Deep-Sea Coral Garden Invertebrates and Their Associated Fungi Are Genetic Resources for Chronic Disease Drug Discovery
Abstract
1. Introduction
2. Results
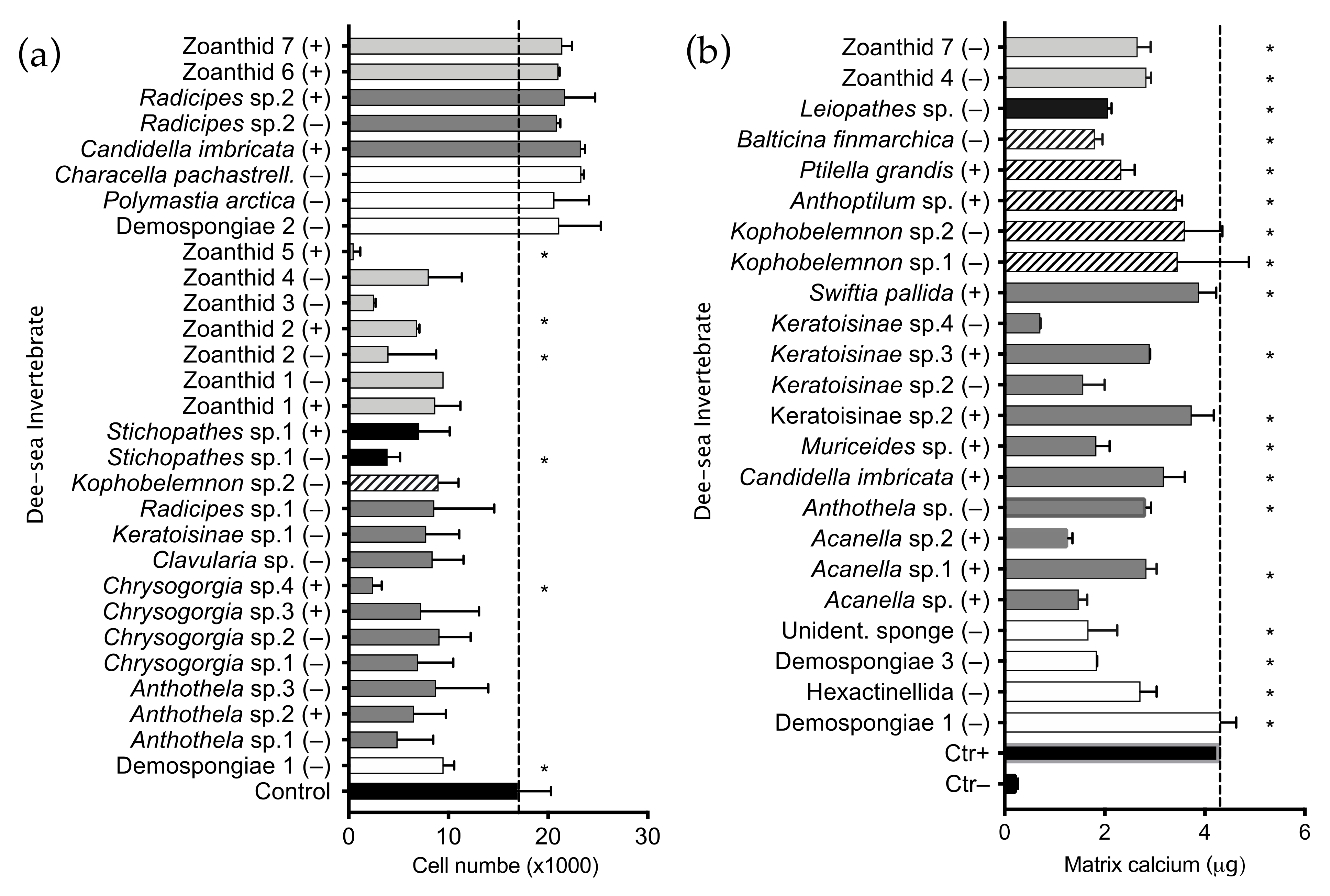
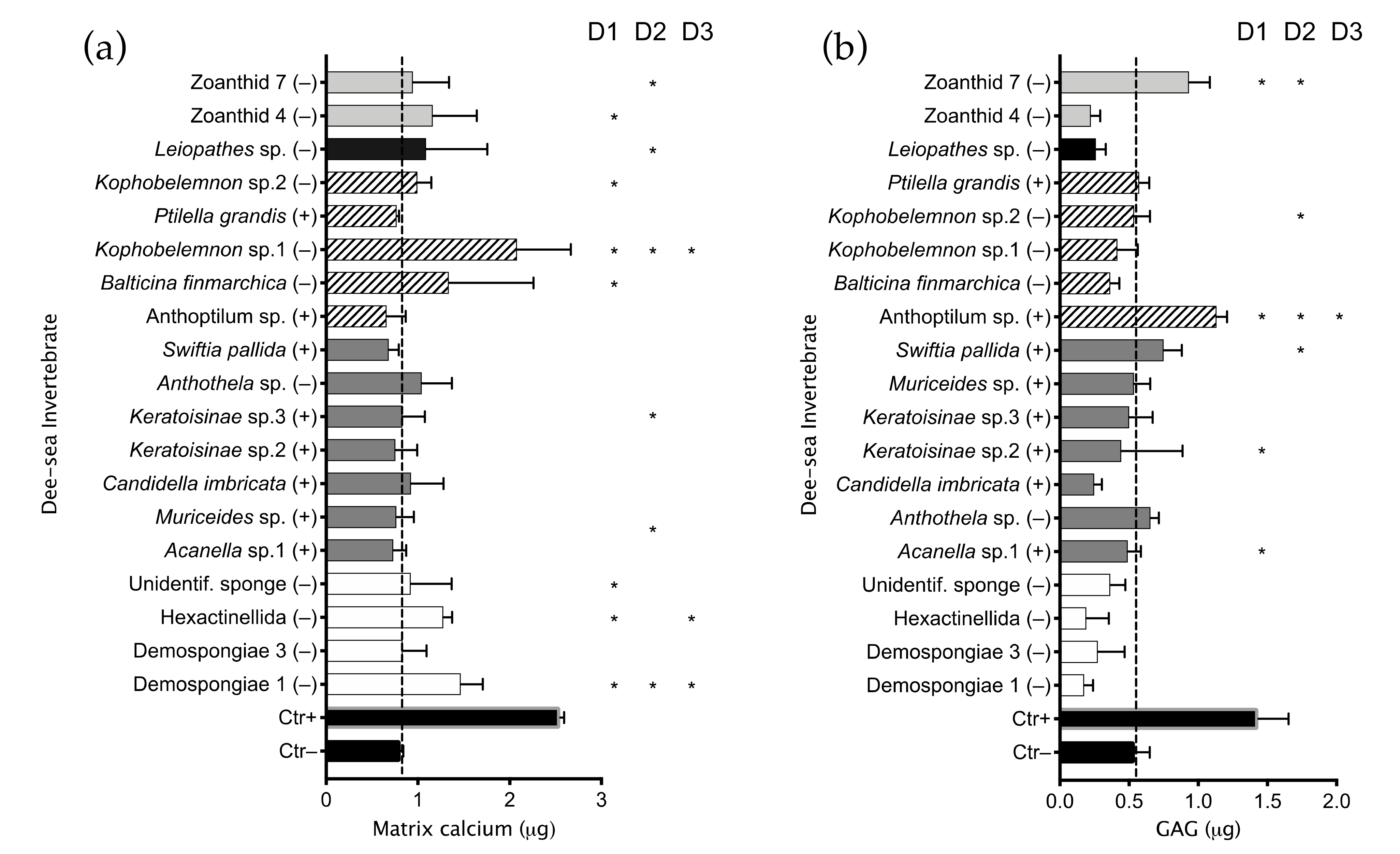
2.1. Invertebrate Extract Bioactivity toward hMSCs
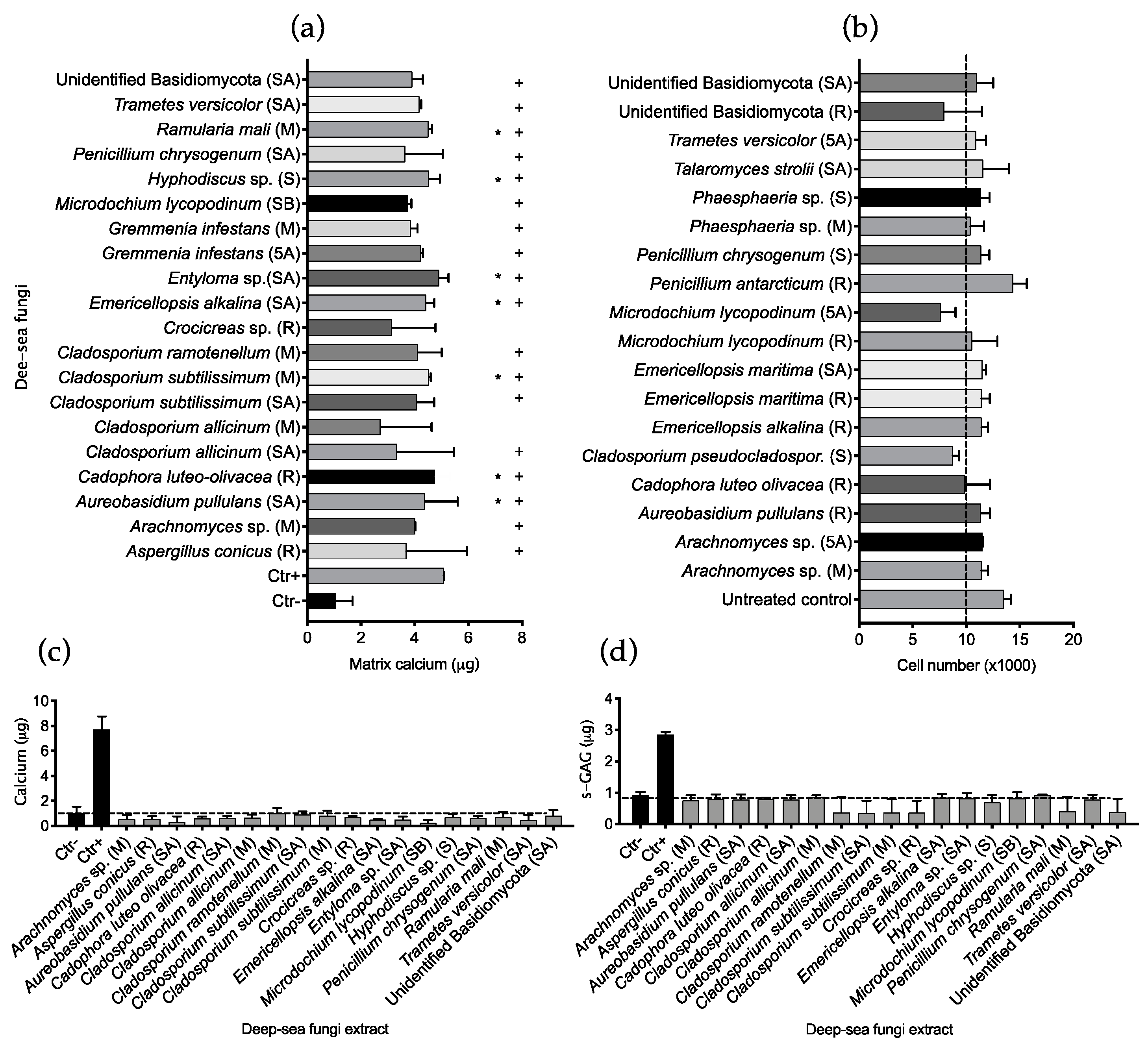
2.2. Fungal Extract Bioactivity toward hMSCs
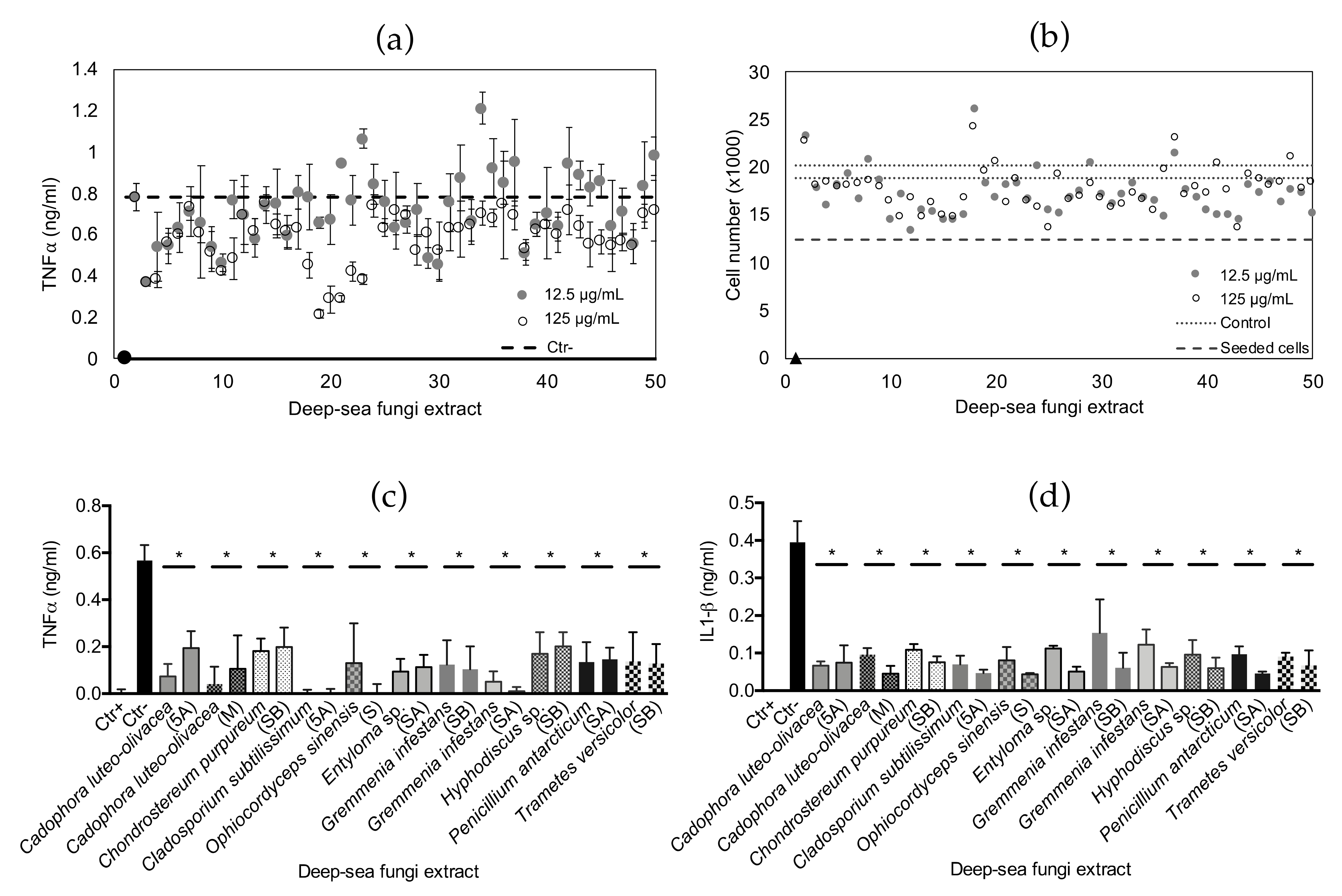
2.3. Anti-Inflammatory Bioactivity
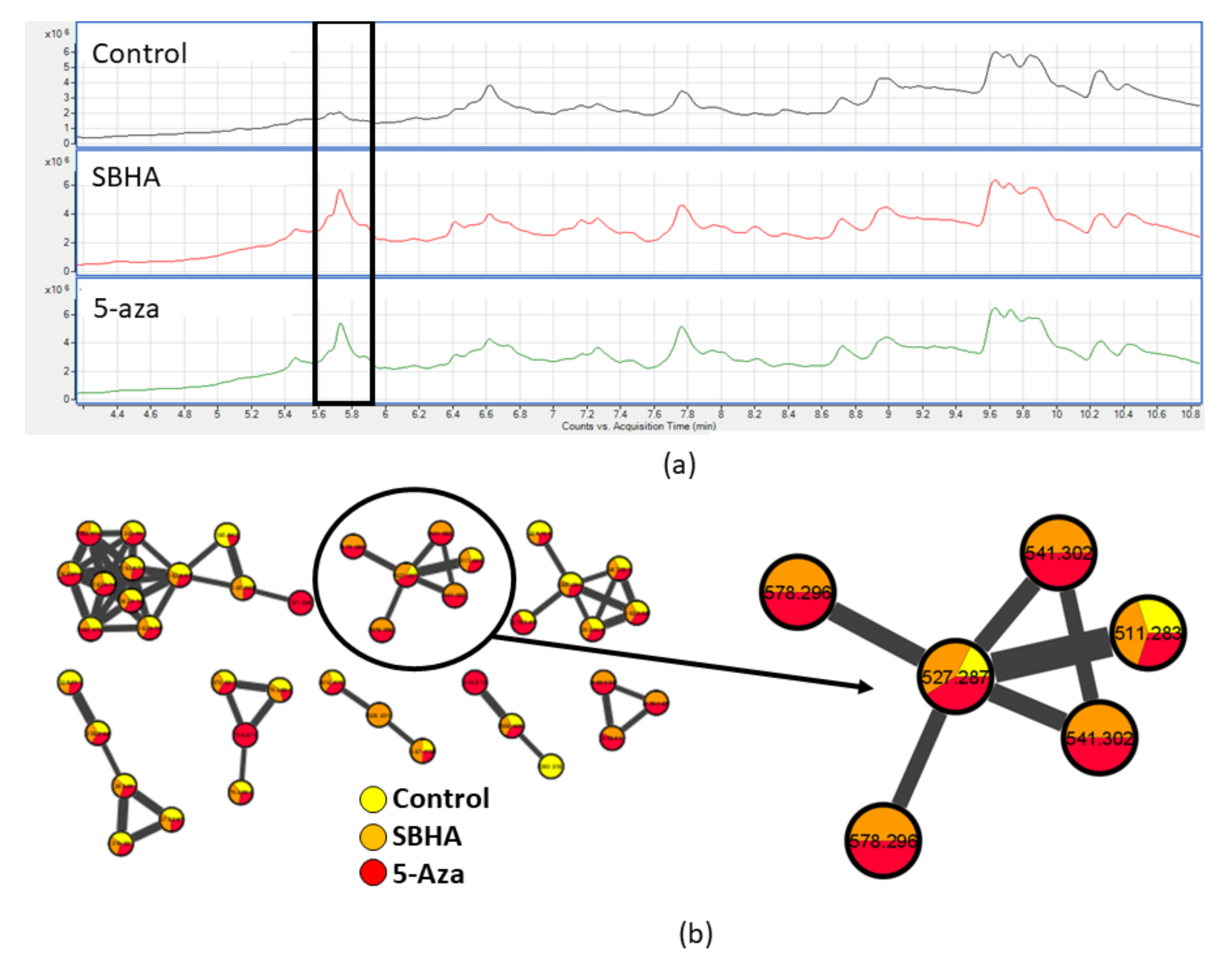
3. Discussion
4. Materials and Methods
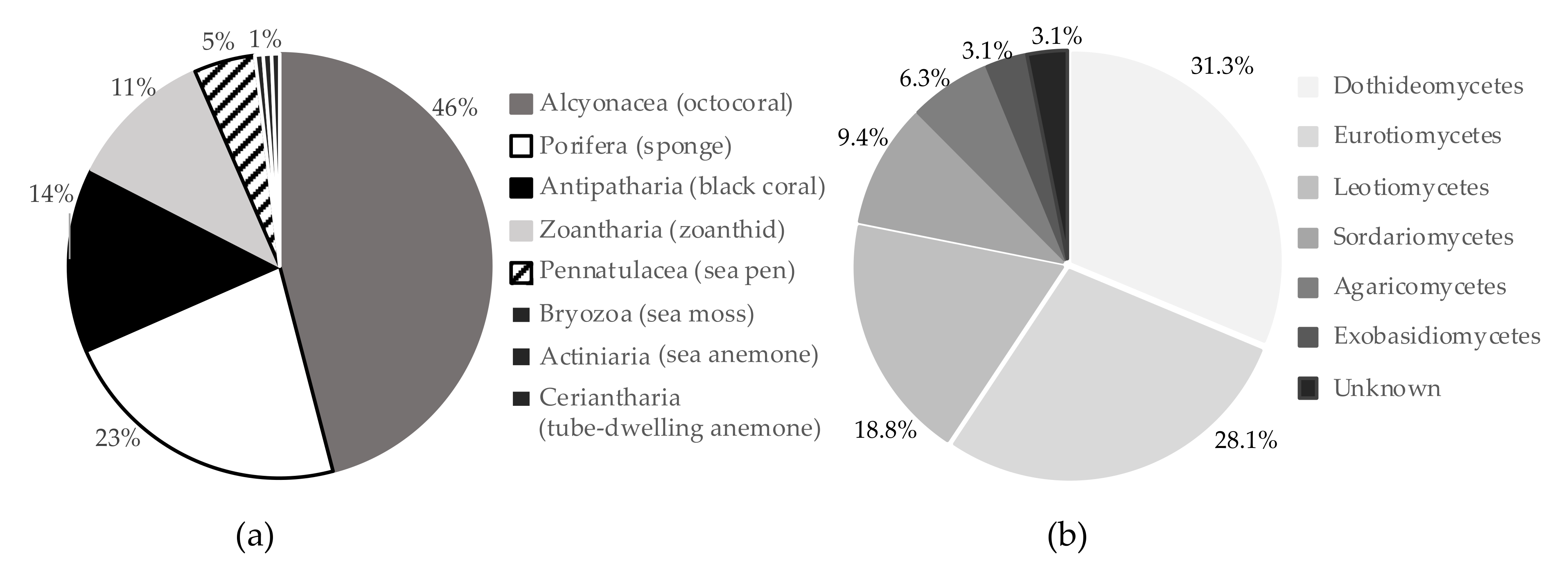
4.1. Deep-Sea Organism Collection
4.2. Deep-Sea Benthic Invertebrate Metabolite Extraction
4.3. Deep-Sea Fungal Culture and Metabolite Extraction
4.4. Culture of Human Mesenchymal Stem Cells and THP1 Macrophages
4.5. Marine Extract Library Screening
4.6. High-Throughput Osteogenic Assay
4.7. Chondrogenic Assay
4.8. High-Throughput Anti-Inflammatory Assay
4.9. LC-MS Analysis of Extracts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Barry, F.; Boynton, R.E.; Liu, B.; Murphy, M. Chondrogenic Differentiation of Mesenchymal Stem Cells from Bone Marrow: Differentiation-Dependent Gene Expression of Matrix Components. Exp. Cell Res. 2001, 268, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Sacchetti, B.; Funari, A.; Michienzi, S.; Di Cesare, S.; Piersanti, S.; Saggio, I.; Tagliafico, E.; Ferrari, S.; Robey, P.; Riminucci, M.; et al. Self-Renewing Osteoprogenitors in Bone Marrow Sinusoids Can Organize a Hematopoietic Microenvironment. Cell 2007, 131, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Friendenstein, A.J.; Chailakhyan, R.K.; Gerasimov, U.V. Bone marrow osteogenic stem cells. In Calcium Regulation and Bone Metabolism. Basic and Clinical Aspects; Cohn, D.V., Glorieux, F.H., Martin, T.J., Eds.; Excerpta Medica: Amsterdam, The Netherlands, 1990; Volume 10, pp. 19–29. [Google Scholar]
