Polar Lipids Composition, Antioxidant and Anti-Inflammatory Activities of the Atlantic Red Seaweed Grateloupia turuturu
Abstract
:1. Introduction
2. Results
2.1. Lipids Content and Carbon/Nitrogen Ratio
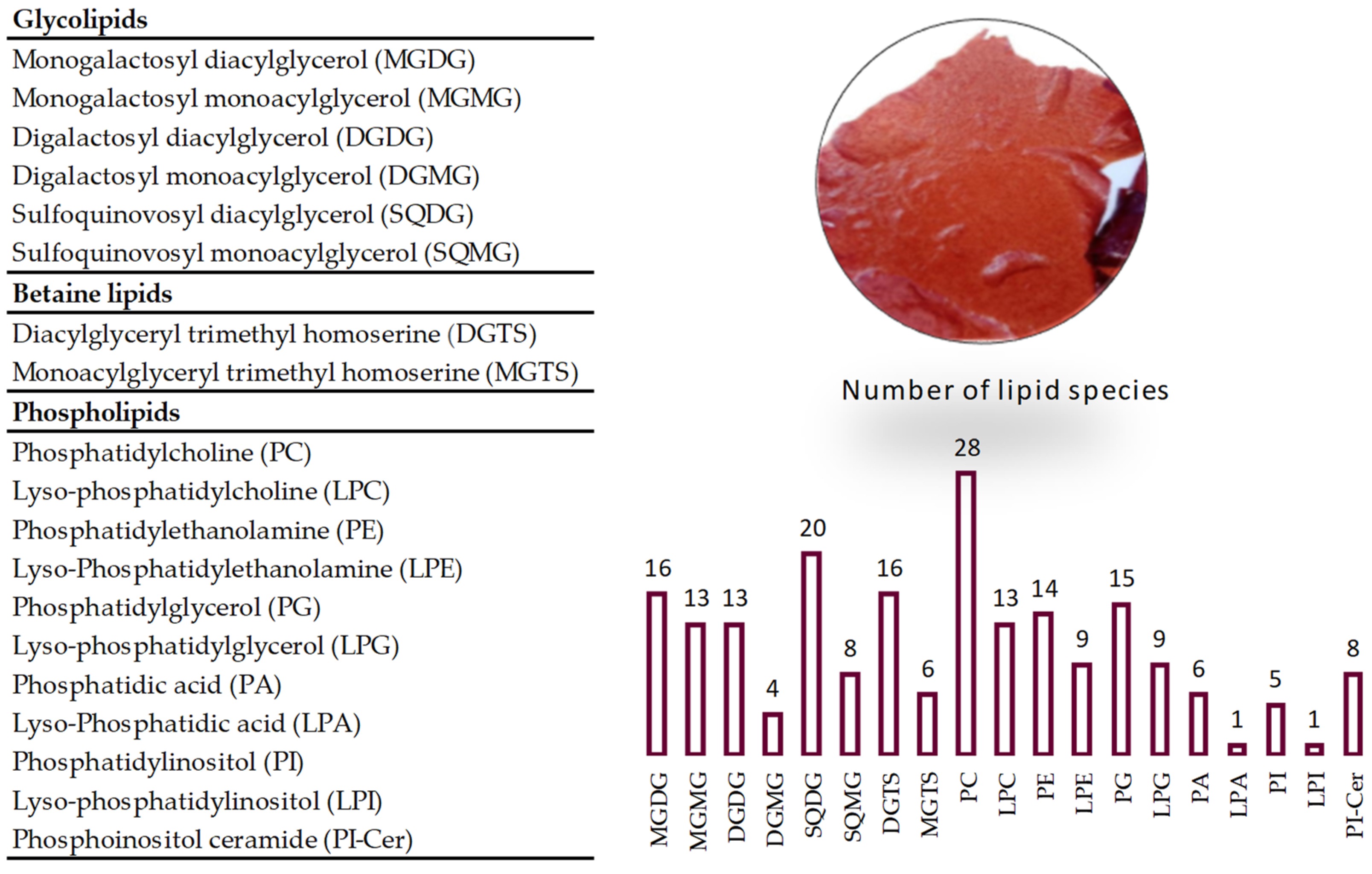
2.2. Lipidome of Grateloupia turuturu
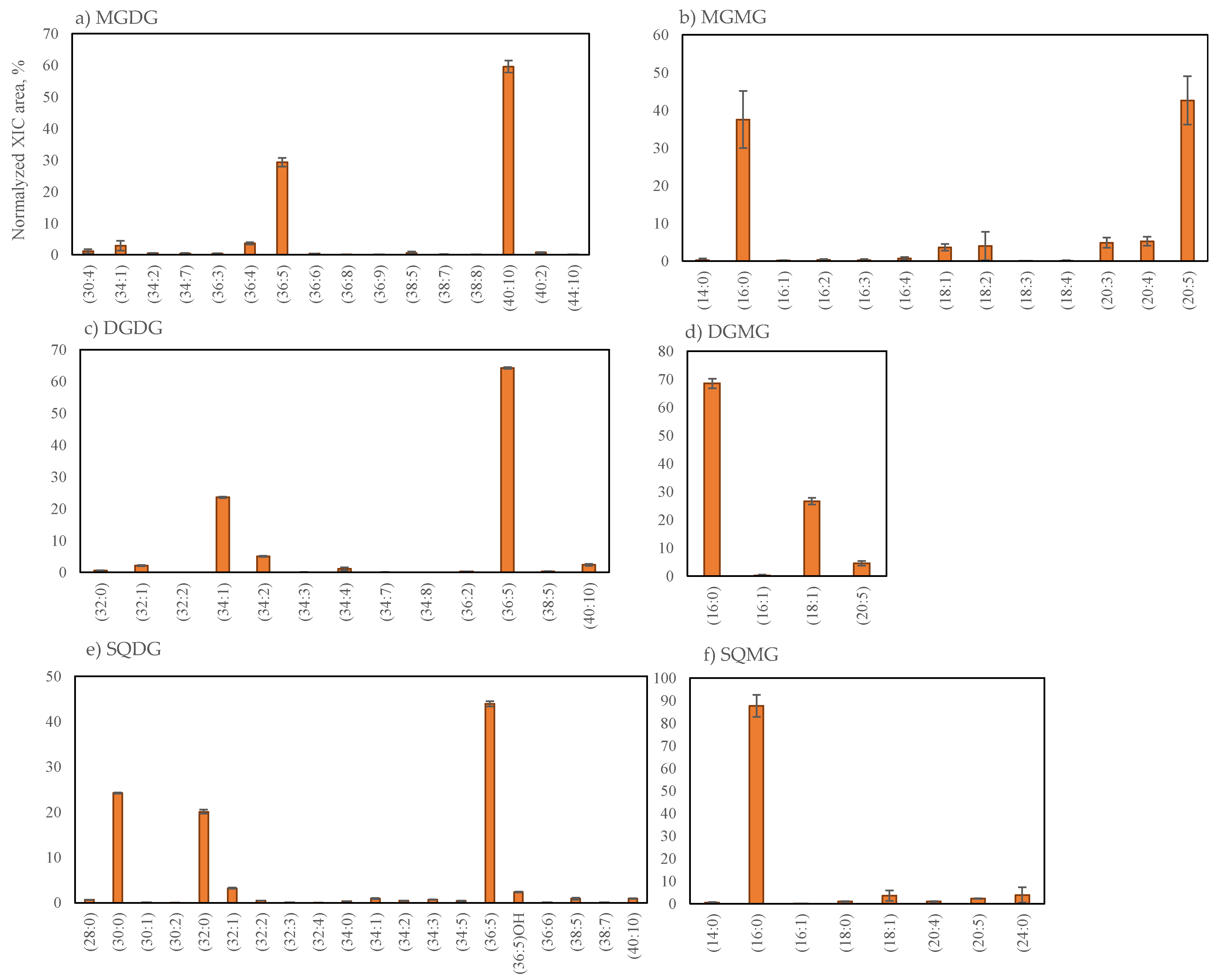
2.2.1. Glycolipids Profile of Grateloupia turuturu
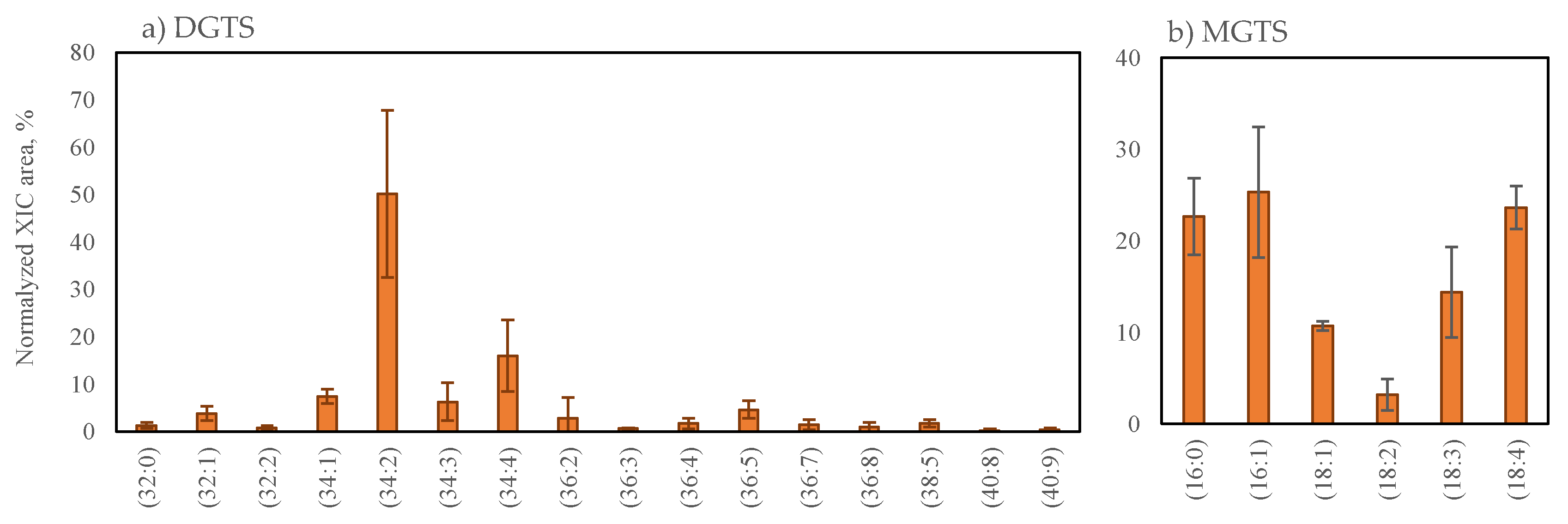
2.2.2. Betaine Lipids Profile of Grateloupia turuturu
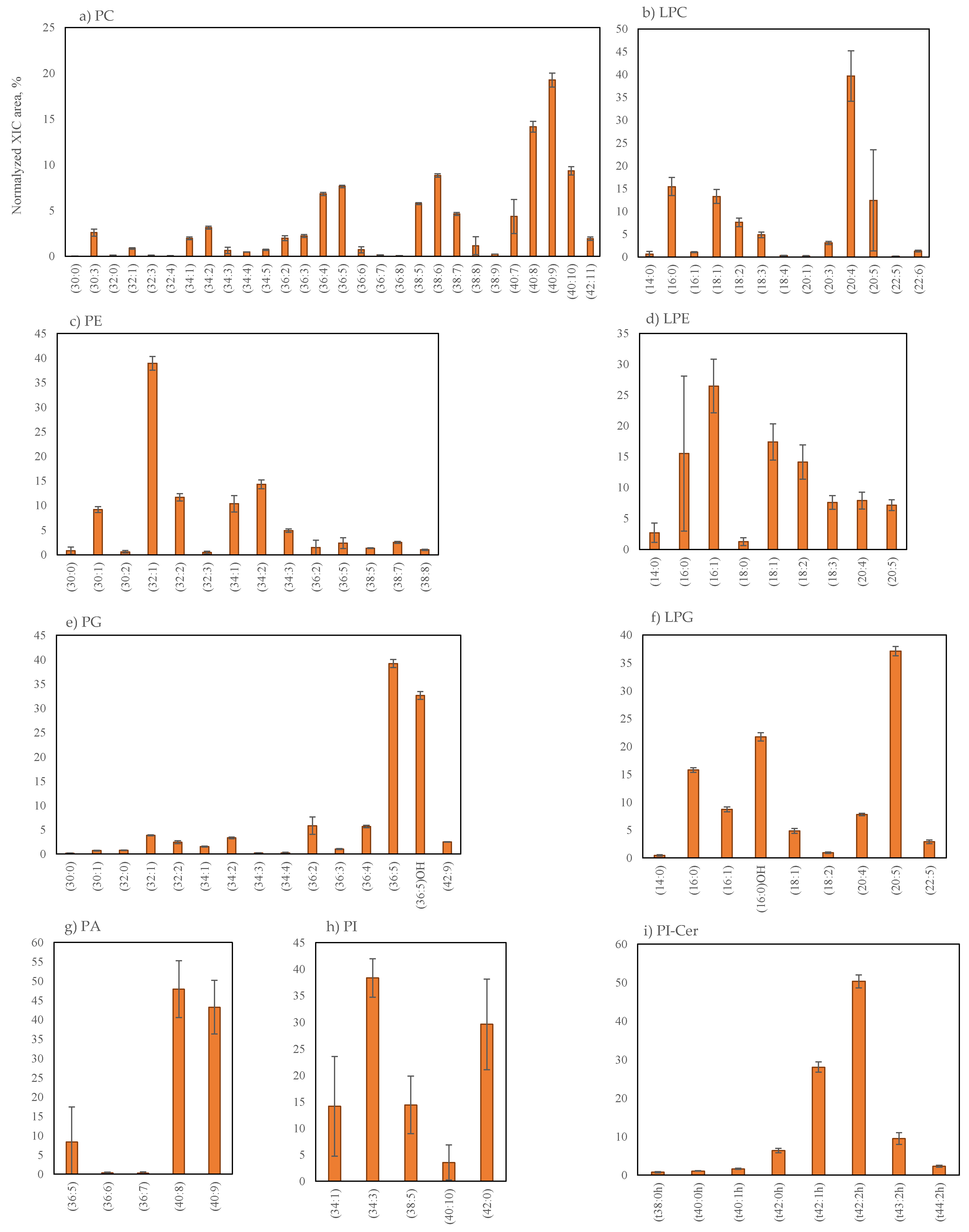
2.2.3. Phospholipids Profile of Grateloupia turuturu
2.2.4. Fatty Acids Profile of Grateloupia turuturu
2.3. Antioxidant Activity of the Lipid Extracts of Grateloupia turuturu
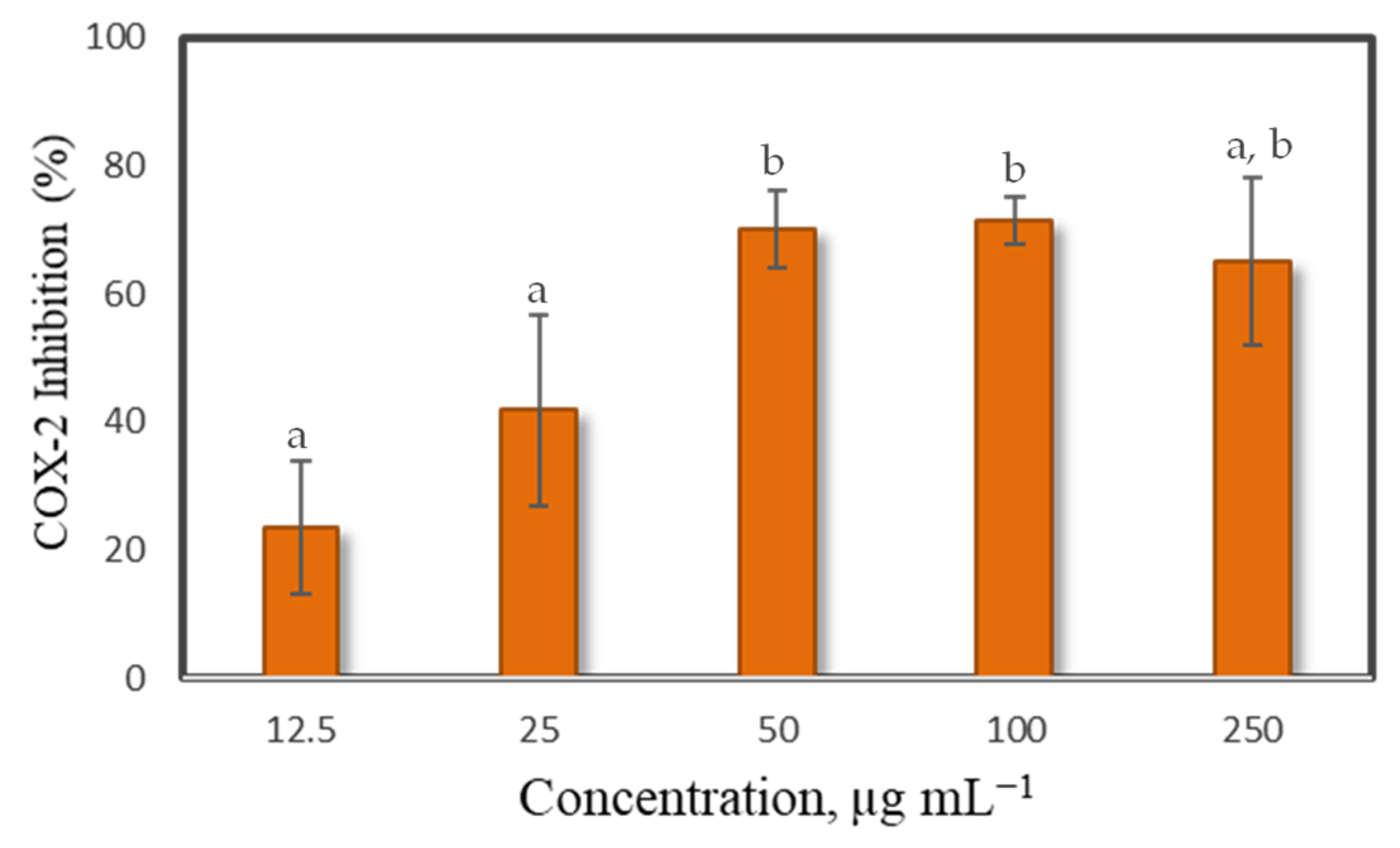
2.4. Cyclooxygenase 2 (COX 2) Enzyme Inhibitory Capacity of the Lipids Extracts of Grateloupia turuturu
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Collection of Macroalgae and Preparation of Biomass
4.3. Proximate Elemental Composition Analysis
4.4. Lipid Extraction
4.5. Hydrophilic Interaction Liquid Chromatography−High Resolution Mass Spectrometry (HILIC−MS) and Tandem Mass Spectrometry (MS/MS)
4.6. Data Analysis and Lipids Identification
4.7. Analysis of Fatty acid by Gas Chromatography–Mass Spectrometry (GC–MS)
4.8. 2,2′-Azino-bis-3-Ethylbenzothiazoline-6-Sulfonic Acid Radical Cation Assay—ABTS Radical Scavenging Activity
4.9. 2,2-Diphenyl-1-Picrylhydrazyl Radical Assay—DPPH Radical Scavenging Activity
4.10. Cyclooxygenase 2 (COX 2) Enzyme Inhibitory Capacity of Lipid Extract
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Klinger, T. The role of seaweeds in the modern ocean. Perspect. Phycol. 2015, 2, 31–39. [Google Scholar] [CrossRef]
- Doumeizel, V.; Aass, K.; McNevin, A.; Cousteau, A.; Yap, A.Y.; Cai, J.; Cottier-Cook, E.J.; Giercksky, E.; Chen, H.; Skjermo, J.; et al. Seaweed Revolution: A Manifesto for a Sustainable Future; Lloyd’s Register Foundation: London, UK, 2020; pp. 1–16. [Google Scholar]
- Rebours, C.; Marinho-Soriano, E.; Zertuche-González, J.A.; Hayashi, L.; Vásquez, J.A.; Kradolfer, P.; Soriano, G.; Ugarte, R.; Abreu, M.H.; Bay-Larsen, I.; et al. Seaweeds: An opportunity for wealth and sustainable livelihood for coastal communities. J. Appl. Phycol. 2014, 26, 1939–1951. [Google Scholar] [CrossRef] [Green Version]
- Chapman, A.S.; Stévant, P.; Larssen, W.E. Food or fad? Challenges and opportunities for including seaweeds in a Nordic diet. Bot. Mar. 2015, 58, 423–433. [Google Scholar] [CrossRef]
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Pacheco, D.; Araújo, G.S.; Cotas, J.; Gaspar, R.; Neto, J.M.; Pereira, L. Invasive Seaweeds in the Iberian Peninsula: A Contribution for Food Supply. Mar. Drugs 2020, 18, 560. [Google Scholar] [CrossRef]
- Cotas, J.; Leandro, A.; Pacheco, D.; Gonçalves, A.M.M.; Pereira, L. A comprehensive review of the nutraceutical and therapeutic applications of red seaweeds (Rhodophyta). Life 2020, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Mathieson, A.C.; Dawes, C.J.; Pederson, J.; Gladych, R.A.; Carlton, J.T. The Asian red seaweed Grateloupia turuturu (Rhodophyta) invades the Gulf of Maine. Biol. Invasions 2008, 10, 985–988. [Google Scholar] [CrossRef] [Green Version]
- Bárbara, I.; Cremades, J. Grateloupia lanceola versus Grateloupia doryphora (Gigartinales, Rhodophyta) en las costas de la Península Ibérica. Anales del Jardín Botánico de Madrid 2004, 61. [Google Scholar] [CrossRef]
- Araújo, R.; Violante, J.; Pereira, R.; Abreu, H.; Arenas, F.; Sousa-Pinto, I. Distribution and population dynamics of the introduced seaweed Grateloupia turuturu (halymeniaceae, rhodophyta) along the Portuguese coast. Phycologia 2011, 50, 392–402. [Google Scholar] [CrossRef]
- Mulas, M.; Bertocci, I. Devil’s tongue weed (Grateloupia turuturu Yamada) in northern Portugal: Passenger or driver of change in native biodiversity? Mar. Environ. Res. 2016, 118, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Petrocelli, A.; Alabiso, G.; Cecere, E.; Ricci, P.; Carlucci, R. Invasive or not? The case of Grateloupia turuturu (Rhodophyta, Halymeniales) in the Northern Ionian Sea (Mediterranean Sea). Mar. Pollut. Bull. 2020, 161, 111748. [Google Scholar] [CrossRef] [PubMed]
- Denis, C.; Morançais, M.; Li, M.; Deniaud, E.; Gaudin, P.; Wielgosz-Collin, G.; Barnathan, G.; Jaouen, P.; Fleurence, J. Study of the chemical composition of edible red macroalgae Grateloupia turuturu from Brittany (France). Food Chem. 2010, 119, 913–917. [Google Scholar] [CrossRef]
- Perfeto, P.N.M. Relation between chemical composition of Grateloupia doryphora (montagne) howe, Gymnogongrus griffithsiae (turner) martius, and abiotic parameters. Acta Bot. Brasilica 1998, 12, 77–88. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, D.; Freitas, A.C.; Pereira, L.; Rocha-Santos, T.A.P.; Vasconcelos, M.W.; Roriz, M.; Rodríguez-Alcalá, L.M.; Gomes, A.M.P.; Duarte, A.C. Chemical composition of red, brown and green macroalgae from Buarcos bay in Central West Coast of Portugal. Food Chem. 2015, 183, 197–207. [Google Scholar] [CrossRef]
