mantle, P.G.; Weedon, C.M. Biosynthesis and transformation of tremorgenic indole-diterpenoids by Penicillium paxilli and Acremonium lolii. Phytochemistry 1994, 36, 1209–1217. [Google Scholar] [CrossRef]
- Belofsky, G.N.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. Antiinsectan alkaloids: Shearinines A–C and a new paxilline derivative from the ascostromata of Eupenicillium shearii. Tetrahedron 1995, 51, 3959–3968. [Google Scholar] [CrossRef]
- Springer, J.P.; Clardy, J.; Wells, J.M.; Cole, R.J.; Kirksey, J.W. Structure of paxilline, a tremorgenic metabolite of Penicillium paxilli. Tetrahedron Lett. 1975, 16, 2531–2534. [Google Scholar] [CrossRef]
- Gao, S.S.; Li, X.M.; Williams, K.; Proksch, P.; Ji, N.Y.; Wang, B.G. Rhizovarins A-F, indole-diterpenes from the mangrove-derived endophytic fungus Mucor Irregularis QEN-189. J. Nat. Prod. 2016, 79, 2066–2074. [Google Scholar] [CrossRef] [PubMed]
- Mc, G.J. The filter paper disc method of assaying antibiotics. J. Bacteriol. 1945, 50, 717. [Google Scholar]



| No. | 1 | 2 | ||
|---|---|---|---|---|
| 1H | 13C | 1H | 13C | |
| 1 | 7.74, s | 7.77, s | ||
| 2 | 150.8, C | 150.6, C | ||
| 3 | 53.1, C | 53.1, C | ||
| 4 | 39.3, C | 39.3, C | ||
| 5 | 1.94, dd (11.3, 5.8) 1.62, m | 33.6, CH2 | 2.05, dd (13.7,4.6) 2.01, dd (12.5, 5.4) | 33.1, CH2 |
| 6 | 1.89, dd (14.7, 2.7) 1.87, m | 26.9, CH2 | 1.90, m | 23.8, CH2 |
| 7 | 3.59, dd (9.8, 5.9) | 72.8, CH | 4.80, dd (10.2, 5.9) | 75.5, CH |
| 9 | 3.68, dd (8.3, 4.6) | 84.1, CH | 3.57, dd (8.5, 4.3) | 83.6, CH |
| 10 | 1.34, dd (12.1, 5.6) 1.32, m | 22.2, CH2 | 1.59, m | 22.6, CH2 |
| 11 | 1.71, td (12.9, 3.0) 1.41, m | 33.9, CH2 | 1.55, m 1.32, m | 34.4, CH2 |
| 12 | 39.3, C | 40.2, C | ||
| 13 | 1.69, s | 41.4, CH | 1.78, dd (12.7, 3.1) | 40.4, CH |
| 14 | 1.81, d (12.6) 1.64, brs | 25.2, CH2 | 1.82, m 1.65, td (12.9, 4.1) | 25.1, CH2 |
| 15 | 1.71, m 1.49, td (12.4, 4.4) | 23.0, CH2 | 1.59, m | 22.6, CH2 |
| 16 | 2.78, m | 47.8, CH | 2.78, m | 48.7, CH |
| 17 | 2.70, dd (13.2, 6.5) 2.35, dd (13.2, 10.6) | 27.5, CH2 | 2.70, dd (13.2, 6.4) 2.36, dd (13.2, 10.6) | 27.5, CH2 |
| 18 | 118.4, C | 118.3, C | ||
| 19 | 125.4, C | 125.1, C | ||
| 20 | 7.45, ddd (6.6, 3.2, 1.0) | 118.3, CH | 7.44, m | 118.4, CH |
| 21 | 7.09, m | 119.6, CH | 7.09, m | 119.6, CH |
| 22 | 7.10, m | 120.5, CH | 7.09, m | 120.5, CH |
| 23 | 7.32, ddd (6.4, 2.5, 0.8) | 111.4, CH | 7.32, m | 111.5, CH |
| No. | 3 | 4 | 5 | |||
|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | |
| 1 | 7.78, s | 7.70, s | 7.79, s | |||
| 2 | 151.0, C | 151.9, C | 149.3, C | |||
| 3 | 50.5, C | 50.7, C | 50.5, C | |||
| 4 | 43.1, C | 43.1, C | 42.4, C | |||
| 5 | 2.80, td (13.5, 4.9) 1.49, m | 28.1, CH | 2.78, td (13.7, 4.4) 1.47, m | 28.0, CH | 2.15, td (13.7, 6.4) 1.58, m | 31.4, CH |
| 6 | 2.31, m 1.95, m | 28.3, CH2 | 2.31, m 1.93, m | 28.3, CH2 | 2.44, m 1.90, dd (14.6, 4.3) | 28.8, CH2 |
| 7 | 4.84, m | 72.9, CH | 4.84, m | 72.9, CH | 96.5, C | |
| 9 | 4.85, brs | 80.4, CH | 4.85, brs | 80.4, CH | 4.06, s | 76.9, CH |
