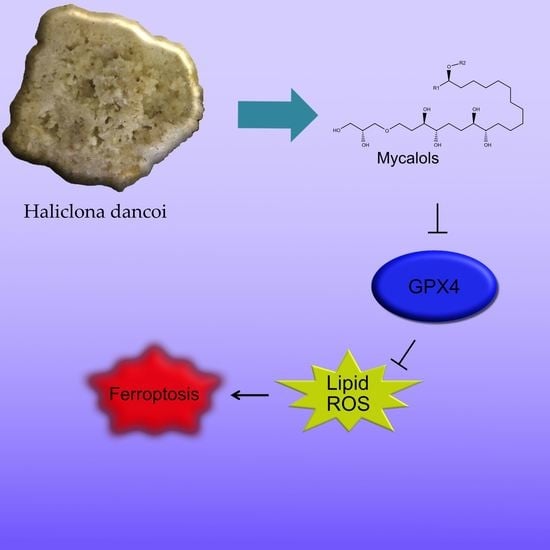
Bioactivity Screening of Antarctic Sponges Reveals Anticancer Activity and Potential Cell Death via Ferroptosis by Mycalols
Abstract
:1. Introduction
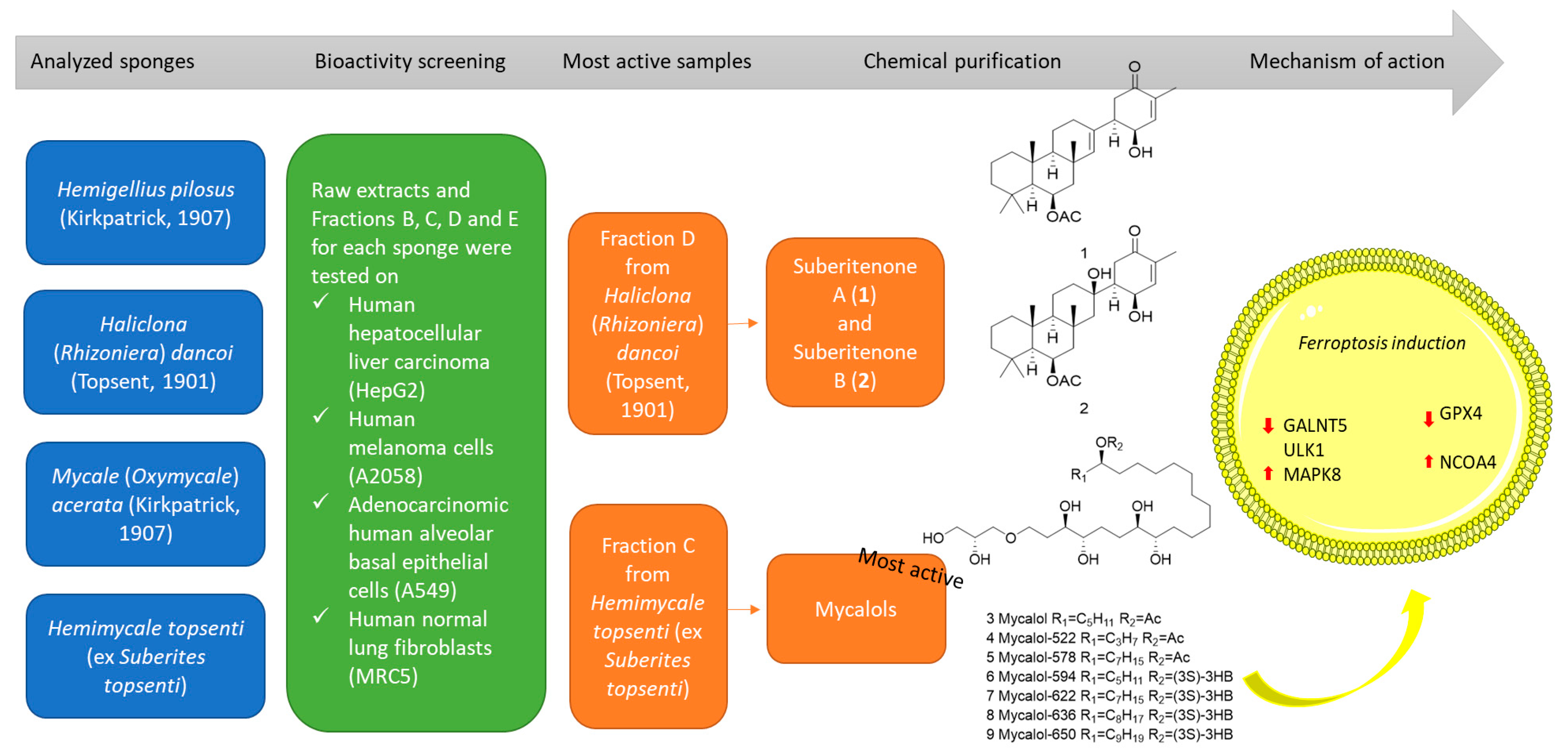
2. Results
2.1. Species Identification of Specimen C7
2.2. Chemical Fractionation and Cell Viability on Cancer Cell Lines
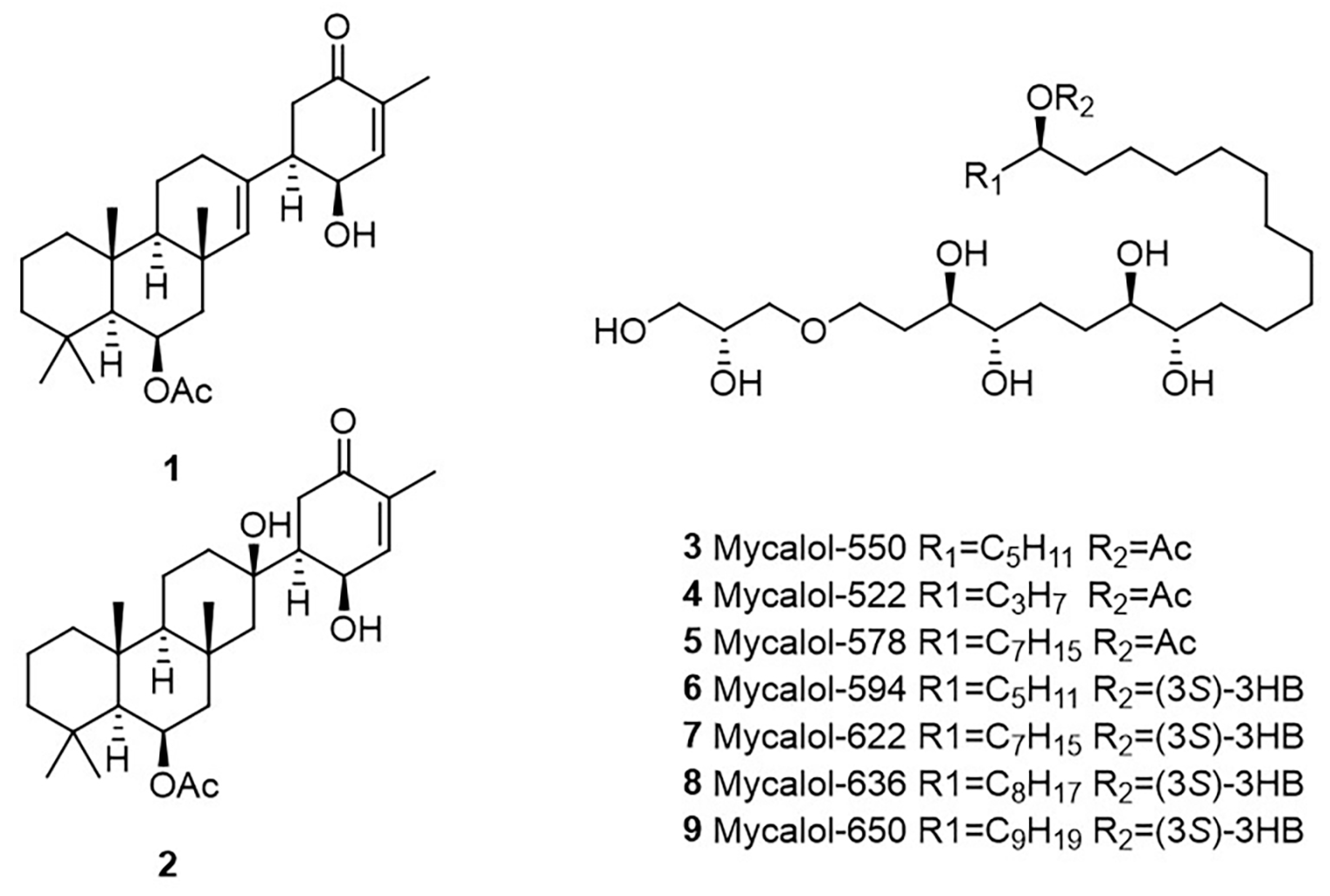
2.3. Identification of Compounds in Most Active Fractions
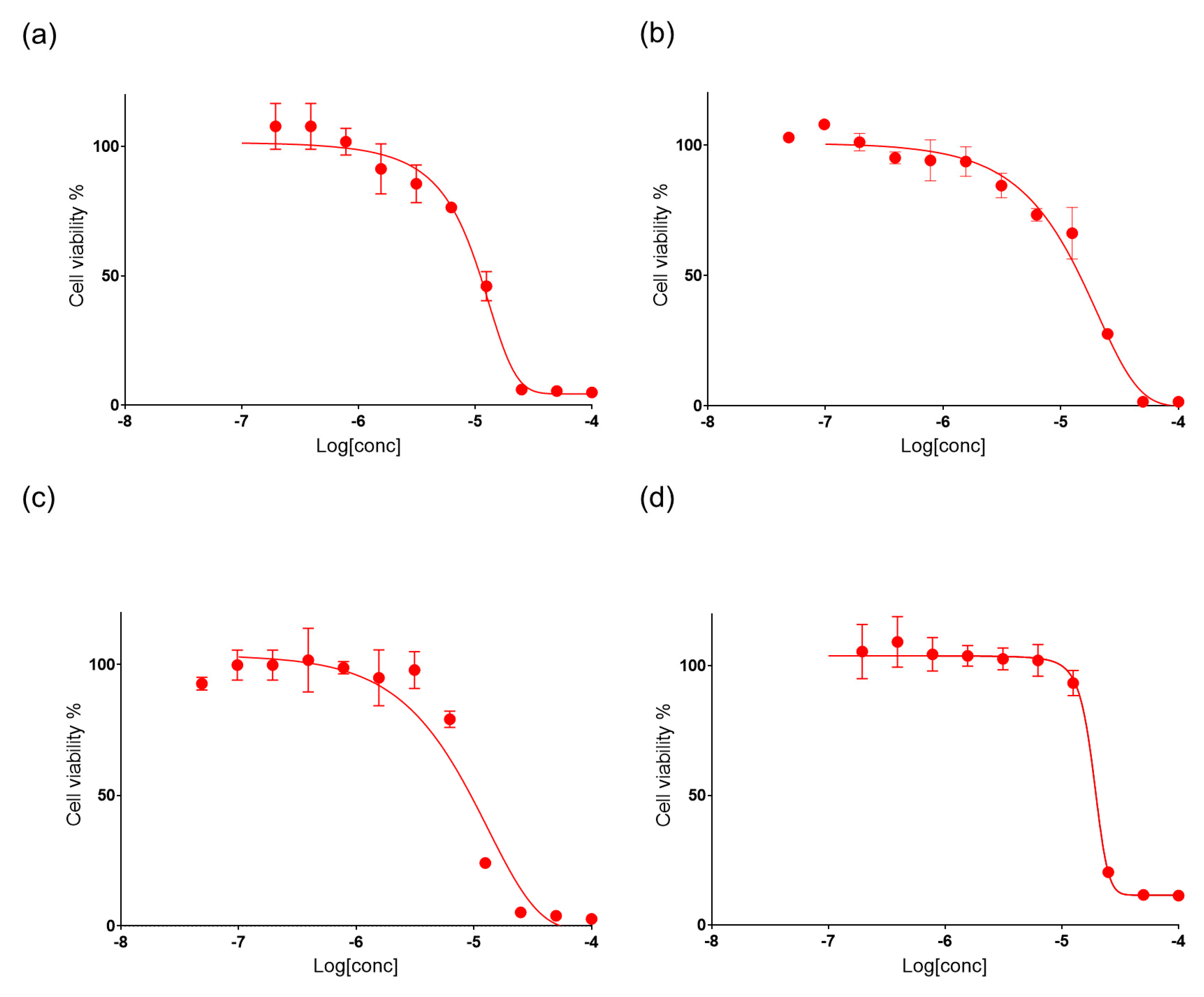
2.4. Bioactivity of Suberitenones A and B from Hemimycale topsenti and Mycalols from Haliclona (Rhizoniera) dancoi on Cancer Cell Lines
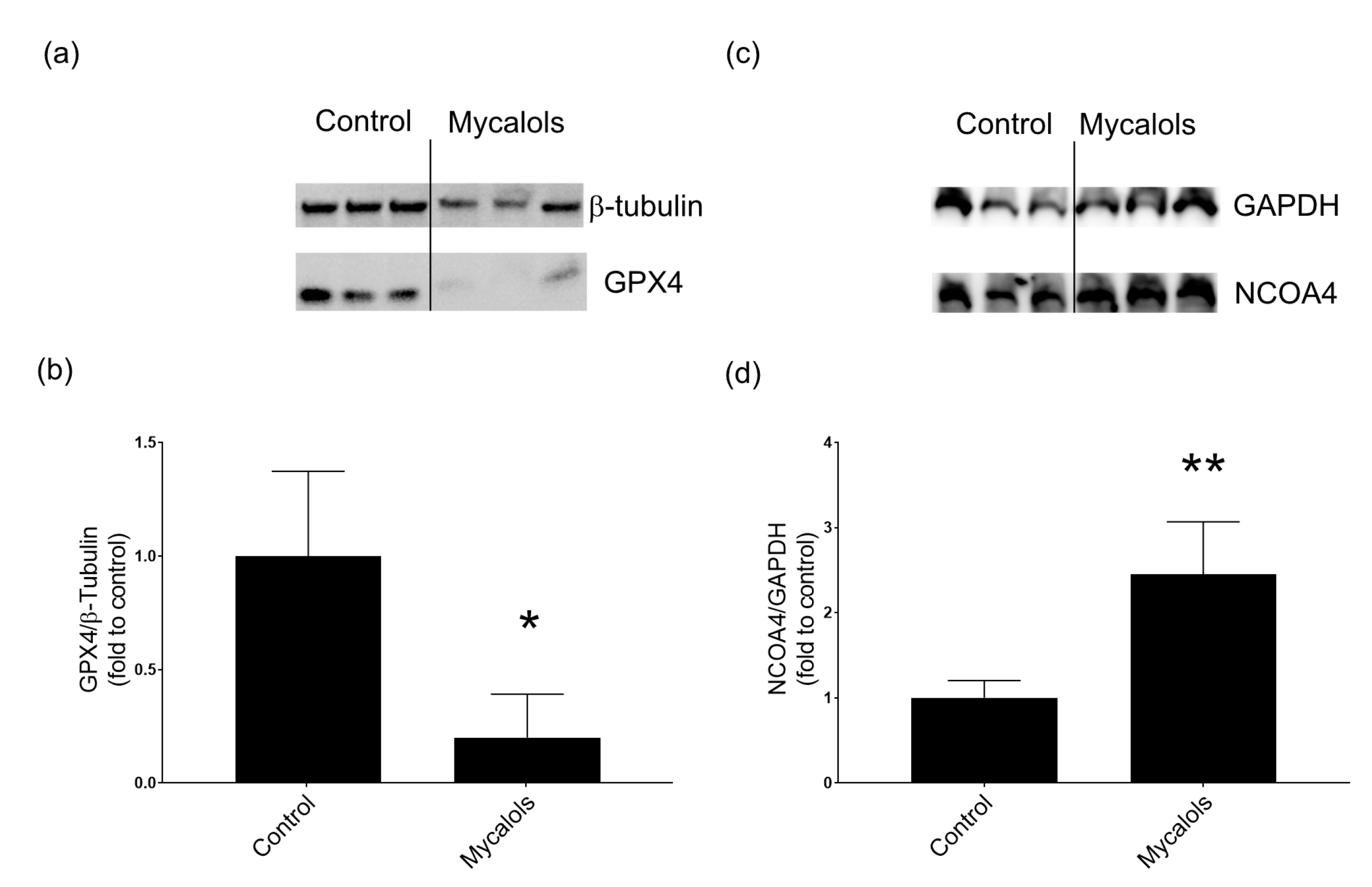
2.4.1. Mechanism of Action
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sampling
5.2. Morphological Analysis and Polymerase Chain Reaction (PCR) of 18S/28S rRNA and CO1 Markers
- for 18S and 28S, a denaturation step at 95 °C for 2 min, [35 cycles denaturation step at 95 °C for 1 min, annealing step at 52 °C (18S1/18S2; [48]), 55 °C (18S-AF/18S-BR, NL4F/NL4R; [49,50]), 57 °C (C2/D2; [51]), for 1 min and 72 °C of primer extension for 2 min], a final extension step at 72 °C for 10 min;
5.3. Chemical Extraction and SPE Fractionation
5.4. Purification and Characterization of Active Compounds
5.4.1. Haliclona (Rhizoniera) dancoi (H. d.)
5.4.2. Hemimycale topsenti (H. t.)
5.5. Cell Lines
5.6. Antibody
5.7. In Vitro Cell Viability Studies
5.8. RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
5.9. Protein Extraction and Western Blotting Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thakur, N.L.; Thakur, A.N.; Müller, W.E.G. Article RP 18 Marine natural products in drug discovery. NISCAIR Online Period. Repos. 2005, 4, 471–477. [Google Scholar]
- Berne, S.; Kalauz, M.; Lapat, M.; Savin, L.; Janussen, D.; Kersken, D.; Ambrožič Avguštin, J.; Zemljič Jokhadar, Š.; Jaklič, D.; Gunde-Cimerman, N.; et al. Screening of the Antarctic marine sponges (Porifera) as a source of bioactive compounds. Polar Biol. 2016, 39, 947–959. [Google Scholar] [CrossRef]
- Giordano, D.; Costantini, M.; Coppola, D.; Lauritano, C.; Núñez Pons, L.; Ruocco, N.; di Prisco, G.; Ianora, A.; Verde, C. Biotechnological Applications of Bioactive Peptides From Marine Sources. In Advances in Microbial Physiology; Academic Press: London, UK, 2018; Volume 73, pp. 171–220. ISBN 9780128151907. [Google Scholar]
