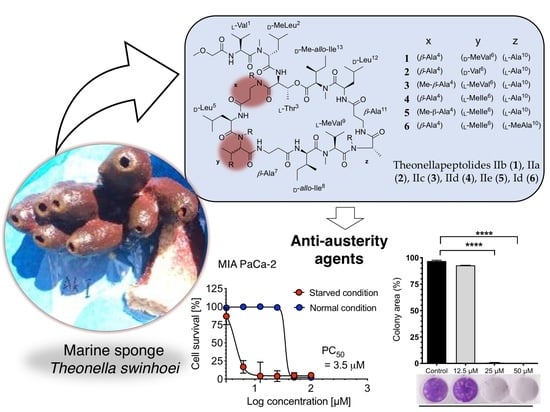
New Theonellapeptolides from Indonesian Marine Sponge Theonella swinhoei as Anti-Austerity Agents
Abstract
1. Introduction
2. Results and Discussion
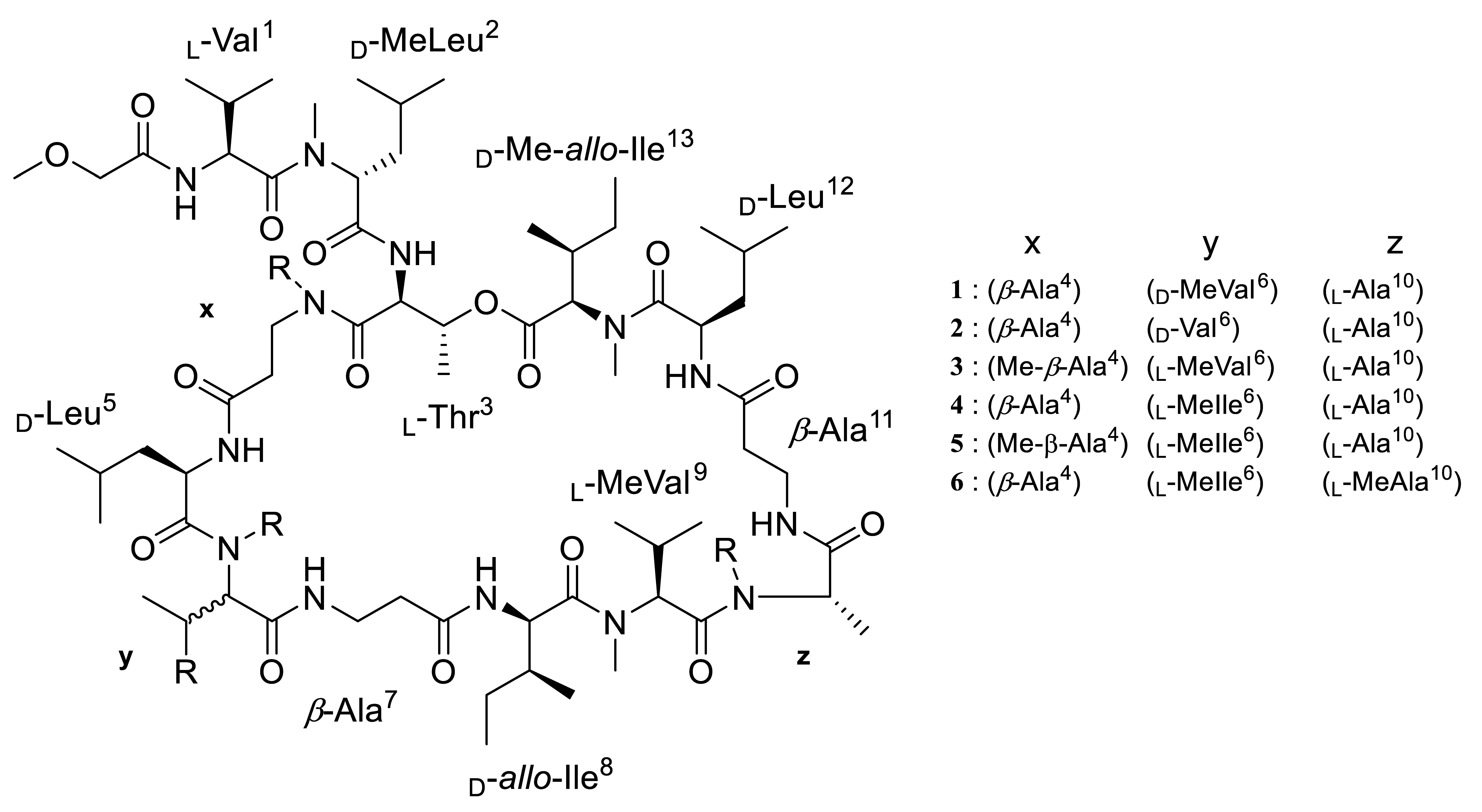
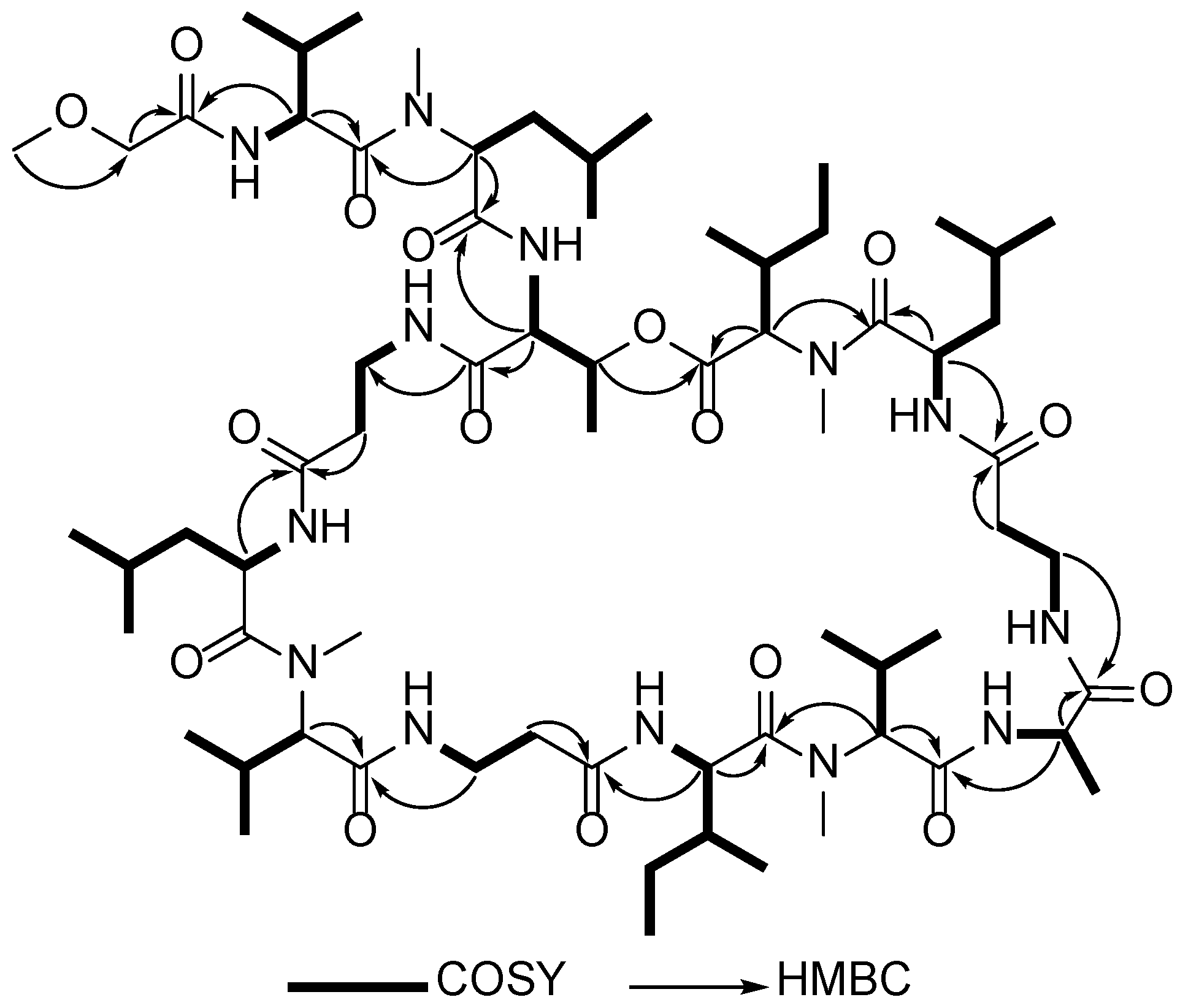
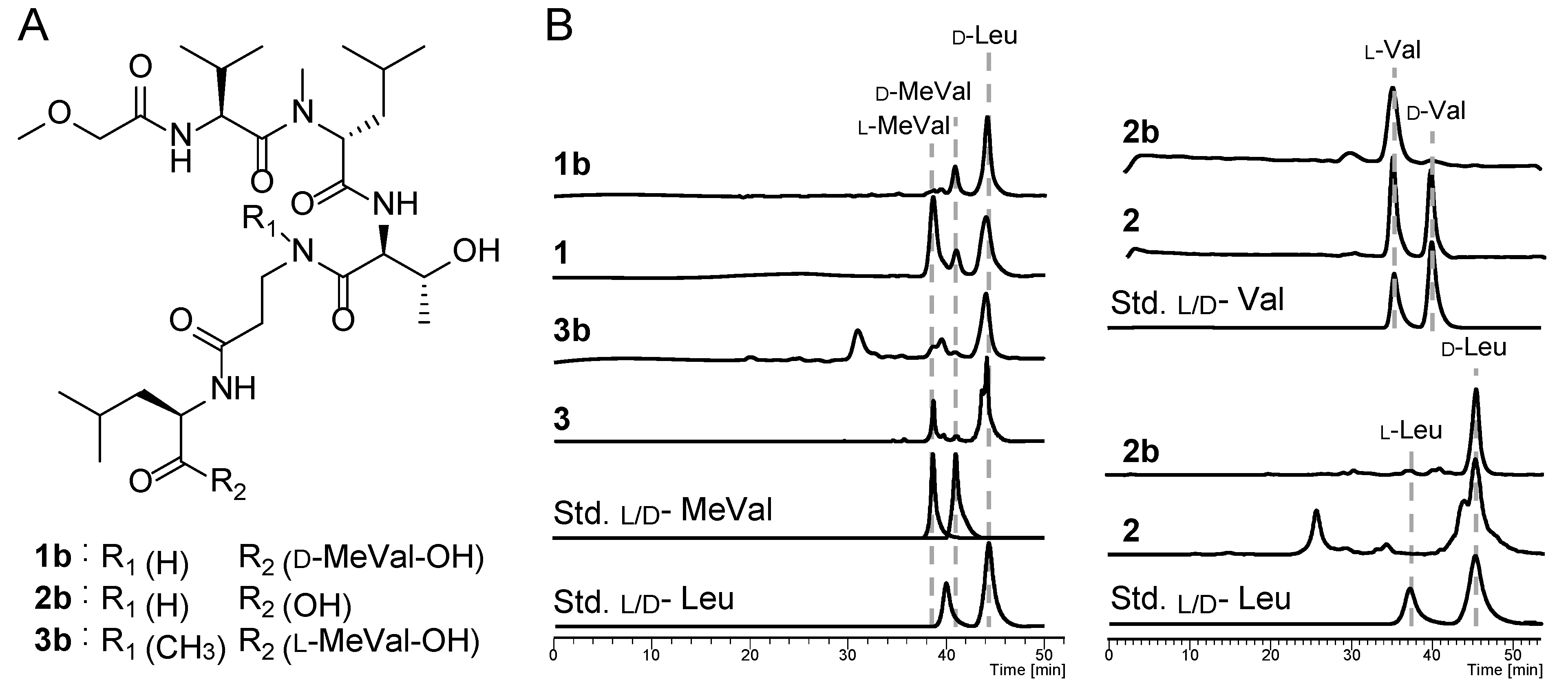
2.1. Structure Elucidation of Theonellapeptolides 1–3
2.2. Investigation of Biological Activities
2.2.1. Antibacterial Activity
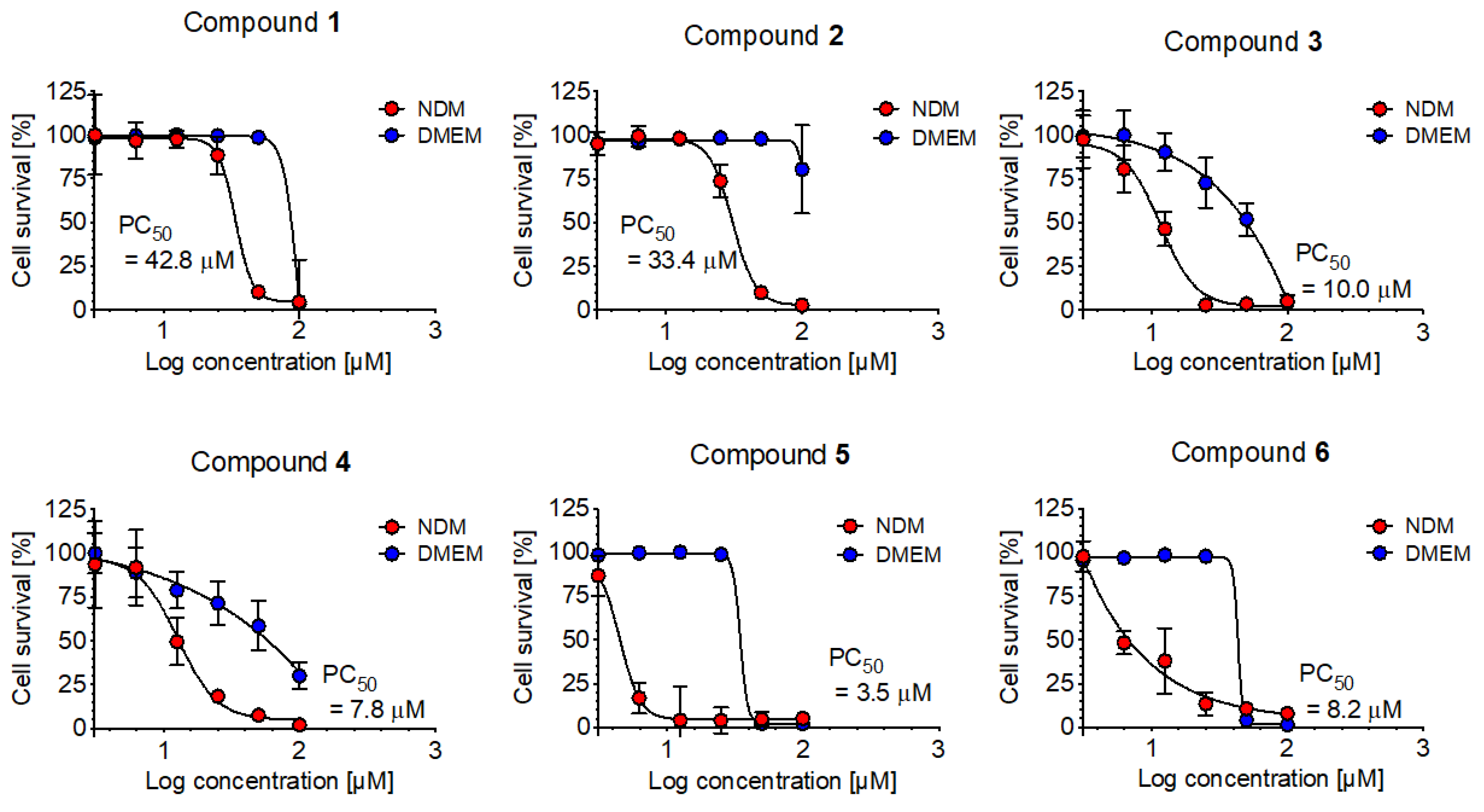
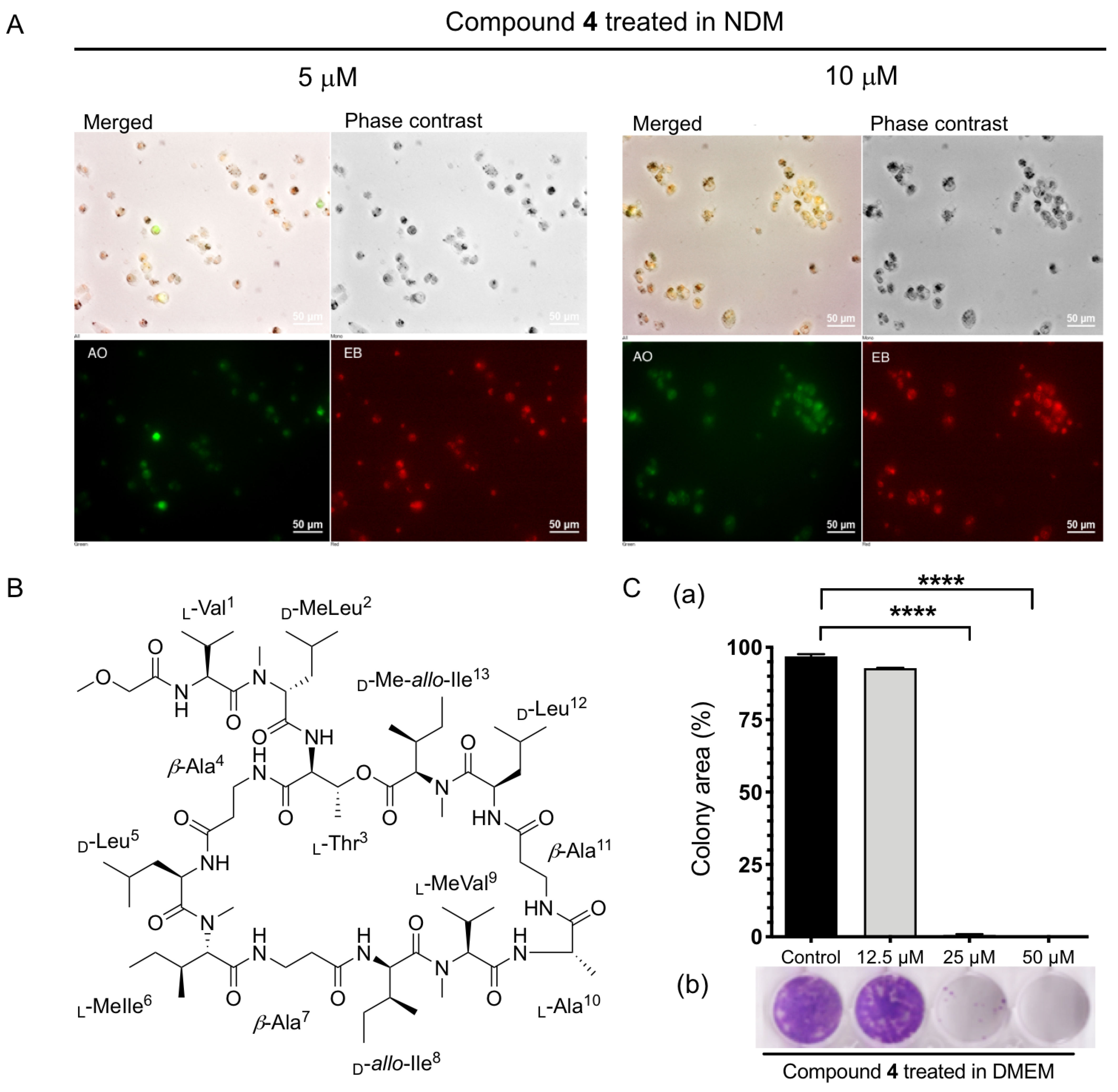
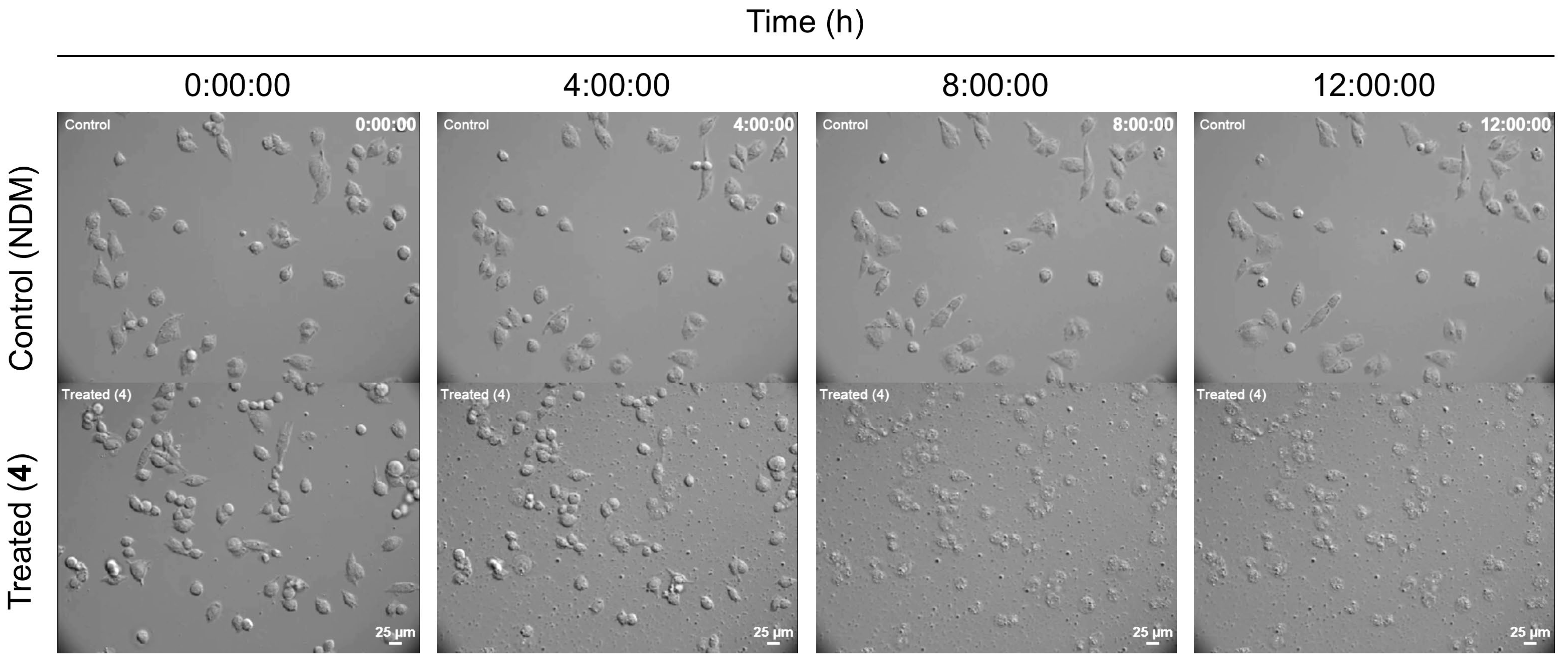
2.2.2. Anti-austerity Activity against the MIA PaCa-2 Cell Line
2.2.3. Anti-Austerity Activity against the HepG2 (Human Liver Cancer Cells) and MCF-7 (Human Breast Cancer Cells)
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Animal Material
3.3. Extraction and Isolation
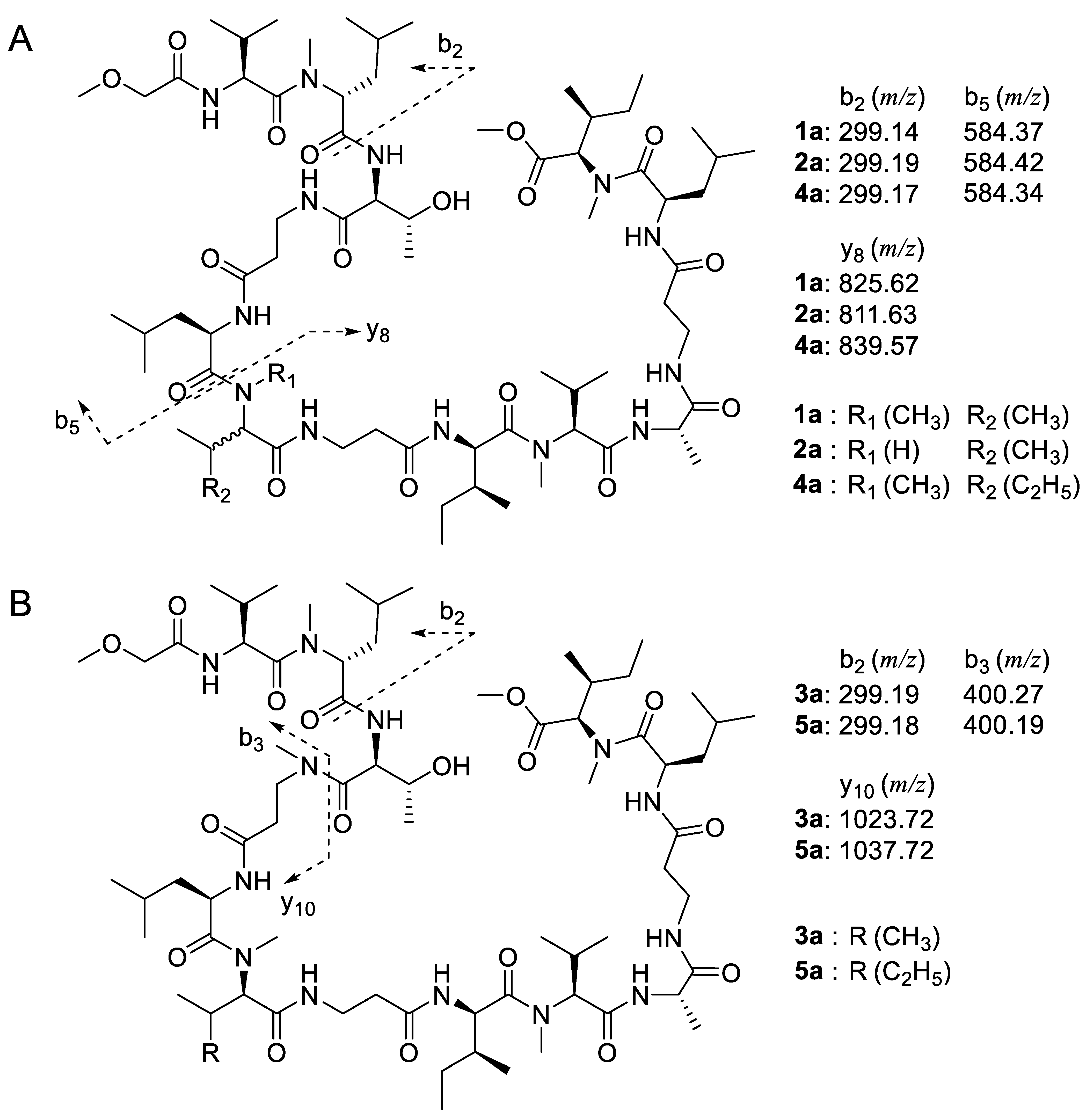
3.4. Determination of Amino Acid Sequence by Tandem MS Analysis
3.5. Total Hydrolysis
3.6. Partial Hydrolysis
3.7. Amino Acid Analysis by Marfey’s Method
3.8. Antibacterial Activity
3.9. Preferential Cytotoxic Activity Assay against MIA PaCa-2, HepG2, and MCF-7 Cell Lines
3.10. Morphological Analysis