- Somoza, R.; Welter, J.F.; Correa, D.; Caplan, A.I. Chondrogenic Differentiation of Mesenchymal Stem Cells: Challenges and Unfulfilled Expectations. Tissue Eng. Part B Rev. 2014, 20, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.; Walsh, P.; Maggs, C.; Buchanan, F. Designs from the deep: Marine organisms for bone tissue engineering. Biotechnol. Adv. 2011, 29, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Haynesworth, S.E.; Caplan, A.I.; Bruder, S.P. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J. Cell. Biochem. 1997, 64, 295–312. [Google Scholar] [CrossRef]
- Friedman, M.S.; Long, M.W.; Hankenson, K.D. Osteogenic differentiation of human mesenchymal stem cells is regulated by bone morphogenetic protein-6. J. Cell. Biochem. 2006, 98, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Fiorentini, E.; Granchi, D.; Leonardi, E.; Baldini, N.; Ciapetti, G. Effects of osteogenic differentiation inducers on in vitro ex-panded adult mesenchymal stromal cells. Int. J. Artif. Organs 2011, 34, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, B.; Hering, T.M.; Caplan, A.; Goldberg, V.M.; Yoo, J.U. In Vitro Chondrogenesis of Bone Marrow-Derived Mesenchymal Progenitor Cells. Exp. Cell Res. 1998, 238, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Crecente-Campo, J.; Borrajo, E.; Vidal, A.; Garcia-Fuentes, M. New scaffolds encapsulating TGF-beta3/BMP-7 combinations driving strong chondrogenic differentiation. Eur. J. Pharm. Biopharm. 2017, 114, 69–78. [Google Scholar] [CrossRef]
- Friedlaender, G.E.; Perry, C.R.; Cole, J.D.; Cook, S.D.; Cierny, G.; Muschler, G.F.; Zych, G.A.; Calhoun, J.H.; LaForte, A.J.; Yin, S. Osteogenic Protein-1 (Bone Morphogenetic Protein-7) in the Treatment of Tibial Nonunions: A Prospective, Random-ized Clinical Trial Comparing rhOP-1 with Fresh Bone Autograft. J. Bone Jt. Surg. 2001, 83-A, 151–158. [Google Scholar] [CrossRef]