- Romano, G.; Ianora, A.; Costantini, M.; Sansone, C.; Lauritano, C.; Ruocco, N.; Ianora, A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar. Environ. Res. 2016, 128, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef] [Green Version]
- Jaspars, M.; De Pascale, D.; Andersen, J.H.; Reyes, F.; Crawford, A.D.; Ianora, A. The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. UK 2016, 96, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Ruocco, N.; Esposito, R.; Bertolino, M.; Zazo, G.; Sonnessa, M.; Andreani, F.; Coppola, D.; Giordano, D.; Nuzzo, G.; Lauritano, C.; et al. A metataxonomic approach reveals diversified bacterial communities in antarctic sponges. Mar. Drugs 2021, 19, 173. [Google Scholar] [CrossRef]
- Lauritano, C.; Rizzo, C.; Giudice, A.L.; Saggiomo, M. Physiological and molecular responses to main environmental stressors of microalgae and bacteria in polar marine environments. Microorganisms 2020, 8, 1957. [Google Scholar] [CrossRef] [PubMed]
- Peck, L.S. Antarctic marine biodiversity: Adaptations, Environments and Responses to Change. An Annu. Rev. 2018, 56, 105–236. [Google Scholar]
- McClintock, J.B.; Amsler, C.D.; Baker, B.J.; Van Soest, R.W.M. Ecology of antarctic marine sponges: An overview. Integr. Comp. Biol. 2005, 45, 359–368. [Google Scholar] [CrossRef]
- de Broyer, C.; Koubbi, P.; Griffiths, H.; Raymond, B.; d’Acoz, C.d.; van de Putte, A.; Danis, B.; David, B.; Grant, S.; Gutt, J.; et al. Biogeographic Atlas of the Southern Ocean; XII; Scientific Committee on Antarctic Research: Cambridge, UK, 2014; ISBN 978-0-948277-28-3. [Google Scholar]
- Papaleo, M.C.; Fondi, M.; Maida, I.; Perrin, E.; Lo Giudice, A.; Michaud, L.; Mangano, S.; Bartolucci, G.; Romoli, R.; Fani, R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012, 30, 272–293. [Google Scholar] [CrossRef]
- Von Salm, J.L.; Witowski, C.G.; Fleeman, R.M.; McClintock, J.B.; Amsler, C.D.; Shaw, L.N.; Baker, B.J. Darwinolide, a New Diterpene Scaffold That Inhibits Methicillin-Resistant Staphylococcus aureus Biofilm from the Antarctic Sponge Dendrilla membranosa. Org. Lett. 2016, 18, 2596–2599. [Google Scholar] [CrossRef]