3.11. Colony Formation Assay
3.12. Live Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef]
- Morrow, C.; Cárdenas, P. Proposal for a revised classification of the Demospongiae (Porifera). Front Zool. 2015, 12, 7. [Google Scholar] [CrossRef]
- Fusetani, N.; Matsunaga, S. Bioactive sponge peptides. Chem. Rev. 1993, 93, 1793–1806. [Google Scholar] [CrossRef]
- Winder, P.L.; Pomponi, S.A.; Wright, A.E. Natural products from the Lithistida: A review of the literature since 2000. Mar. Drugs 2011, 9, 2643–2682. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, A.S.; Bugni, T.S.; Feng, X.; Harper, M.K.; Skalicky, J.J.; Mohammed, K.A.; Andjelic, C.D.; Barrows, L.R.; Ireland, C.M. Theopapuamide, a cyclic depsipeptide from a Papua New Guinea Lithistid sponge Theonella swinhoei. J. Nat. Prod. 2006, 69, 1582–1586. [Google Scholar] [CrossRef]
- Araki, T.; Matsunaga, S.; Nakao, Y.; Furihata, K.; West, L.; Faulkner, D.J.; Fusetani, N. Koshikamide B, a cytotoxic peptide lactone from a marine sponge Theonella sp. J. Org. Chem. 2008, 73, 7889–7894. [Google Scholar] [CrossRef]