- Kendel, M.; Couzinet-Mossion, A.; Viau, M.; Fleurence, J.; Barnathan, G.; Wielgosz-Collin, G. Seasonal composition of lipids, fatty acids, and sterols in the edible red alga Grateloupia turuturu. J. Appl. Phycol. 2012, 25, 425–432. [Google Scholar] [CrossRef]
- Floreto, E.A.T.; Teshima, S. The fatty acid composition of seaweed exposed to different levels of light intensity and salinity. Bot. Mar. 1998, 41, 467–481. [Google Scholar] [CrossRef]
- Munier, M.; Dumay, J.; Morançais, M.; Jaouen, P.; Fleurence, J. Variation in the biochemical composition of the edible seaweed Grateloupia turuturu yamada harvested from two sampling sites on the Brittany coast (France): The influence of storage method on the extraction of the seaweed pigment r-phycoerythrin. J. Chem. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Kendel, M.; Barnathan, G.; Fleurence, J.; Rabesaotra, V.; Wielgosz-Collin, G. Non-methylene interrupted and hydroxy fatty acids in polar lipids of the alga Grateloupia turuturu over the four seasons. Lipids 2013, 48, 535–545. [Google Scholar] [CrossRef]
- Calder, P.C. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 2006, 83, 1505S–1519S. [Google Scholar] [CrossRef]
- Simopoulos, A.P. Essential fatty acids in health and chronic disease. Am. J. Clin. Nutr. 1999, 70, 560S–569S. [Google Scholar] [CrossRef] [Green Version]
- Bowen, K.J.; Harris, W.S.; Kris-Etherton, P.M. Omega-3 fatty acids and cardiovascular disease: Are there benefits? Curr. Treat. Options Cardiovasc. Med. 2016, 18. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-C.; Hou, M.-F.; Huang, H.-W.; Chang, F.-R.; Yeh, C.-C.; Tang, J.-Y.; Chang, H.-W. Marine algal natural products with anti-oxidative, anti-inflammatory, and anti-cancer properties. Cancer Cell Int. 2013, 13, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Roos, B.; Mavrommatis, Y.; Brouwer, I.A. Long-chain n-3 polyunsaturated fatty acids: New insights into mechanisms relating to inflammation and coronary heart disease. Br. J. Pharmacol. 2009, 158, 413–428. [Google Scholar] [CrossRef] [Green Version]
- Harwood, J.L.; Guschina, I.A. The versatility of algae and their lipid metabolism. Biochimie 2009, 91, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Vaskovsky, V.E.; Khotimchenko, S.V.; Svetlana, K.; Bangmei, X.; Hefang, L. Polar lipids and fatty acids of some marine macrophytes from Yellow Sea. Phytochemistry 1996, 42, 1347–1356. [Google Scholar] [CrossRef]
- Melo, T.; Alves, E.; Azevedo, V.; Martins, A.S.; Neves, B.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingues, M.R. Lipidomics as a new approach for the bioprospecting of marine macroalgae—Unraveling the polar lipid and fatty acid composition of Chondrus crispus. Algal Res. 2015, 8, 181–191. [Google Scholar] [CrossRef]
- Da Costa, E.; Melo, T.; Moreira, A.S.P.A.; Bernardo, C.; Helguero, L.; Ferreira, I.; Cruz, M.T.M.; Rego, A.M.A.; Domingues, P.; Calado, R.; et al. Valorization of lipids from Gracilaria sp. through lipidomics and decoding of antiproliferative and anti-Inflammatory activity. Mar. Drugs 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Costa, E.; Azevedo, V.; Melo, T.; Rego, A.M.; Evtuguin, D.V.; Domingues, P.; Calado, R.; Pereira, R.; Abreu, M.H.; Domingues, M.R. High-Resolution lipidomics of the early life stages of the red seaweed Porphyra dioica. Molecules 2018, 23, 187. [Google Scholar] [CrossRef] [Green Version]
- Lopes, D.; Melo, T.; Rey, F.; Costa, E.; Moreira, A.S.P.; Abreu, M.H.; Domingues, P.; Lillebø, A.I.; Calado, R.; Rosário Domingues, M. Insights of species-specific polar lipidome signatures of seaweeds fostering their valorization in the blue bioeconomy. Algal Res. 2021, 55. [Google Scholar] [CrossRef]
- Lee, H.-G.; Lu, Y.-A.; Je, J.-G.; Jayawardena, T.U.; Kang, M.-C.; Lee, S.-H.; Kim, T.-H.; Lee, D.-S.; Lee, J.-M.; Yim, M.-J.; et al. Effects of ethanol extracts from Grateloupia elliptica, a red seaweed, and Its chlorophyll derivative on 3T3-L1 adipocytes: Suppression of lipid accumulation through dDownregulation of adipogenic protein expression. Mar. Drugs 2021, 19, 91. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, L. Effects of UV-B irradiation on isoforms of antioxidant enzymes and their activities in red alga Grateloupia filicina (Rhodophyta). Chinese J. Oceanol. Limnol. 2014, 32, 1364–1372. [Google Scholar] [CrossRef]
- Ferreira, J.; Marques, A.; Abreu, H.; Pereira, R.; Rego, A.; Pacheco, M.; Gaivão, I. Red seaweeds Porphyra umbilicalis and Grateloupia turuturu display antigenotoxic and longevity-promoting potential in Drosophila melanogaster. Eur. J. Phycol. 2019, 54, 519–530. [Google Scholar] [CrossRef]