| 10 | 195.3, C | 195.3, C | 198.5, C | |||
| 11 | 5.85, d (2.1) | 120.1, CH | 5.84, d (2.0) | 120.1, CH | 5.81, d (2.0) | 122.1, CH |
| 12 | 166.1, C | 166.1, C | 165.1, C | |||
| 13 | 77.5, C | 77.5, C | 2.83, m | 42.0, CH | ||
| 14 | 2.08, m 1.66, m | 34.4, CH2 | 2.07, m 1.67, m | 34.4, CH2 | 1.72, m 1.52, m | 25.5, CH2 |
| 15 | 2.07, m | 20.9, CH2 | 2.05, m 1.80, m | 20.9, CH2 | 1.86, m | 24.1, CH2 |
| 16 | 2.89, d (2.8) | 49.6, CH | 2.85, m | 49.6, CH | 2.85, m | 49.0, CH |
| 17 | 2.91, d (6.2) 2.62, m | 29.0, CH2 | 2.74, dd (13.0, 6.3) 2.45, dd (13.2, 10.9) | 27.2, CH2 | 2.74, dd (13.4, 6.4) 2.45, m | 27.3, CH2 |
| 18 | 116.9, C | 117.1, C, | 118.4, C | |||
| 19 | 124.4, C | 125.6, C | 125.0, C | |||
| 20 | 133.1, C | 7.25 (brs) | 117.6, CH | 7.47, dd (6.8, 2.2) | 118.5, CH | |
| 21 | 6.88, dt (7.3, 0.8) | 119.0, CH | 133.3, C | 7.12, m | 120.8, CH | |
| 22 | 7.03, dd (8.1, 7.3) | 121.0, CH | 6.94, dd (8.2, 1.7) | 121.4, CH | 7.12, m | 119.8, CH |
| 23 | 7.17, dd (8.1, 1.0) | 109.3, CH | 7.23, dd (8.1, 1.0) | 111.3, CH | 7.33, dd (6.8, 2.2) | 111.4, CH |
| 24 | 139.7, C | 138.2, C | 140.0, C | |||
| 25 | 1.34, s | 16.2, CH3 | 1.33, s | 16.2, CH3 | 1.12, s | 14.7, CH3 |
| 26 | 1.08, s | 19.8, CH3 | 1.05, s | 19.7, CH3 | 1.03, s | 15.9, CH3 |
| 27 | 81.9, C | 81.9, C | 72.4, C | |||
| 28 | 1.46, s | 22.8, CH3 | 1.46, s | 22.8, CH3 | 1.29, s | 24.0, CH3 |
| 29 | 1.68, s | 23.8, CH3 | 1.68, s | 23.8, CH3 | 1.35, s | 26.7, CH3 |
| 30 | 170.8, C | 170.8, C | 3.40, s | 49.1, CH3 | ||
| 31 | 2.06, s | 22.3, CH3 | 2.06, s | 22.3, CH3 | ||
| 32 | 3.64, d (7.3) | 32.0, CH2 | 3.43, d (7.3) | 34.5, CH2 | ||
| 33 | 5.43, m | 123.7, CH | 5.40, m | 124.6, CH | ||
| 34 | 131.8, C | 131.4, C | ||||
| 35 | 1.76, s | 25.8, CH3 | 1.76, s | 25.8, CH3 | ||
| 36 | 1.77, s | 17.8, CH3 | 1.77, s | 17.8, CH3 | ||
| HepG-2 | U2OS | MCF 7 | JeKo-1 | HL-60 | |
|---|---|---|---|---|---|
| Compound 1 | 55.1 | 56.1 | 56.4 | 71.2 | 65.8 |
| Compound 6 | 52.4 | 83.4 | 47.5 | 72.4 | 60.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, W.; Li, H.; Wu, Z.; Su, J.; Zhang, Z.; Yang, L.; Deng, X.; Xu, Q. Paspalines C–D and Paxillines B–D: New Indole Diterpenoids from Penicillium brefeldianum WZW-F-69. Mar. Drugs 2022, 20, 684. https://doi.org/10.3390/md20110684
Lin W, Li H, Wu Z, Su J, Zhang Z, Yang L, Deng X, Xu Q. Paspalines C–D and Paxillines B–D: New Indole Diterpenoids from Penicillium brefeldianum WZW-F-69. Marine Drugs. 2022; 20(11):684. https://doi.org/10.3390/md20110684
Chicago/Turabian StyleLin, Weiwen, Hanpeng Li, Zhiwen Wu, Jingyi Su, Zehong Zhang, Li Yang, Xianming Deng, and Qingyan Xu. 2022. "Paspalines C–D and Paxillines B–D: New Indole Diterpenoids from Penicillium brefeldianum WZW-F-69" Marine Drugs 20, no. 11: 684. https://doi.org/10.3390/md20110684
APA StyleLin, W., Li, H., Wu, Z., Su, J., Zhang, Z., Yang, L., Deng, X., & Xu, Q. (2022). Paspalines C–D and Paxillines B–D: New Indole Diterpenoids from Penicillium brefeldianum WZW-F-69. Marine Drugs, 20(11), 684. https://doi.org/10.3390/md20110684