- Govender, S.; Csimma, C.; Genant, H.K.; Valentin-Opran, A.; Amit, Y.; Arbel, R.; Aro, H.; Atar, D.; Bishay, M.; Börner, M.G.; et al. Recombinant human bone morphogenetic protein-2 for treatment of open tibial fractures: A prospective, controlled, randomized study of four hundred and fifty patients. J. Bone Jt. Surg. 2002, 84, 2123–2134. [Google Scholar] [CrossRef]
- Blaney Davidson, E.N.; Vitters, E.L.; van Beuningen, H.M.; van de Loo, F.A.; van den Berg, W.B.; van der Kraan, P.M. Re-semblance of osteophytes in experimental osteoarthritis to transforming growth factor beta-induced osteophytes: Limited role of bone morphogenetic protein in early osteoarthritic osteophyte formation. Arthritis Rheum. 2007, 56, 4065–4073. [Google Scholar] [CrossRef]
- Walsh, S.; Jordan, G.R.; Jefferiss, C.; Stewart, K.; Beresford, J.N. High concentrations of dexamethasone suppress the prolifer-ation but not the differentiation or further maturation of human osteoblast precursors in vitro: Relevance to glucocorti-coid-induced osteoporosis. Rheumatology 2001, 40, 70–83. [Google Scholar] [CrossRef]
- Goldring, S.R.; Goldring, M. The Role of Cytokines in Cartilage Matrix Degeneration in Osteoarthritis. Clin. Orthop. Relat. Res. 2004, 427, S27–S36. [Google Scholar] [CrossRef]
- Blom, A.B.; Van Der Kraan, P.M.; Van Den Berg, W.B. Cytokine Targeting in Osteoarthritis. Curr. Drug Targets 2007, 8, 283–292. [Google Scholar] [CrossRef]
- Doherty, T.M.; Asotra, K.; Fitzpatrick, L.A.; Qiao, J.-H.; Wilkin, D.J.; Detrano, R.C.; Dunstan, C.; Shah, P.K.; Rajavashisth, T.B. Calcification in atherosclerosis: Bone biology and chronic inflammation at the arterial crossroads. Proc. Natl. Acad. Sci. USA 2003, 100, 11201–11206. [Google Scholar] [CrossRef]