- Kim, C.-K.; Wang, D.; Bokesch, H.R.; Fuller, R.W.; Smith, E.; Henrich, C.J.; Durrant, D.E.; Morrison, D.K.; Bewley, C.A.; Gustafson, K.R. Swinhopeptolides A and B: Cyclic depsipeptides from the sponge Theonella swinhoei that inhibit Ras/Raf interaction. J. Nat. Prod. 2020, 83, 1288–1294. [Google Scholar] [CrossRef]
- Kobayashi, M.; Lee, N.K.; Shibuya, H.; Momose, T.; Kitagawa, I. Marine natural products. XXVI. Biologically active tridecapeptide lactones from the Okinawan marine sponge Theonella swinhoei (Theonellidae). (2). Structures of theonellapeptolides Ia, Ib, Ic, and Ie. Chem. Pharm. Bull. 1991, 39, 1177–1184. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kanzaki, K.; Katayama, S.; Ohashi, K.; Okada, H.; Ikegami, S.; Kitagawa, I. Marine natural products. XXXIII. Theonellapeptolide IId, a new tridecapeptide lactone from the Okinawan marine sponge Theonella swinhoei. Chem. Pharm. Bull. 1994, 42, 1410–1415. [Google Scholar] [CrossRef]
- Sinisi, A.; Calcinai, B.; Cerrano, C.; Dien, H.A.; Zampella, A.; D’Amore, C.; Renga, B.; Fiorucci, S.; Taglialatela-Scafati, O. New tridecapeptides of the theonellapeptolide family from the Indonesian sponge Theonella swinhoei. Beilstein J. Org. Chem. 2013, 9, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, I.; Lee, N.K.; Kobayashi, M.; Shibuya, H. Marine