- Di, X.; Rouger, C.; Hardardottir, I.; Freysdottir, J.; Molinski, T.F.; Tasdemir, D.; Omarsdottir, S. 6-Bromoindole derivatives from the icelandic marine sponge Geodia barretti: Isolation and anti-inflammatory activity. Mar. Drugs 2018, 16, 437. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Peifer, C.; Janussen, D.; Tasdemir, D. New discorhabdin alkaloids from the antarctic deep-sea sponge Latrunculia biformis. Mar. Drugs 2019, 17, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilling, A.J.; Witowski, C.G.; Maschek, J.A.; Azhari, A.; Vesely, B.A.; Kyle, D.E.; Amsler, C.D.; McClintock, J.B.; Baker, B.J. Spongian Diterpenoids Derived from the Antarctic Sponge Dendrilla antarctica Are Potent Inhibitors of the Leishmania Parasite. J. Nat. Prod. 2020, 83, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Pons, L.; Shilling, A.; Verde, C.; Baker, B.J.; Giordano, D. Marine terpenoids from polar latitudes and their potential applications in biotechnology. Mar. Drugs 2020, 18, 401. [Google Scholar] [CrossRef] [PubMed]
- Giordano, D. Bioactive Molecules from Extreme Environments. Mar. Drugs 2020, 18, 640. [Google Scholar] [CrossRef]
- Ciaglia, E.; Malfitano, A.M.; Laezza, C.; Fontana, A.; Nuzzo, G.; Cutignano, A.; Abate, M.; Pelin, M.; Sosa, S.; Bifulco, M.; et al. Immuno-Modulatory and Anti-Inflammatory Effects of Dihydrogracilin A, a Terpene Derived from the Marine Sponge Dendrilla membranosa. Int. J. Mol. Sci. 2017, 18, 1643. [Google Scholar] [CrossRef] [Green Version]
- Cutignano, A.; Nuzzo, G.; D’Angelo, D.; Borbone, E.; Fusco, A.; Fontana, A. Mycalol: A natural lipid with promising cytotoxic properties against human anaplastic thyroid carcinoma cells. Angew. Chemie Int. Ed. 2013, 52, 9256–9260. [Google Scholar] [CrossRef]
- Cutignano, A.; Seetharamsingh, B.; D’Angelo, D.; Nuzzo, G.; Khairnar, P.V.; Fusco, A.; Reddy, D.S.; Fontana, A. Identification and Synthesis of Mycalol Analogues with Improved Potency against Anaplastic Thyroid Carcinoma Cell Lines. J. Nat. Prod. 2017, 80, 1125–1133. [Google Scholar] [CrossRef]