- Leszczynska, A.; O’Doherty, A.; Farrell, E.; Pindjakova, J.; O’Brien, F.J.; O’Brien, T.; Barry, F.; Murphy, J.M. Differentiation of Vascular Stem Cells Contributes to Ectopic Calcification of Atherosclerotic Plaque. Stem Cells 2016, 34, 913–923. [Google Scholar] [CrossRef]
- Goldring, M.B. Anticytokine therapy for osteoarthritis. Expert Opin. Biol. Ther. 2001, 1, 817–829. [Google Scholar] [CrossRef]
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nat. Cell Biol. 2001, 409, 1092–1101. [Google Scholar] [CrossRef]
- Wright, P.C.; Westacott, R.E.; Burja, A.M. Piezotolerance as a metabolic engineering tool for the biosynthesis of natural products. Biomol. Eng. 2003, 20, 325–331. [Google Scholar] [CrossRef]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef]
- Schupp, P.J.; Kohlert-Schupp, C.; Whitefield, S.; Engemann, A.; Rohde, S.; Hemscheidt, T.; Pezzuto, J.M.; Kondratyuk, T.P.; Park, E.J.; Marler, L.; et al. Cancer Chemopreventive and Anticancer Evaluation of Extracts and Fractions from Marine Macro-and Micro-organisms Collected from Twilight Zone Waters Around Guam. Nat. Prod. Commun. 2009, 4, 1717. [Google Scholar]
- Dumdei, E.J.; Blunt, J.W.; Munro, M.H.G.; Pannell, L.K. Isolation of Calyculins, Calyculinamides, and Swinholide H from the New Zealand Deep-Water Marine Sponge Lamellomorpha strongylata. J. Org. Chem. 1997, 62, 2636–2639. [Google Scholar] [CrossRef]
- Pilkington, L.I. A Chemometric Analysis of Deep-Sea Natural Products. Molecules 2019, 24, 3942. [Google Scholar] [CrossRef]
- Thao, N.P.; Luyen, B.T.T.; Lee, S.H.; Jang, H.D.; Kiem, P.V.; Minh, C.V.; Kim, Y.H. Antiosteoporotic and antioxidant activi-ties of diterpenoids from the Vietnamese soft corals Sinularia maxima and Lobophytum crassum. Med. Chem. Res. 2015, 24, 3551–3560. [Google Scholar] [CrossRef]