natural products. XXV. Biologically active tridecapeptide lactones from the Okinawan marine sponge Theonella swinhoei (Theonellidae). Structure of theonellapeptolide Id. Tetrahedron 1991, 47, 2169–2180. [Google Scholar] [CrossRef]
- Roy, M.C.; Ohtani, I.; Ichiba, T.; Tanaka, J.; Satari, R.; Higa, T. New cyclic peptides from the Indonesian sponge Theonella swinhoei. Tetrahedron 2000, 56, 9079–9092. [Google Scholar] [CrossRef]
- Tsuda, M.; Shimbo, K.; Kubota, T.; Mikami, Y.; Kobayashi, J. Two theonellapeptolide congeners from marine sponge Theonella sp. Tetrahedron 1999, 55, 10305–10314. [Google Scholar] [CrossRef]
- Doi, M.; Ishida, T.; Kobayashi, M.; Deschamps, J.R.; Flippen-Anderson, J.L. The highly solvated structure of theonellapeptolide Id, a tridecapeptide lactone from the Okinawa marine sponge Theonella swinhoei. Acta Cryst. 1999, 55, 796–798. [Google Scholar] [CrossRef]
- Kitagawa, I.; Ohashi, K.; Kawanishi, H.; Shibuya, H.; Shinkai, K.; Akedo, H. Ionophoretic activities of oligopeptide lactones and resin-glycosides in human erythrocytes. Chem. Pharm. Bull. 1989, 37, 1679–1681. [Google Scholar] [CrossRef]