- Cutignano, A.; Nuzzo, G.; Ianora, A.; Luongo, E.; Romano, G.; Gallo, C.; Sansone, C.; Aprea, S.; Mancini, F.; D’Oro, U.; et al. Development and application of a novel SPE-method for bioassay-guided fractionation of marine extracts. Mar. Drugs 2015, 13, 5736–5749. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Seo, Y.; Rho, J.R.; Baek, E.; Kwon, B.M.; Jeong, T.S.; Bok, S.H. Suberitenones a and B: Sesterterpenoids of an Unprecedented Skeletal Class from the Antarctic Sponge Suberites sp. J. Org. Chem. 1995, 60, 7582–7588. [Google Scholar] [CrossRef]
- Seetharamsingh, B.; Rajamohanan, P.R.; Reddy, D.S. Total synthesis and structural revision of mycalol, an anticancer natural product from the marine source. Org. Lett. 2015, 17, 1652–1655. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.G.; Hurley, J.H. Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 2016, 39, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Rajak, S.; Iannucci, L.F.; Zhou, J.; Anjum, B.; George, N.; Singh, B.K.; Ghosh, S.; Yen, P.M.; Sinha, R.A. Loss of ULK1 Attenuates Cholesterogenic Gene Expression in Mammalian Hepatic Cells. Front. Cell Dev. Biol. 2020, 8, 523550. [Google Scholar] [CrossRef]
- Sinha, R.A.; Singh, B.K.; Zhou, J.; Xie, S.; Farah, B.L.; Lesmana, R.; Ohba, K.; Tripathi, M.; Ghosh, S.; Hollenberg, A.N.; et al. Loss of ULK1 increases RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells. Autophagy 2017, 13, 169–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Chen, P.; Zhai, B.; Zhang, M.; Xiang, Y.; Fang, J.; Xu, S.; Gao, Y.; Chen, X.; Sui, X.; et al. The emerging role of ferroptosis in inflammation. Biomed. Pharmacother. 2020, 127, 110108. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.l.; Huang, Z.j.; Lin, Z.t.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.f.; Zou, T.; Tuo, Q.Z.; Xu, S.; Li, H.; Belaidi, A.A.; Lei, P. Ferroptosis: Mechanisms and links with diseases. Signal Transduct. Target. Ther. 2021, 6, 49. [Google Scholar] [CrossRef]
- Detarya, M.; Sawanyawisuth, K.; Aphivatanasiri, C.; Chuangchaiya, S.; Saranaruk, P.; Sukprasert, L.; Silsirivanit, A.; Araki, N.; Wongkham, S.; Wongkham, C. The O-GalNAcylating enzyme GALNT5 mediates carcinogenesis and progression of cholangiocarcinoma via activation of AKT/ERK signaling. Glycobiology 2020, 30, 312–324. [Google Scholar] [CrossRef]