- Allemand, D.; Tambutté, É.; Zoccola, D.; Tambutté, S. Coral Calcification, Cells to Reefs. In Coral Reefs: An Ecosystem in Transition; Springer: Berlin/Heidelberg, Germany, 2011; pp. 119–150. [Google Scholar]
- Gentili, C.; Cancedda, R. Cartilage and Bone Extracellular Matrix. Curr. Pharm. Design 2009, 15, 1334–1348. [Google Scholar] [CrossRef]
- Zoccola, D.; Moya, A.; Beranger, G.E.; Tambutte, E.; Allemand, D.; Carle, G.F.; Tambutte, S. Specific expression of BMP2/4 ortholog in biomineralizing tissues of corals and action on mouse BMP receptor. Mar. Biotechnol. 2009, 11, 260–269. [Google Scholar] [CrossRef]
- Kim, H.K.; Cho, S.G.; Kim, J.H.; Doan, T.K.P.; Hu, Q.S.; Ulhaq, R.; Song, E.K.; Yoon, T.R. Mevinolin enhances osteogenic genes (ALP, type I collagen and osteocalcin), CD44, CD47 and CD51 expression during osteogenic differentiation. Life Sci. 2009, 84, 290–295. [Google Scholar] [CrossRef]
- Xu, J.; Yi, M.; Ding, L.; He, S. A Review of Anti-Inflammatory Compounds from Marine Fungi, 2000–2018. Mar. Drugs 2019, 17, 636. [Google Scholar] [CrossRef]
- Marchese, P.; Mahajan, N.; O’Connell, E.; Fearnhead, H.; Tuohy, M.; Krawczyk, J.; Thomas, O.P.; Barry, F.; Murphy, M.J. A Novel High-Throughput Screening Platform Identifies Itaconate Derivatives from Marine Penicillium antarcticum as Inhibi-tors of Mesenchymal Stem Cell Differentiation. Mar. Drugs 2020, 18, 192. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Llodra, E.; Brandt, A.; Danovaro, R.; De Mol, B.; Escobar, E.; German, C.R.; Levin, L.A.; Martinez Arbizu, P.; Me-not, L.; Buhl-Mortensen, P.; et al. Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Harden-Davies, H. Deep-sea genetic resources: New frontiers for science and stewardship in areas beyond national jurisdic-tion. Deep Sea Res. Part II Top. Stud. Oceanogr. 2017, 137, 504–513. [Google Scholar] [CrossRef]