- Ismail, M.M.; Alotaibi, B.S.; EL-Sheekh, M.M. Therapeutic uses of red macroalgae. Molecules 2020, 25, 4411. [Google Scholar] [CrossRef]
- Athukorala, Y.; Lee, K.-W.; Song, C.; Ahn, C.B.; Shin, T.-S.; Cha, Y.-J.; Shahidi, F.; Jeon, Y.-J. Potential antioxidant activity of marine red alga Grateloupia filicina extracts. J. Food Lipids 2003, 10, 251–265. [Google Scholar] [CrossRef]
- Athukorala, Y.; Lee, K.W.; Park, E.J.; Heo, M.S.; Yeo, I.K.; Lee, Y.D.; Jeon, Y.J. Reduction of lipid peroxidation and H2O2-mediated DNA damage by a red alga (Grateloupia filicina) methanolic extract. J. Sci. Food Agric. 2005, 85, 2341–2348. [Google Scholar] [CrossRef]
- Yang, E.-J.; Moon, J.-Y.; Kim, M.-J.; Kim, D.S.; Kim, C.-S.; Lee, W.J.; Lee, N.H.; Hyun, C.-G. Inhibitory effect of Jeju endemic seaweeds on the production of pro-inflammatory mediators in mouse macrophage cell line RAW 264.7. J. Zhejiang Univ. Sci. B 2010, 11, 315–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Bueno, N.; Turpin, V.; Cognie, B.; Dumay, J.; Morançais, M.; Amat, M.; Pédron, J.M.; Atucha, A.M.; Fleurence, J.; Decottignies, P. Can the european abalone Haliotis tuberculata survive on an invasive algae? A comparison of the nutritional value of the introduced Grateloupia turuturu and the native Palmaria palmata, for the commercial european abalone industry. J. Appl. Phycol. 2016, 28, 2427–2433. [Google Scholar] [CrossRef]
- Huang, W.; Fujita, Y.; Ninomiya, M.; Ohno, M. Seed production and cultivation of Grateloupia turuturu (Cryptonemiales, Rhodophyta) by callus culture. Bull. Mar. Sci. Fish. 1999, 19, 1–7. [Google Scholar]
- Fleurence, J. Seaweed proteins: Biochemical, nutritional aspects and potential uses. Trends Food Sci. Technol. 1999, 10, 25–28. [Google Scholar] [CrossRef]
- Cian, R.E.; Drago, S.R.; De Medina, F.S.; Martínez-Augustin, O. Proteins and carbohydrates from red seaweeds: Evidence for beneficial effects on gut function and microbiota. Mar. Drugs 2015, 13, 5358–5383. [Google Scholar] [CrossRef] [Green Version]
- Fleurence, J.; Morançais, M.; Dumay, J.; Decottignies, P.; Turpin, V.; Munier, M.; Garcia-Bueno, N.; Jaouen, P. What are the prospects for using seaweed in human nutrition and for marine animals raised through aquaculture? Trends Food Sci. Technol. 2012, 27, 57–61. [Google Scholar] [CrossRef]
- Kendel, M.; Wielgosz-collin, G.; Bertrand, S.; Roussakis, C.; Bourgougnon, N.; Bedoux, G. Lipid composition, fatty acids and sterols in the seaweeds Ulva armoricana, and Solieria chordalis from Brittany (France): An analysis from nutritional, chemotaxonomic, and antiproliferative activity perspectives. Mar. Drugs 2015, 13, 5606–5628. [Google Scholar] [CrossRef]
- Hotimchenko, S.V. Fatty acid composition of algae from habitats with varying amounts of illumination. Russ. J. Mar. Biol. 2002, 28, 218–220. [Google Scholar] [CrossRef]
- Kumari, P.; Kumar, M.; Gupta, V.; Reddy, C.R.K.; Jha, B. Tropical marine macroalgae as potential sources of nutritionally important PUFAs. Food Chem. 2010, 120, 749–757. [Google Scholar] [CrossRef]
- Lopes, D.; Melo, T.; Meneses, J.; Abreu, M.H.; Pereira, R.; Domingues, P.; Lillebø, A.I.; Calado, R.; Rosário Domingues, M. A New Look for the Red Macroalga Palmaria palmata: A seafood with polar lipids rich in EPA and with antioxidant properties. Mar. Drugs 2019, 17, 533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleurence, J.; Gutbier, G.; Mabeau, S.; Leray, C. Fatty acids from 11 marine macroalgae of the French Brittany coast. J. Appl. Phycol. 1994, 6, 527–532. [Google Scholar] [CrossRef]
- Wells, M.L.; Potin, P.; Craigie, J.S.; Raven, J.A.; Merchant, S.S.; Helliwell, K.E.; Smith, A.G.; Camire, M.E.; Brawley, S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017, 29, 949–982. [Google Scholar] [CrossRef] [PubMed]
- Harrysson, H.; Hayes, M.; Eimer, F.; Carlsson, N.G.; Toth, G.B.; Undeland, I. Production of protein extracts from Swedish red, green, and brown seaweeds, Porphyra umbilicalis Kützing, Ulva lactuca Linnaeus, and Saccharina latissima (Linnaeus) J. V. Lamouroux using three different methods. J. Appl. Phycol. 2018, 30, 3565–3580. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Hurd, C.L. Nitrogen ecophysiology of intertidal seaweeds from New Zealand: N uptake, storage and utilisation in relation to shore position and season. Mar. Ecol. Prog. Ser. 2003, 264, 31–48. [Google Scholar] [CrossRef]
- Roleda, M.Y.; Hurd, C.L. Seaweed nutrient physiology: Application of concepts to aquaculture and bioremediation. Phycologia 2019, 58, 552–562. [Google Scholar] [CrossRef] [Green Version]