- Farley, C.M.; Dibwe, D.F.; Ueda, J.Y.; Hall, E.A.; Awale, S.; Magolan, J. Evaluation of synthetic coumarins for antiausterity cytotoxicity against pancreatic cancers. Bioorg. Med. Chem. Lett. 2016, 26, 1471–1474. [Google Scholar] [CrossRef]
- Dibwe, D.F.; Sun, S.; Ueda, J.Y.; Balachandran, C.; Matsumoto, K.; Awale, S. Discovery of potential antiausterity agents from the Japanese cypress Chamaecyparis obtusa. Bioorg. Med. Chem. Lett. 2017, 27, 4898–4903. [Google Scholar] [CrossRef]
- Sun, S.; Phrutivorapongkul, A.; Dibwe, D.F.; Balachandran, C.; Awale, S. Chemical constituents of Thai Citrus hystrix and their antiausterity activity against the PANC-1 human pancreatic cancer cell line. J. Nat. Prod. 2018, 81, 1877–1883. [Google Scholar] [CrossRef]
- Awale, S.; Dibwe, D.F.; Balachandran, C.; Fayez, S.; Feineis, D.; Lombe, B.K.; Bringmann, G. Ancistrolikokine E3, a 5,8′-coupled naphthylisoquinoline alkaloid, eliminates the tolerance of cancer cells to nutrition starvation by inhibition of the Akt/mTOR/Autophagy signaling pathway. J. Nat. Prod. 2018, 81, 2282–2291. [Google Scholar] [CrossRef]