- Macreadie, P.I.; McLean, D.; Thomson, P.G.; Partridge, J.C.; Jones, D.O.; Gates, A.; Benfield, M.C.; Collin, S.P.; Booth, D.; Smith, L.L.; et al. Eyes in the sea: Unlocking the mysteries of the ocean using industrial, remotely operated vehicles (ROVs). Sci. Total. Environ. 2018, 634, 1077–1091. [Google Scholar] [CrossRef]
- Bullimore, R.D.; Foster, N.L.; Howell, K.L. Coral-characterized benthic assemblages of the deep Northeast Atlantic: Defining “Coral Gardens” to support future habitat mapping efforts. ICES J. Mar. Sci. 2013, 70, 511–522. [Google Scholar] [CrossRef][Green Version]
- Marchese, P.; Garzoli, L.; Young, R.; Allcock, L.; Barry, F.; Tuohy, M.; Murphy, M. Fungi populate deep-sea coral gardens as well as marine sediments in the Irish Atlantic Ocean. Environ. Microbiol. 2021. [Google Scholar] [CrossRef]
- Mancuso, P.; Raman, S.; Glynn, A.; Barry, F.; Murphy, J.M. Mesenchymal Stem Cell Therapy for Osteoarthritis: The Critical Role of the Cell Secretome. Front. Bioeng. Biotechnol. 2019, 7, 9. [Google Scholar] [CrossRef]
- Demers, D.H.; Knestrick, M.A.; Fleeman, R.; Tawfik, R.; Azhari, A.; Souza, A.; Vesely, B.; Netherton, M.; Gupta, R.; Colon, B.; et al. Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery. Mar. Drugs 2018, 16, 376. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzat-to-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecu-lar Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Brey, D.M.; Motlekar, N.A.; Diamond, S.L.; Mauck, R.L.; Garino, J.P.; Burdick, J.A. High-throughput screening of a small molecule library for promoters and inhibitors of mesenchymal stem cell osteogenic differentiation. Biotechnol. Bioeng. 2011, 108, 163–174. [Google Scholar] [CrossRef]