- Jin, M.; Shi, C.; Li, T.; Wu, Y.; Hu, C.; Huang, G. Solasonine promotes ferroptosis of hepatoma carcinoma cells via glutathione peroxidase 4-induced destruction of the glutathione redox system. Biomed. Pharmacother. 2020, 129, 110282. [Google Scholar] [CrossRef]
- Wu, C.; Zhao, W.; Yu, J.; Li, S.; Lin, L.; Chen, X. Induction of ferroptosis and mitochondrial dysfunction by oxidative stress in PC12 cells. Sci. Rep. 2018, 8, 574. [Google Scholar] [CrossRef] [Green Version]
- Del Rey, M.Q.; Mancias, J.D. NCOA4-mediated ferritinophagy: A potential link to neurodegeneration. Front. Neurosci. 2019, 13, 238. [Google Scholar] [CrossRef] [Green Version]
- Abdelaleem, E.R.; Samy, M.N.; Desoukey, S.Y.; Liu, M.; Quinn, R.J.; Abdelmohsen, U.R. Marine natural products from sponges (Porifera) of the order Dictyoceratida (2013 to 2019); a promising source for drug discovery. RSC Adv. 2020, 10, 34959–34976. [Google Scholar] [CrossRef]
- Solanki, H.; Angulo-Preckler, C.; Calabro, K.; Kaur, N.; Lasserre, P.; Cautain, B.; de la Cruz, M.; Reyes, F.; Avila, C.; Thomas, O.P. Suberitane sesterterpenoids from the Antarctic sponge Phorbas areolatus (Thiele, 1905). Tetrahedron Lett. 2018, 59, 3353–3356. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.X.; Huang, X.; Li, Y.; Sun, T.; Zang, S.; Guan, K.L.; Xiong, Y.; Liu, J.; Yuan, H.X. Targeting ferroptosis alleviates methionine-choline deficient (MCD)-diet induced NASH by suppressing liver lipotoxicity. Liver Int. 2020, 40, 1378–1394. [Google Scholar] [CrossRef]
- Kist, M.; Vucic, D. Cell death pathways: Intricate connections and disease implications. EMBO J. 2021, 40, e106700. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, E.A. Regulated cell death signaling pathways and marine natural products that target them. Mar. Drugs 2019, 17, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.T.; Bow, Y.D.; Fu, P.J.; Li, C.Y.; Wu, C.Y.; Chang, Y.H.; Teng, Y.N.; Li, R.N.; Lu, M.C.; Liu, Y.C.; et al. A Marine Terpenoid, Heteronemin, Induces Both the Apoptosis and Ferroptosis of Hepatocellular Carcinoma Cells and Involves the ROS and MAPK Pathways. Oxid. Med. Cell. Longev. 2021, 2021, 7689045. [Google Scholar] [CrossRef]