- Geider, R.J.; La Roche, J. Redfield revisited: Variability of C:N:P in marine microalgae and its biochemical basis. Eur. J. Phycol. 2002, 37, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Rodríguez, N.; Beltrán, S.; Jaime, I.; de Diego, S.M.; Sanz, M.T.; Carballido, J.R. Production of omega-3 polyunsaturated fatty acid concentrates: A review. Innov. Food Sci. Emerg. Technol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Calder, P.C.; Albers, R.; Antoine, J.-M.; Blum, S.; Ferns, G.A.; Folkerts, G.; Bourdet-Sicard, R.; Friedmann, P.S.; Frost, G.S.; Guarner, F.; et al. Inflammatory disease processes and interactions with nutrition. Br. J. Nutr. 2009, 101, S2–S14. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. Omega-3 fatty acids and inflammatory processes: From molecules to man. Biochem. Soc. Trans. 2017, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wołoszyn, J.; Haraf, G.; Okruszek, A.; Wereńska, M.; Goluch, Z.; Teleszko, M. Fatty acid profiles and health lipid indices in the breast muscles of local Polish goose varieties. Poult. Sci. 2020, 99, 1216–1224. [Google Scholar] [CrossRef]
- Fernandes, C.E.; Vasconcelos, M.A.D.S.; De Almeida Ribeiro, M.; Sarubbo, L.A.; Andrade, S.A.C.; Filho, A.B.D.M. Nutritional and lipid profiles in marine fish species from Brazil. Food Chem. 2014, 160, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Plouguerné, E.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Glycolipids from seaweeds and their potential biotechnological applications. Front. Cell. Infect. Microbiol. 2014, 4, 1–3. [Google Scholar] [CrossRef]
- Küllenberg, D.; Taylor, L.A.; Schneider, M.; Massing, U. Health effects of dietary phospholipids. Lipids Health Dis. 2012, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arao, T.; Yamada, M. Positional distribution of fatty acids in galactolipids of algae. Phytochemistry 1989, 28, 805–810. [Google Scholar] [CrossRef]
- Khotimchenko, S.V.; Vaskovsky, V.E. An inositol-containing sphingolipid from the red alga Gracilaria verrucosa. Russ. J. Bioorganic Chem. 2004, 30, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Khotimchenko, S.V.; Klochkova, N.G.; Vaskovsky, V.E. Polar lipids of marine macrophytic algae as chemotaxonomic markers. Biochem. Syst. Ecol. 1990, 18, 93–101. [Google Scholar] [CrossRef]
- Lordan, R.; Tsoupras, A.; Zabetakis, I. Phospholipids of animal and marine origin: Structure, function, and anti-inflammatory properties. Molecules 2017, 22, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lordan, S.; Ross, R.P.; Stanton, C. Marine bioactives as functional food ingredients: Potential to reduce the incidence of chronic diseases. Mar. Drugs 2011, 9, 1056–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lordan, R.; Redfern, S.; Tsoupras, A.; Zabetakis, I. Inflammation and cardiovascular disease: Are marine phospholipids the answer? Food Funct. 2020, 11, 2861–2885. [Google Scholar] [CrossRef]
- Burri, L.; Hoem, N.; Banni, S.; Berge, K. Marine omega-3 phospholipids: Metabolism and biological activities. Int. J. Mol. Sci. 2012, 13, 15401–15419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, D.; Melo, T.; Rey, F.; Meneses, J.; Monteiro, F.L.; Helguero, L.A.; Abreu, M.H.; Lillebø, A.I.; Calado, R.; Domingues, M.R. Valuing bioactive lipids from Green, Red and Brown macroalgae from aquaculture, to foster functionality and biotechnologicala. Molecules 2020, 25, 3883. [Google Scholar] [CrossRef] [PubMed]
- Arulselvan, P.; Fard, M.T.; Tan, W.S.; Gothai, S.; Fakurazi, S.; Norhaizan, M.E.; Kumar, S.S. Role of antioxidants and natural products in inflammation. Oxid. Med. Cell. Longev. 2016, 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kebede, M.; Admassu, S. Application of antioxidants in food processing industry: Options to improve the extraction yields and market value of natural products. Adv. Food Technol. Nutr. Sci. Open J. 2019, 5, 38–49. [Google Scholar] [CrossRef]
- Sanches-Silva, A.; Costa, D.; Albuquerque, T.G.; Buonocore, G.G.; Ramos, F.; Castilho, M.C.; Machado, A.V.; Costa, H.S. Trends in the use of natural antioxidants in active food packaging: A review. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2014, 31, 374–395. [Google Scholar] [CrossRef] [PubMed]
- da Costa, E.; Amaro, H.M.; Melo, T.; Guedes, A.C.; Domingues, M.R. Screening for polar lipids, antioxidant, and anti-inflammatory activities of Gloeothece sp. lipid extracts pursuing new phytochemicals from cyanobacteria. J. Appl. Phycol. 2020, 32, 3015–3030. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.P.; Craigie, J.S.; Hafting, J.T.; Critchley, A.T. Polar lipids from the marine macroalga Palmaria palmata inhibit lipopolysaccharide-induced nitric oxide production in RAW264.7 macrophage cells. Phytochemistry 2014, 101, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.A.H.; Stefanova, R.; Sperker, S.; Lall, S.; Craigie, J.S.J.S.; Hafting, J.T.J.T. Lipids isolated from the cultivated red alga Chondrus crispus inhibit nitric oxide production. J. Appl. Phycol. 2014, 26, 1565–1571. [Google Scholar] [CrossRef]