- Alves, H.; Dechering, K.; Van Blitterswijk, C.; De Boer, J. High-throughput assay for the identification of compounds regu-lating osteogenic differentiation of human mesenchymal stromal cells. PLoS ONE 2011, 6, e26678. [Google Scholar] [CrossRef]
- Roberts, J.M.; Murray, F.; Anagnostou, E.; Hennige, S.; Gori, A.; Henry, L.-A.; Fox, A.; Kamenos, N.; Foster, G.L. Cold-Water Corals in an Era of Rapid Global Change: Are These the Deep Ocean’s Most Vulnerable Ecosystems? In The Cnidaria, Past, Present and Future; Springer: Berlin/Heidelberg, Germany, 2016; pp. 593–606. [Google Scholar]
- Fernandez-Arcaya, U.; Ramirez-Llodra, E.; Aguzzi, J.; Allcock, A.L.; Davies, J.S.; Dissanayake, A.; Harris, P.; Howell, K.; Huvenne, V.A.I.; Macmillan-Lawler, M.; et al. Ecological Role of Submarine Canyons and Need for Canyon Conservation: A Review. Front. Mar. Sci. 2017, 4, 4. [Google Scholar] [CrossRef]
- Daly, E.; Johnson, M.P.; Wilson, A.M.; Gerritsen, H.D.; Kiriakoulakis, K.; Allcock, A.L.; White, M. Bottom trawling at Whit-tard Canyon: Evidence for seabed modification, trawl plumes and food source heterogeneity. Prog. Oceanogr. 2018, 169, 227–240. [Google Scholar] [CrossRef]
| Organism | Polarity | Bioactivity | ||||
|---|---|---|---|---|---|---|
| Pro-Osteogenic | Pro-Chondrogenic | Proliferative | Anti-prolif. | Cytotoxic | ||
| Acanella sp.1 | + | x | ||||
| Anthoptilum sp. | + | x | ||||
| Anthothela sp. | − | x | ||||
| Anthothela sp.1 | − | x | ||||
| Anthothela sp.2 | + | x | ||||
| Anthothela sp.3 | − | x | ||||
| Balticina finmarchica | − | x | ||||
| Candidella imbricata | + | x | ||||
| Characella pachastrell. | − | x | ||||
| Chrysogorgia sp.1 | − | x | ||||
| Chrysogorgia sp.2 | − | x | ||||