- Tang, M.; Chen, Z.; Wu, D.; Chen, L. Ferritinophagy/ferroptosis: Iron-related newcomers in human diseases. J. Cell. Physiol. 2018, 233, 9179–9190. [Google Scholar] [CrossRef] [PubMed]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.-M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell 2021, 56, 1363–1393. [Google Scholar] [CrossRef] [PubMed]
- Mishchenko, T.A.; Balalaeva, I.V.; Vedunova, M.V.; Krysko, D.V. Ferroptosis and Photodynamic Therapy Synergism: Enhancing Anticancer Treatment. Trends Cancer 2021, 7, 484–487. [Google Scholar] [CrossRef]
- Mangano, S.; Michaud, L.; Caruso, C.; Brilli, M.; Bruni, V.; Fani, R.; Lo Giudice, A. Antagonistic interactions between psychrotrophic cultivable bacteria isolated from Antarctic sponges: A preliminary analysis. Res. Microbiol. 2009, 160, 27–37. [Google Scholar] [CrossRef]
- Rützler, K. Sponges in coral reefs. In Coral Reefs: Research Methods, Monographs on Oceanographic Methodology; Stoddart, D.R., Johannes, R.E., Eds.; Unesco: Paris, France, 1978; pp. 299–313. [Google Scholar]
- Hooper, J.N.A. ‘Sponguide’. Guide to Sponge Collection and Identification; Queensland Museum: South Brisbane, Australia, 2000. [Google Scholar]
- Soest, V.; Rob, W.M.; Boury-Esnault, N.; Hooper, J.N.A.; Rützler, K.; de Voogd, N.J.; Alvarez, B.; Hajdu, E.; Pisera, A.B.; Manconi, R.; et al. World Porifera Database; World Register of Marine Species: Ostend, Belgium, 2014. [Google Scholar]
- Manuel, M.; Borchiellini, C.; Alivon, E.; Le Parco, Y.; Vacelet, J.; Boury-Esnault, N. Phylogeny and evolution of calcareous sponges: Monophyly of calcinea and calcaronea, high level of morphological homoplasy, and the primitive nature of axial symmetry. Syst. Biol. 2003, 52, 311–333. [Google Scholar] [CrossRef]
- Collins, A.G. Phylogeny of medusozoa and the evolution of cnidarian life cycles. J. Evol. Biol. 2002, 15, 418–432. [Google Scholar] [CrossRef] [Green Version]
- Dohrmann, M.; Janussen, D.; Reitner, J.; Collins, A.G.; Wörheide, G. Phylogeny and evolution of glass sponges (Porifera, Hexactinellida). Syst. Biol. 2008, 57, 388–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chombard, C.; Boury-Esnault, N.; Tillier, S. Reassessment of homology of morphological characters in Tetractinellid sponges based on molecular data. Syst. Biol. 1998, 47, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.P.; Geller, J.B.; Paulay, G. Fine scale endemism on coral reefs: Archipelagic differentiation in turbinid gastropods. Evolution 2005, 59, 113–125. [Google Scholar] [CrossRef]