- Sahaka, M.; Amara, S.; Wattanakul, J.; Gedi, M.A.; Aldai, N.; Parsiegla, G.; Christeller, J.T.; Gray, D.; Gontero, B.; Villeneuve, P. The digestion of galactolipids and its ubiquitous function in Nature for the uptake of the essential α-linolenic acid. Food Funct. 2020. [Google Scholar] [CrossRef] [PubMed]
- Araújo, R.; Vázquez Calderón, F.; Sánchez López, J.; Azevedo, I.C.; Bruhn, A.; Fluch, S.; Garcia Tasende, M.; Ghaderiardakani, F.; Ilmjärv, T.; Laurans, M.; et al. Current Status of the algae production industry in Europe: An emerging sector of the Blue Bioeconomy. Front. Mar. Sci. 2021. [Google Scholar] [CrossRef]
- Aued-Pimentel, S.; Lago, J.H.G.; Chaves, M.H.; Kumagai, E.E. Evaluation of a methylation procedure to determine cyclopropenoids fatty acids from Sterculia striata St. Hil. Et Nauds seed oil. J. Chromatogr. A 2004, 1054, 235–239. [Google Scholar] [CrossRef]
- Santos, F.; Monteiro, J.P.; Duarte, D.; Melo, T.; Lopes, D.; da Costa, E.; Domingues, M.R. Unraveling the lipidome and antioxidant activity of native Bifurcaria bifurcata and invasive Sargassum muticum seaweeds: A lipid perspective on how systemic intrusion may present an opportunity. Antioxidants 2020, 9, 642. [Google Scholar] [CrossRef]

| Compound | Content |
|---|---|
| Lipid content (g 100 g−1 DW) a | 0.88 ± 0.25 |
| C (g 100 g−1 DW) b | 34.88 ± 0.31 |
| N (g 100 g−1 DW) b | 4.20 ± 0.11 |
| C/N | 8.30 ± 0.19 |
| Protein content (N × 6.25) (g 100 g−1 DW) | 26.26 ± 0.69 |
| Lipid Species (C:N) | Calculated m/z | Observed m/z | Error (ppm) | Fatty Acyl Chains (C:N) | Formula |
|---|---|---|---|---|---|
| MGDG identified as [M + NH4]+ | |||||
| MGDG(30:4) | 712.5000 | 712.5028 | 3.9169 | a | C39H70NO10 |
| MGDG(34:7) | 762.5156 | 762.5158 | 0.1854 | 18:3_16:4 | C43H72NO10 |
| MGDG(34:2) | 772.5933 | 772.5896 | −4.8099 | a | C43H82NO10 |
| MGDG(34:1) | 774.609 | 774.6103 | 1.6896 | 18:1_16:0 | C43H84NO10 |
| MGDG(36:9) | 786.5156 | 786.5177 | 2.6952 | a | C45H72NO10 |
| MGDG(36:8) | 788.5313 | 788.5308 | −0.5953 | 20:4_16:4 | C45H74NO10 |
| MGDG(36:6) | 792.5625 | 792.5619 | −0.7124 | a | C45H78NO10 |
| MGDG(36:5) | 794.5782 | 794.5780 | −0.1981 | 20:5_16:0 | C45H80NO10 |
| MGDG(36:4) | 796.5933 | 796.5926 | −0.8679 | 20:4_16:0 | C45H82NO10 |
| MGDG(36:3) | 798.6095 | 798.6074 | −2.6311 | 20:3_16:0 | C45H84NO10 |
| MGDG(38:8) | 816.5626 | 816.5621 | −0.5736 | a | C47H78NO10 |
| MGDG(38:7) | 818.5782 | 818.5800 | 2.135 | a | C47H80NO10 |
| MGDG(38:5) | 822.6095 | 822.6100 | 0.5639 | a | C47H84NO10 |
| MGDG(40:10) | 840.5626 | 840.5633 | 0.8243 | 20:5/20:5 | C49H78NO10 |
| MGDG(40:2) | 856.6878 | 856.6879 | 0.2035 | a | C49H94NO10 |
| MGDG(44:10) | 894.6095 | 894.6102 | 0.7878 | a | C53H84NO10 |
| MGMG identified as [M + NH4]+ | |||||
| MGMG(14:0) | 482.3329 | 482.333 | 0.2488 | a | C23H48NO9 |
| MGMG(16:4) | 502.3016 | 502.3015 | −0.2407 | a | C25H44NO9 |
| MGMG(16:3) | 504.3173 | 504.319 | 3.3957 | a | C25H46NO9 |
| MGMG(16:2) | 506.3329 | 506.3321 | −1.6151 | a | C25H48NO9 |
| MGMG(16:1) | 508.3486 | 508.349 | 0.9149 | a | C25H50NO9 |
| MGMG(16:0) | 510.3642 | 510.3637 | −0.8972 | 16:0 | C25H52NO9 |
| MGMG(18:4) | 530.3329 | 530.3342 | 2.3898 | a | C27H48NO9 |
| MGMG(18:3) | 532.3486 | 532.3497 | 2.1221 | a | C27H50NO9 |
| MGMG 18:2) | 534.3642 | 534.3639 | −0.5996 | a | C27H52NO9 |
| MGMG(18:1) | 536.3799 | 536.3793 | −1.1276 | a | C27H54NO9 |
| MGMG(20:5) | 556.3486 | 556.3488 | 0.3521 | 20:5 | C29H50NO9 |
| MGMG(20:4) | 558.3642 | 558.3646 | 0.7432 | a | C29H52NO9 |
| MGMG(20:3) | 560.3799 | 560.3807 | 1.5291 | a | C29H54NO9 |
| DGDG identified as [M + NH4]+ | |||||
| DGDG(32:2) | 906.6154 | 906.6158 | 0.4273 | 16:2_16:0 | C47H88O15N |
| DGDG(32:1) | 908.6310 | 908.6315 | 0.5304 | 18:1_14:0; 16:1_16:0 | C47H90O15N |
| DGDG(32:0) | 910.6467 | 910.6465 | −0.2435 | a | C47H92O15N |
| DGDG(34:8) | 922.5528 | 922.5488 | −4.3802 | a | C49H80O15N |
| DGDG(34:7) | 924.5684 | 924.5639 | −4.8671 | a | C49H82O15N |
| DGDG(34:4) | 930.6154 | 930.6146 | −0.824 | a | C49H88O15N |
| DGDG(34:3) | 932.6310 | 932.6304 | −0.6395 | 18:3_16:0 | C49H90O15N |
| DGDG(34:2) | 934.6467 | 934.6468 | 0.0566 | 18:2_16:0 | C49H92O15N |
| DGDG(34:1) | 936.6623 | 936.6627 | 0.422 | 18:1_16:0 | C49H94O15N |
| DGDG(36:5) | 956.6310 | 956.6315 | 0.5293 | 20:5_16:0 | C51H90O15N |
| DGDG(36:2) | 962.6780 | 962.6777 | −0.2759 | 18:1/18:1 | C51H96O15N |
| DGDG(38:5) | 984.6623 | 984.6647 | 2.4117 | a | C53H94O15N |
| DGDG(40:10) | 1002.6154 | 1002.6164 | 0.9974 | 20:5/20:5 | C55H88O15N |
| DGMG identified as [M + NH4]+ | |||||
| DGMG(16:1) | 670.4014 | 670.4020 | 0.8999 | a | C31H60NO14 |
| DGMG(16:0) | 672.4170 | 672.4174 | 0.5239 | 16:0 | C31H62NO14 |
| DGMG(18:1) | 698.4327 | 698.4332 | 0.7083 | 18:1 | C33H64NO14 |