- Alexander, B.E.; Sun, S.; Palframan, M.J.; Kociok-Köhn, G.; Dibwe, D.F.; Watanabe, S.; Caggiano, L.; Awale, S.; Lewis, S. Sidechain diversification of grandifloracin allows identification of analogues with enhanced anti-austerity activity against human PANC-1 pancreatic cancer cells. Chem. Med. Chem. 2020, 15, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Alilou, M.; Dibwe, D.F.; Schwaiger, S.; Khodami, M.; Troppmair, J.; Awale, S.; Stuppner, H. Antiausterity activity of secondary metabolites from the roots of Ferula hezarlalehzarica against the PANC-1 human pancreatic cancer cell line. J. Nat. Prod. 2020, 83, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Takagi, A.; Usuguchi, K.; Takashima, I.; Okuda, K. Total synthesis of antiausterity agent (±)-uvaridacol l by regioselective axial diacylation of a myo-inositol orthoester. Org. Lett. 2021, 23, 4083–4087. [Google Scholar] [CrossRef] [PubMed]
- Kotoku, N.; Ishida, R.; Matsumoto, H.; Arai, M.; Toda, K.; Setiawan, A.; Muraoka, O.; Kobayashi, M. Biakamides A–D, unique polyketides from a marine sponge, act as selective growth inhibitors of tumor cells adapted to nutrient starvation. J. Org. Chem. 2017, 82, 1705–1718. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Naime, W.A.; Kimishima, A.; Setiawan, A.; Fahim, J.R.; Fouad, M.A.; Kamel, M.S.; Arai, M. Mitochondrial targeting in an anti-austerity approach involving bioactive metabolites isolated from the marine-derived fungus Aspergillus sp. Mar. Drugs. 2020, 18, 555. [Google Scholar] [CrossRef]
- Kuranaga, T.; Enomoto, A.; Tan, H.; Fujita, K.; Wakimoto, T. Total synthesis of theonellapeptolide Id. Org. Lett. 2017, 19, 1366–1369. [Google Scholar] [CrossRef]
- Roepstorff, P.; Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984, 11, 601. [Google Scholar] [CrossRef]
- Harrison, A.G.; Young, A.B. Fragmentation of protonated oligoalanines: Amide bond cleavage and beyond. J. Am. Soc. Mass. Spectrom. 2004, 15, 1810–1819. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D -amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Izuishi, K.; Kato, K.; Ogura, T.; Kinoshita, T.; Esumi, H. Remarkable tolerance of tumor cells to nutrient deprivation: Possible new biochemical target for cancer therapy. Cancer Res. 2000, 60, 6201–6207. Available online: https://aacrjournals.org/cancerres/article/60/21/6201/506825/Remarkable-Tolerance-of-Tumor-Cells-to-Nutrient (accessed on 5 November 2021).