- Rot, C.; Goldfarb, I.; Ilan, M.; Huchon, D. Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol. Biol. 2006, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Riccio, G.; Bottone, S.; La Regina, G.; Badolati, N.; Passacantilli, S.; Rossi, G.B.; Accardo, A.; Dentice, M.; Silvestri, R.; Novellino, E.; et al. A Negative Allosteric Modulator of WNT Receptor Frizzled 4 Switches into an Allosteric Agonist. Biochemistry 2018, 57, 839–851. [Google Scholar] [CrossRef] [PubMed]
| Species Name | Abbreviation | Sample IDs | MNA Code |
|---|---|---|---|
| Hemigellius pilosus (Kirkpatrick, 1907) | H. p. | D4 | 13266 |
| Haliclona (Rhizoniera) dancoi (Topsent, 1901) | H. d. | C6 | 13265 |
| Mycale (Oxymycale) acerata (Kirkpatrick, 1907) | M. a. | B4 | 13264 |
| Hemimycale topsenti (ex Suberites topsenti) | H. t. | C7 | 13860 |
| Compounds | A549 | A2058 | HepG2 | MRC5 |
|---|---|---|---|---|
| Mycalols | 10.1 | 15.3 | 9.0 | 21.3 |
| Suberitenones A (1) | 28.5 | 10.2 | 17.6 | 7.4 |
| Suberitenones B (2) | 80.7 | 14.6 | 19.2 | 8.5 |
| Unigene | RefSeq | Symbol | Description | Fold | SD |
|---|---|---|---|---|---|
| Genes down-regulated by mycalols treatment | |||||
| Hs.47061 | NM_003565 | ULK1 | Unc-51-like kinase 1 (C. elegans) | −12.03 | 0.025 |
| Hs.269027 | NM_014568 | GALNT5 | UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | −10.71 | 0.001 |
| Hs.2490 | NM_033292 | CASP1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | −6.85 | 0.005 |
| Hs.484111 | NM_002546 | TNFRSF11 | Tumor necrosis factor receptor superfamily, member 11b | −5.57 | 0.001 |
| Hs.81791 | NM_014592 | KCNIP1 | Kv channel interacting protein 1 | −2.57 | 0.001 |
| Hs.160562 | NM_000618 | IGF1 | Insulin-like growth factor 1 (somatomedin C) | −2.41 | 0.002 |
| Hs.552567 | NM_001160 | APAF1 | Apoptotic peptidase activating factor 1 | −2.03 | 0.026 |
| Genes up-regulated by mycalols treatment | |||||
| Hs.513667 | NM_003946 | NOL3 | Nucleolar protein 3 (apoptosis repressor with CARD domain) | 7.40 | 0.029 |
| Hs.227817 | NM_004049 | BCL2A1 | BCL2-related protein A1 | 5.14 | 0.002 |