| DGMG(20:5) | 718.4014 | 718.4012 | −0.3122 | a | C35H60NO14 |
| SQDG identified as [M − H]− | |||||
| SQDG(28:0) | 737.4510 | 737.4518 | 1.0919 | 12:0_16:0 | C37H69O12S |
| SQDG(30:2) | 761.4510 | 761.4519 | 1.2214 | a | C39H69O12S |
| SQDG(30:1) | 763.4666 | 763.4668 | 0.2246 | 14:0_16:1 | C39H71O12S |
| SQDG(30:0) | 765.4823 | 765.4828 | 0.7155 | 14:0_16:0 | C39H73O12S |
| SQDG(32:4) | 785.4510 | 785.453 | 2.5189 | 14:0_18:4 | C41H69O12S |
| SQDG(32:3) | 787.4666 | 787.4658 | −1.0089 | a | C41H71O12S |
| SQDG(32:2) | 789.4823 | 789.4833 | 1.2813 | 16:2_16:0 | C41H73O12S |
| SQDG(32:1) | 791.4979 | 791.4991 | 1.4932 | 16:1_16:0 | C41H75O12S |
| SQDG(32:0) | 793.5136 | 793.5147 | 1.3591 | 16:0/16:0 | C41H77O12S |
| SQDG(34:5) | 811.4666 | 811.4701 | 4.2331 | 14:0_20:5 | C43H71O12S |
| SQDG(34:3) | 815.4979 | 815.4986 | 0.8394 | 18:3_16:0 | C43H75O12S |
| SQDG(34:2) | 817.5136 | 817.5151 | 1.9049 | a | C43H77O12S |
| SQDG(34:1) | 819.5292 | 819.5308 | 1.9621 | 18:1_16:0 | C43H79O12S |
| SQDG(34:0) | 821.5449 | 821.5459 | 1.2441 | a | C43H81O12S |
| SQDG(36:6) | 837.4823 | 837.4844 | 2.5124 | b | C45H73O12S |
| SQDG(36:5) | 839.4979 | 839.4989 | 1.1462 | 20:5_16:0 | C45H75O12S |
| SQDG(36:5)OH | 855.4939 | 855.4943 | 0.4953 | 20:5−OH_16:0 | C45H75O12SO |
| SQDG(38:7) | 863.4979 | 863.4996 | 1.9862 | 20:4_18:3 | C47H75O12S |
| SQDG(38:5) | 867.5292 | 867.5298 | 0.7138 | 20:5_18:0 | C47H79O12S |
| SQDG(40:10) | 885.4823 | 885.4837 | 1.5835 | 20:5/20:5 | C49H73O12S |
| SQMG identified as [M − H]− | |||||
| SQMG(14:0) | 527.2526 | 527.2540 | 2.7046 | a | C23H43O11S |
| SQMG(16:1) | 553.2683 | 553.2701 | 3.267 | a | C25H45O11S |
| SQMG(16:0) | 555.2839 | 555.2849 | 1.7351 | 16:0 | C25H47O11S |
| SQMG(18:1) | 581.2996 | 581.3024 | 4.8607 | a | C27H49O11S |
| SQMG(18:0) | 583.3152 | 583.3166 | 2.3402 | 18:0 | C27H51O11S |
| SQMG(20:5) | 601.2683 | 601.2696 | 2.2023 | a | C29H45O11S |
| SQMG(20:4) | 603.2839 | 603.2851 | 2.0425 | a | C29H47O11S |
| SQMG(24:0) | 667.4091 | 667.4117 | 3.9262 | a | C33H63O11S |
| Lipid Species (C:N) | Calculated m/z | Observed m/z | Error (ppm) | Fatty Acyl Chains (C:N) | Formula |
|---|---|---|---|---|---|
| DGTS identified as [M+H]+ | |||||
| DGTS(32:2) | 708.5778 | 708.5792 | 1.9979 | a | C42H78O7N |
| DGTS(32:1) | 710.5935 | 710.5939 | 0.5111 | a | C42H80O7N |
| DGTS(32:0) | 712.6091 | 712.6083 | −1.1556 | a | C42H82O7N |
| DGTS(34:4) | 732.5778 | 732.5784 | 0.7925 | 16:0_18:4 | C44H78O7N |
| DGTS(34:3) | 734.5935 | 734.5923 | −1.6029 | a | C44H80O7N |
| DGTS(34:2) | 736.6091 | 736.6101 | 1.3137 | 18:1_16:1 | C44H82O7N |
| DGTS(34:1) | 738.6248 | 738.6238 | −1.4067 | a | C44H84O7N |
| DGTS(36:8) | 752.5465 | 752.5482 | 2.2335 | a | C46H74O7N |
| DGTS(36:7) | 754.5622 | 754.5614 | −1.0468 | a | C46H76O7N |
| DGTS(36:5) | 758.5935 | 758.5942 | 0.9613 | a | C46H80O7N |
| DGTS(36:4) | 760.6091 | 760.6072 | −2.4467 | a | C46H82O7N |
| DGTS(36:3) | 762.6248 | 762.6238 | −1.2584 | a | C46H84O7N |
| DGTS(36:2) | 764.6404 | 764.6413 | 1.2089 | 18:1/18:1 | C46H86O7N |
| DGTS(38:5) | 786.6248 | 786.6239 | −1.1346 | a | C48H84O7N |
| DGTS(40:9) | 806.5935 | 806.5917 | −2.2229 | a | C50H80O7N |
| DGTS(40:8) | 808.6091 | 808.6098 | 0.8873 | a | C50H82O7N |
| MGTS identified as [M+H]+ | |||||
| MGTS(16:1) | 472.3638 | 472.3631 | −1.6157 | 16:1 | C26H50O6N |
| MGTS(16:0) | 474.3795 | 474.3795 | −0.0942 | 16:0 | C26H52O6N |
| MGTS(18:4) | 494.3482 | 494.3486 | 0.9760 | 18:4 | C28H48O6N |
| MGTS(18:3) | 496.3638 | 496.364 | 0.2825 | 18:3 | C28H50O6N |
| MGTS(18:2) | 498.3795 | 498.3794 | −0.1732 | a | C28H52O6N |
| MGTS(18:1) | 500.3951 | 500.3961 | 1.9549 | a | C28H54O6N |
| Lipid Species (C:N) | Calculated m/z | Observed m/z | Error (ppm) | Fatty Acyl Chains (C:N) | Formula |
|---|---|---|---|---|---|
| PC identified as [M+H]+ | |||||
| PC(30:3) | 700.4917 | 700.4902 | −2.1598 | a | C38H71NO8P |
| PC(30:0) | 706.5387 | 706.5383 | −0.506 | b | C38H77NO8P |
| PC(32:4) | 726.5074 | 726.5092 | 2.4576 | a | C40H73NO8P |
| PC(32:3) | 728.5230 | 728.524 | 1.3945 | a | C40H75NO8P |
| PC(32:1) | 732.5543 | 732.5519 | −3.3249 | 16:0/16:1; 14:0/18:1 | C40H79NO8P |
| PC(32:0) | 734.5700 | 734.5701 | 0.1221 | a | C40H81NO8P |
| PC(34:5) | 752.5230 | 752.5205 | −3.3016 | 14:0/20:5 | C42H75NO8P |
| PC(34:4) | 754.5387 | 754.5394 | 0.9047 | 14:0/20:4 | C42H77NO8P |
| PC(34:3) | 756.5543 | 756.5535 | −1.0593 | 16:0/18:3 | C42H79NO8P |
| PC(34:2) | 758.5700 | 758.5674 | −3.4063 | 16:0/18:2 | C42H81NO8P |
| PC(34:1) | 760.5856 | 760.5849 | −0.9624 | 16:0/18:1 | C42H83NO8P |
| PC(36:8) | 774.5074 | 774.5039 | −4.519 | a | C44H73NO8P |
| PC(36:7) | 776.5230 | 776.5222 | −1.038 | b | C44H75NO8P |
| PC(36:6) | 778.5387 | 778.5372 | −1.9005 | 20:5/16:1; 18:3/18:3 | C44H77NO8P |