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
| Pos. | δC, Type | δH, (Mult) | Pos. | δC, Type | δH, (Mult) | Pos. | δC, Type | δH, (Mult) |
|---|---|---|---|---|---|---|---|---|
| Methoxy acetyl | D-Leucine5 | C2 | 62.3, CH | 4.65, (m) | ||||
| C1 | 170.1, C | C1 | n.r., C | C3 | 26.7, CH | 2.22, (m) | ||
| C2 | 71.5, CH2 | 3.89, (m) | C2 | 47.7, CH | 4.89, (m) | C4 | 18.6, CH3 | 0.75, (m) |
| C3 | 70.2, CH3 | 3.59, (m) | C3 | 41.0, CH2 | a: 1.32, (m) | C5 | 19.6, CH3 | 0.90, (m) |
| L-Valine1 | b: 1.44, (m) | NMe | 31.0, CH3 | 2.99, (m) | ||||
| C1 | 173.1, C | C4 | 24.6, CH | 1.49, (m) | L-Alanine10 | |||
| C2 | 53.9, CH | 4.72, (m) | C5 | 23.0, CH3 | 0.85, (m) | C1 | 172.7, C | |
| C3 | 30.9, CH | 1.99, (m) | C6 | 23.1, CH3 | 0.88, (m) | C2 | 48.7, CH | 4.22, (m) |
| C4 | 19.3, CH3 | 0.90, (m) | NH | 7.57, (m) | C3 | 17.2, CH3 | 1.15, (m) | |
| C5 | 17.3, CH3 | 0.85, (m) | D-Methyl Valine6 | NH | 7.45, (m) | |||
| NH | 7.20, (m) | C1 | 170.4, C | β-Alanine11 | ||||
| D-Methyl Leucine2 | C2 | 62.1, CH | 4.72, (m) | C1 | 171.4, C | |||
| C1 | 171.2, C | C3 | 25.7, CH | 2.15, (m) | C2 | 34.9, CH2 | a: 2.38, (m) | |
| C2 | 54.9, CH | 5.08, (m) | C4 | 19.8, CH3 | 0.91, (m) | b: 2.29, (m) | ||
| C3 | 37.1, CH2 | a: 1.58, (m) | C5 | 18.8, CH3 | 0.75, (m) | C3 | 36.2, CH2 | a: 3.43, (m) |
| b: 1.72, (m) | NMe | 31.3, CH3 | 3.01, (m) | b: 3.35, (m) | ||||
| C4 | 24.8, CH | 1.35, (m) | β-Alanine7 | NH | 7.52, (m) | |||
| C5 | 21.1, CH3 | 0.82, (m) | C1 | 172.5, C | D-Leucine12 | |||
| C6 | 21.5, CH3 | 0.87, (m) | C2 | 35.8, CH2 | a: 2.36, (m) | C1 | 173.1, C | |
| NMe | 30.8, CH3 | 3.05, (m) | b: n.r. | C2 | 47.8, CH | 4.92, (m) | ||
| L-Threonine3 | C3 | 36.0, CH2 | a: 3.60, (m) | C3 | 41.6, CH2 | a: 1.28, (m) | ||
| C1 | 168.2, C | b: 3.19, (m) | b: 1.43, (m) | |||||
| C2 | 55.7, CH | 4.39, (m) | NH | 7.52, (m) | C4 | 24.6, CH | 1.50, (m) | |
| C3 | 70.2, CH | 5.08, (m) | D-allo Isoleucine8 | C5 | 23.1, CH3 | 0.83, (m) | ||
| C4 | 15.9, CH3 | 1.05, (m) | C1 | 173.7, C | C6 | 23.2, CH3 | 0.89, (m) | |
| NH | 7.59, (m) | C2 | 60.2, CH | 4.80, (m) | NH | 7.78, (m) | ||
| β-Alanine4 | C3 | 24.8, CH | 2.05, (m) | D-Methyl allo Isoleucine13 | ||||
| C1 | 171.3, C | C4 | 24.6, CH | a: 1.24, (m) | C1 | 173.09, C | ||
| C2 | 35.8, CH2 | a: 2.46, (m) | b: 0.96, (m) | C2 | 54.7, CH | 5.15, (m) | ||
| b: 2.19, (m) | C5 | 15.2, CH3 | 0.91, (m) | C3 | 36.6, CH | 1.71, (m) | ||
| C3 | 36.0, CH2 | a: 3.47, (m) | C6 | 15.1, CH3 | 0.83, (m) | C4 | 26.2, CH | a: 1.38, (m) |
| b: 3.26, (m) | NH | 7.83, (m) | b: 1.16, (m) | |||||
| NH | 7.25, (m) | L-Methyl Valine9 | C5 | 17.9, CH3 | 0.88, (m) | |||
| C1 | 169.5, C | C6 | 17.2, CH3 | 0.85, (m) | ||||
| NMe | 30.6, CH3 | 2.98, (m) | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haedar, J.R.; Uria, A.R.; Lallo, S.; Dibwe, D.F.; Wakimoto, T. New Theonellapeptolides from Indonesian Marine Sponge Theonella swinhoei as Anti-Austerity Agents. Mar. Drugs 2022, 20, 661. https://doi.org/10.3390/md20110661
Haedar JR, Uria AR, Lallo S, Dibwe DF, Wakimoto T. New Theonellapeptolides from Indonesian Marine Sponge Theonella swinhoei as Anti-Austerity Agents. Marine Drugs. 2022; 20(11):661. https://doi.org/10.3390/md20110661
Chicago/Turabian StyleHaedar, Jabal Rahmat, Agustinus Robert Uria, Subehan Lallo, Dya Fita Dibwe, and Toshiyuki Wakimoto. 2022. "New Theonellapeptolides from Indonesian Marine Sponge Theonella swinhoei as Anti-Austerity Agents" Marine Drugs 20, no. 11: 661. https://doi.org/10.3390/md20110661
APA StyleHaedar, J. R., Uria, A. R., Lallo, S., Dibwe, D. F., & Wakimoto, T. (2022). New Theonellapeptolides from Indonesian Marine Sponge Theonella swinhoei as Anti-Austerity Agents. Marine Drugs, 20(11), 661. https://doi.org/10.3390/md20110661