| Hs.587290 | NM_003900 | SQSTM1 | Sequestosome 1 | 4.75 | 1.256 |
| Hs.442337 | NM_176823 | S100A7A | S100 calcium binding protein A7A | 4.15 | 0.001 |
| Hs.553833 | NM_001004467 | OR10J3 | Olfactory receptor, family 10, subfamily J, member 3 | 4.05 | 0.001 |
| Hs.202676 | NM_014258 | SYCP2 | Synaptonemal complex protein 2 | 3.74 | 0.001 |
| Hs.138211 | NM_002750 | MAPK8 | Mitogen-activated protein kinase 8 | 3.71 | 0.132 |
| Hs.519680 | NM_001145805 | IRGM | Immunity-related GTPase family, M | 3.19 | 0.001 |
| Hs.643440 | NM_002361 | MAG | Myelin associated glycoprotein | 3.17 | 0.006 |
| Hs.181301 | NM_004079 | CTSS | Cathepsin S | 2.97 | 0.004 |
| Hs.32949 | NM_005218 | DEFB1 | Defensin, beta 1 | 2.74 | 0.005 |
| Hs.29169 | NM_024610 | HSPBAP1 | HSPB (heat shock 27kDa) associated protein 1 | 2.28 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riccio, G.; Nuzzo, G.; Zazo, G.; Coppola, D.; Senese, G.; Romano, L.; Costantini, M.; Ruocco, N.; Bertolino, M.; Fontana, A.; et al. Bioactivity Screening of Antarctic Sponges Reveals Anticancer Activity and Potential Cell Death via Ferroptosis by Mycalols. Mar. Drugs 2021, 19, 459. https://doi.org/10.3390/md19080459
Riccio G, Nuzzo G, Zazo G, Coppola D, Senese G, Romano L, Costantini M, Ruocco N, Bertolino M, Fontana A, et al. Bioactivity Screening of Antarctic Sponges Reveals Anticancer Activity and Potential Cell Death via Ferroptosis by Mycalols. Marine Drugs. 2021; 19(8):459. https://doi.org/10.3390/md19080459
Chicago/Turabian StyleRiccio, Gennaro, Genoveffa Nuzzo, Gianluca Zazo, Daniela Coppola, Giuseppina Senese, Lucia Romano, Maria Costantini, Nadia Ruocco, Marco Bertolino, Angelo Fontana, and et al. 2021. "Bioactivity Screening of Antarctic Sponges Reveals Anticancer Activity and Potential Cell Death via Ferroptosis by Mycalols" Marine Drugs 19, no. 8: 459. https://doi.org/10.3390/md19080459
APA StyleRiccio, G., Nuzzo, G., Zazo, G., Coppola, D., Senese, G., Romano, L., Costantini, M., Ruocco, N., Bertolino, M., Fontana, A., Ianora, A., Verde, C., Giordano, D., & Lauritano, C. (2021). Bioactivity Screening of Antarctic Sponges Reveals Anticancer Activity and Potential Cell Death via Ferroptosis by Mycalols. Marine Drugs, 19(8), 459. https://doi.org/10.3390/md19080459