| PC(36:5) | 780.5543 | 780.554 | −0.4112 | 16:0/20:5; 18:3/18:2 | C44H79NO8P |
| PC(36:4) | 782.5700 | 782.5697 | −0.342 | 16:0/20:4; 18:2/18:2 | C44H81NO8P |
| PC(36:3) | 784.5856 | 784.5854 | −0.2952 | b | C44H83NO8P |
| PC(36:2) | 786.6013 | 786.6008 | −0.5978 | 18:1/18:1 | C44H85NO8P |
| PC(38:9) | 800.5230 | 800.5217 | −1.674 | b | C46H75NO8P |
| PC(38:8) | 802.5387 | 802.5376 | −1.2955 | 18:3/20:5 | C46H77NO8P |
| PC(38:7) | 804.5543 | 804.5540 | −0.4369 | 18:3/20:4 | C46H79NO8P |
| PC(38:6) | 806.5700 | 806.5695 | −0.627 | 18:1/20:5 | C46H81NO8P |
| PC(38:5) | 808.5856 | 808.5850 | −0.7921 | 18:1/20:4 | C46H83NO8P |
| PC(40:10) | 826.5387 | 826.5389 | 0.261 | 20:5/20:5 | C48H77NO8P |
| PC(40:9) | 828.5543 | 828.5542 | −0.2106 | 20:4/20:5 | C48H79NO8P |
| PC(40:8) | 830.5700 | 830.5672 | −3.4091 | 20:4/20:4 | C48H81NO8P |
| PC(40:7) | 832.5856 | 832.5820 | −4.4128 | b | C48H83NO8P |
| PC(42:11) | 852.5543 | 852.5510 | −3.9274 | a | C50H79NO8P |
| LPC identified as [M+H]+ | |||||
| LPC(14:0) | 468.3090 | 468.3093 | 0.5844 | 14:0 | C22H47NO7P |
| LPC(16:1) | 494.3247 | 494.3235 | −2.3082 | 16:1 | C24H49NO7P |
| LPC(16:0) | 496.3403 | 496.3400 | −0.7179 | 16:0 | C24H51NO7P |
| LPC(18:4) | 516.3090 | 516.3074 | −3.2093 | a | C26H47NO7P |
| LPC(18:3) | 518.3247 | 518.3232 | −2.843 | 18:3 | C26H49NO7P |
| LPC(18:2) | 520.3403 | 520.3407 | 0.7228 | 18:2 | C26H51NO7P |
| LPC(18:1) | 522.3560 | 522.3563 | 0.5418 | 18:1 | C26H53NO7P |
| LPC(20:5) | 542.3247 | 542.3244 | −0.5427 | 20:5 | C28H49NO7P |
| LPC(20:4) | 544.3403 | 544.3406 | 0.4892 | 20:4 | C28H51NO7P |
| LPC(20:3) | 546.3560 | 546.3540 | −3.5931 | 20:3 | C28H53NO7P |
| LPC(20:1) | 550.3873 | 550.3881 | 1.5545 | a | C28H57NO7P |
| LPC(22:6) | 568.3403 | 568.3397 | −1.1639 | a | C30H51NO7P |
| LPC(22:5) | 570.3560 | 570.3548 | −2.1309 | a | C30H53NO7P |
| PE identified as [M − H]− | |||||
| PE(30:2) | 658.4448 | 658.4464 | 2.4664 | a | C35H65NO8P |
| PE(30:1) | 660.4604 | 660.4620 | 2.4129 | 16:1/14:0 | C35H67NO8P |
| PE(30:0) | 662.4761 | 662.4762 | 0.0882 | a | C35H69NO8P |
| PE(32:3) | 684.4604 | 684.4622 | 2.6111 | a | C37H67NO8P |
| PE(32:2) | 686.4761 | 686.4774 | 1.9274 | 16:1/16:1 | C37H69O8NP |
| PE(32:1) | 688.4917 | 688.4932 | 2.1531 | 16:0/16:1 | C37H71NO8P |
| PE(34:3) | 712.4917 | 712.4934 | 2.3376 | 16:0/18:3 | C39H71O8NP |
| PE(34:2) | 714.5074 | 714.5087 | 1.8928 | 16:1/18:1;16:0/18:2 | C39H73NO8P |
| PE(34:1) | 716.5230 | 716.5231 | 0.1676 | 16:0/18:1 | C39H75NO8P |
| PE(36:5) | 736.4917 | 736.4928 | 1.4659 | 16:0/20:5 | C41H71O8NP |
| PE(36:2) | 742.5387 | 742.5422 | 4.6648 | 18:1/18:1 | C41H77O8NP |
| PE(38:8) | 758.4760 | 758.4780 | 2.641 | 20:5/18:3 | C43H69NO8P |
| PE(38:7) | 760.4920 | 760.4931 | 1.4905 | 20:5/18:2; 20:4/18:3 | C43H71NO8P |
| PE(38:5) | 764.5230 | 764.5230 | −0.0385 | a | C43H75NO8P |
| LPE identified as [M − H]− | |||||
| LPE(14:0) | 424.2464 | 424.2479 | 3.6359 | a | C19H39NO7P |
| LPE(16:1) | 450.2621 | 450.2634 | 2.9407 | 16:1 | C21H41NO7P |
| LPE(16:0) | 452.2777 | 452.2787 | 2.2511 | 16:0 | C21H43NO7P |
| LPE(18:3) | 474.2621 | 474.2631 | 2.3415 | 18:3 | C23H41NO7P |
| LPE(18:2) | 476.2777 | 476.2790 | 2.811 | 18:2 | C23H43NO7P |
| LPE(18:1) | 478.2934 | 478.2947 | 2.7404 | 18:1 | C23H45NO7P |
| LPE(18:0) | 480.3090 | 480.3109 | 3.961 | a | C23H47NO7P |
| LPE(20:5) | 498.2621 | 498.2634 | 2.6089 | a | C25H41NO7P |
| LPE(20:4) | 500.2777 | 500.2793 | 3.2658 | 20:4 | C25H43NO7P |
| PG identified as [M − H]− | |||||
| PG(30:1) | 691.4550 | 691.4562 | 1.7284 | 16:1/14:0 | C36H68O10P |
| PG(30:0) | 693.4707 | 693.4725 | 2.5772 | a | C36H70O10P |
| PG(32:2) | 717.4707 | 717.4725 | 2.4841 | 16:1/16:1; 18:2/14:0 | C38H70O10P |
| PG(32:1) | 719.4863 | 719.4864 | 0.2257 | 16:0/16:1 | C38H72O10P |
| PG(32:0) | 721.5020 | 721.5036 | 2.2107 | a | C38H74O10P |
| PG(34:4) | 741.4707 | 741.4706 | −0.1774 | 20:4/14:0; 16:1/18:3 | C40H70O10P |
| PG(34:3) | 743.4863 | 743.4854 | −1.1754 | 18:3/16:0 | C40H72O10P |
| PG(34:2) | 745.5020 | 745.5046 | 3.4986 | 18:1/16:1 | C40H74O10P |
| PG(34:1) | 747.5176 | 747.5189 | 1.6913 | 18:1/16:0 | C40H76O10P |
| PG(36:5) | 767.4863 | 767.4878 | 1.9572 | 20:5/16:0; 20:4/16:1 | C42H72O10P |
| PG(36:5)OH | 783.4829 | 783.4833 | 0.4648 | 20:5/16:0−OH | C42H72O10PO |
| PG(36:4) | 769.5020 | 769.5039 | 2.4166 | 20:4/ 16:0; 18:2/18:2 | C42H74O10P |
| PG(36:3) | 771.5176 | 771.5181 | 0.6347 | 18:2/18:1 | C42H76O10P |
| PG(36:2) | 773.5333 | 773.5346 | 1.6895 | 18:1/18:1 | C42H78O10P |
| PG(42:9) | 843.5176 | 843.5169 | −0.8453 | a | C48H76O10P |
| LPG identified as [M − H]− | |||||
| LPG(14:0) | 455.2410 | 455.2421 | 2.5057 | 14:0 | C20H40O9P |
| LPG(16:1) | 481.2566 | 481.2576 | 2.0266 | 16:1 | C22H42O9P |
| LPG(16:0) | 483.2723 | 483.2734 | 2.2252 | 16:0 | C22H44O9P |
| LPG(16:0)OH | 499.2677 | 499.2678 | 0.0841 | 16:0−OH | C22H44O10P |
| LPG(18:2) | 507.2723 | 507.2740 | 3.1604 | a | C24H44O9P |
| LPG(18:1) | 509.2879 | 509.2888 | 1.649 | 18:1 | C24H46O9P |
| LPG(20:5) | 529.2566 | 529.2577 | 1.9811 | 20:5 | C26H42O9P |
| LPG(20:4) | 531.2723 | 531.2744 | 4.0749 | 20:4 | C26H44O9P |
| LPG(22:5) | 557.2879 | 557.2877 | −0.3698 | a | C28H46O9P |
| PA identified as [M − H]− | |||||
| PA(36:7) | 689.4182 | 689.4178 | −0.6446 | a | C39H62O8P |
| PA(36:6) | 691.4339 | 691.4359 | 2.941 | 16:1/20:5 | C39H64O8P |
| PA(36:5) | 693.4495 | 693.4508 | 1.8144 | 16:0/20:5 | C39H66O8P |
| PA(40:10) | 739.4339 | 739.4356 | 2.3648 | a | C43H64O8P |
| PA(40:9) | 741.4495 | 741.4496 | 0.0455 | 20:5/20:4 | C43H66O8P |
| PA(40:8) | 743.4652 | 743.4660 | 1.1816 | 20:4/20:4 | C43H68O8P |
| LPA identified as [M − H]− | |||||
| LPA(20:4) | 457.2355 | 457.2362 | 1.5001 | a | C23H38O7P |
| PI identified as [M − H]− | |||||
| PI(34:3) | 831.5024 | 831.5030 | 0.7891 | a | C43H76O13P |
| PI(34:1) | 835.5337 | 835.5307 | −3.5756 | 16:0/18:1 | C43H80O13P |
| PI(38:5) | 883.5337 | 883.5337 | 0.0086 | a | C47H80O13P |
| PI(40:10) | 901.4867 | 901.4870 | 0.2814 | a | C49H74O13P |
| PI(42:0) | 949.6745 | 949.6721 | −2.567 | a | C51H98O13P |
| LPI identified as [M − H]− | |||||
| LPI(16:0) | 571.2883 | 571.2897 | 2.3400 | 16:0 | C25H48O12P |
| PI-Cer identified as [M − H]− | |||||
| PI−Cer(t38:0h) | 868.5915 | 868.5901 | −1.6315 | a | C44H87NO13P |
| PI−Cer(t40:1h) | 894.6077 | 894.6082 | 0.6170 | b | C46H89NO13P |
| PI−Cer(t40:0h) | 896.6228 | 896.6218 | −1.0707 | a | C46H91NO13P |
| PI−Cer(t42:0h) | 924.6541 | 924.6509 | −3.5109 | a | C48H95NO13P |
| PI−Cer(t42:1h) | 922.6390 | 922.6378 | −1.2553 | b | C48H93NO13P |
| PI−Cer(t42:2h) | 920.6233 | 920.6240 | 0.8190 | t18:1/h24:1 | C48H91NO13P |
| PI−Cer(t43:2h) | 934.6390 | 934.6403 | 1.3860 | b | C49H93NO13P |
| PI−Cer(t44:2h) | 948.6546 | 948.6553 | 0.7034 | b | C50H95NO13P |
| Fatty Acid | Mean ± SD Mean | ||
|---|---|---|---|
| (µg mg−1 Extract) | (µg g−1 DW Biomass) | (Relative Percentage, %) | |
| C14:0 | 5.48 ± 0.75 | 57.11 ± 1.37 | 2.00 ± 0.07 |
| C15:0 | 4.62 ± 0.21 | 48.41 ± 3.73 | 1.70 ± 0.13 |
| C16:0 | 60.48 ± 10.44 | 628.89 ± 47.70 | 22.03 ± 0.98 |
| C16:1n-9 | 4.28 ± 0.21 | 44.89 ± 3.43 | 1.58 ± 0.14 |
| C16:1n-7 | 6.7 ± 0.50 | 70.12 ± 4.23 | 2.46 ± 0.12 |
| C16:2n-6 | 4.37 ± 0.13 | 45.94 ± 4.81 | 1.61 ± 0.15 |
| C17:0 | 3.78 ± 0.02 | 39.74 ± 4.75 | 1.39 ± 0.16 |
| C17:1 | 4.83 ± 0.25 | 50.64 ± 3.88 | 1.78 ± 0.12 |
| C18:0 | 15.02 ± 3.77 | 155.51 ± 23.29 | 5.44 ± 0.66 |
| C18:1n-9 | 20.3 ± 1.94 | 212.35 ± 13.59 | 7.44 ± 0.24 |
| C18:1n-7 | 11.2 ± 0.71 | 117.36 ± 8.03 | 4.12 ± 0.24 |
| C18:2n-6 | 10.85 ± 0.78 | 113.8 ± 11.22 | 3.99 ± 0.28 |
| C18:3n-6 | 5.79 ± 0.20 | 60.79 ± 5.51 | 2.13 ± 0.18 |
| C18:3n-3 | 5.15 ± 0.07 | 54.12 ± 6.05 | 1.90 ± 0.22 |
| C18:4n-3 | 5.14 ± 0.07 | 54.07 ± 5.92 | 1.90 ± 0.20 |
| C20:0 | 6.22 ± 0.04 | 65.46 ± 8.23 | 2.30 ± 0.28 |
| C20:3n-6 | 6.73 ± 0.29 | 70.58 ± 5.86 | 2.48 ± 0.19 |
| C20:4n-6 | 35.41 ± 5.76 | 368.19 ± 20.56 | 12.91 ± 0.56 |
| C20:5n-3 | 57.18 ± 8.74 | 595.19 ± 32.17 | 20.86 ± 0.56 |
| Sum | 273.52 ± 34.49 | 2853.16 ± 139.33 | |
| SFA | 95.6 ± 15.09 | 995.12 ± 66.55 | 37.49 ± 5.91 |
| MUFA | 42.48 ± 3.28 | 444.72 ± 27.65 | 16.66 ± 1.29 |
| PUFA | 126.24 ± 15.77 | 1316.74 ± 57.07 | 49.51 ± 6.18 |
| Σ PUFA/Σ SFA | 1.32 ± 0.04 | ||
| Σ n-6 | 58.77 ± 6.93 | 613.35 ± 27.28 | |
| Σ n-3 | 67.47 ± 8.84 | 703.39 ± 30.47 | |
| n-6/n-3 | 0.87 ± 0.01 | ||
| NVI | 0.77 ± 0.03 | ||
| EPA/AA | 1.62 ± 0.02 | ||
| AI | 0.49 ± 0.02 | ||
| TI | 0.32 ± 0.02 | ||
| h/H | 2.4 ± 0.12 | ||
| PI, % | 218.42 ± 30.47 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, E.; Melo, T.; Reis, M.; Domingues, P.; Calado, R.; Abreu, M.H.; Domingues, M.R. Polar Lipids Composition, Antioxidant and Anti-Inflammatory Activities of the Atlantic Red Seaweed Grateloupia turuturu. Mar. Drugs 2021, 19, 414. https://doi.org/10.3390/md19080414
da Costa E, Melo T, Reis M, Domingues P, Calado R, Abreu MH, Domingues MR. Polar Lipids Composition, Antioxidant and Anti-Inflammatory Activities of the Atlantic Red Seaweed Grateloupia turuturu. Marine Drugs. 2021; 19(8):414. https://doi.org/10.3390/md19080414
Chicago/Turabian Styleda Costa, Elisabete, Tânia Melo, Mariana Reis, Pedro Domingues, Ricardo Calado, Maria Helena Abreu, and Maria Rosário Domingues. 2021. "Polar Lipids Composition, Antioxidant and Anti-Inflammatory Activities of the Atlantic Red Seaweed Grateloupia turuturu" Marine Drugs 19, no. 8: 414. https://doi.org/10.3390/md19080414
APA Styleda Costa, E., Melo, T., Reis, M., Domingues, P., Calado, R., Abreu, M. H., & Domingues, M. R. (2021). Polar Lipids Composition, Antioxidant and Anti-Inflammatory Activities of the Atlantic Red Seaweed Grateloupia turuturu. Marine Drugs, 19(8), 414. https://doi.org/10.3390/md19080414