| Chrysogorgia sp.3 | + | x | ||||
| Chrysogorgia sp.4 | + | x | ||||
| Clavularia sp. | − | x | ||||
| Demospongiae 1 | − | x | ||||
| Demospongiae 2 | − | x | ||||
| Hexactinellida | − | x | ||||
| Keratoisinae sp.1 | − | x | ||||
| Keratoisinae sp.2 | + | x | ||||
| Keratoisinae sp.3 | + | x | ||||
| Kophobelemnon sp.1 | − | x | ||||
| Kophobelemnon sp.2 | − | x | x | |||
| Leiopathes sp. | − | x | ||||
| Muriceides sp. | + | x | ||||
| Polymastia arctica | − | x | ||||
| Radicipes sp.1 | − | x | ||||
| Radicipes sp.2 | − | x | ||||
| Radicipes sp.2 | + | x | ||||
| Stichopathes sp.1 | − | x | ||||
| Stichopathes sp.1 | + | x | ||||
| Swiftia pallida | + | x | ||||
| Unidentif. sponge | − | x | ||||
| Zoanthid 1 | − | x | ||||
| Zoanthid 1 | + | x | ||||
| Zoanthid 2 | − | x | ||||
| Zoanthid 2 | + | x | ||||
| Zoanthid 3 | − | x | x | |||
| Zoanthid 4 | − | x | x | |||
| Zoanthid 5 | + | x | ||||
| Zoanthid 6 | + | x | ||||
| Zoanthid 7 | − | x | x | |||
| Zoanthid 7 | + | x | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchese, P.; Young, R.; O’Connell, E.; Afoullouss, S.; Baker, B.J.; Allcock, A.L.; Barry, F.; Murphy, J.M. Deep-Sea Coral Garden Invertebrates and Their Associated Fungi Are Genetic Resources for Chronic Disease Drug Discovery. Mar. Drugs 2021, 19, 390. https://doi.org/10.3390/md19070390
Marchese P, Young R, O’Connell E, Afoullouss S, Baker BJ, Allcock AL, Barry F, Murphy JM. Deep-Sea Coral Garden Invertebrates and Their Associated Fungi Are Genetic Resources for Chronic Disease Drug Discovery. Marine Drugs. 2021; 19(7):390. https://doi.org/10.3390/md19070390
Chicago/Turabian StyleMarchese, Pietro, Ryan Young, Enda O’Connell, Sam Afoullouss, Bill J. Baker, A. Louise Allcock, Frank Barry, and J. Mary Murphy. 2021. "Deep-Sea Coral Garden Invertebrates and Their Associated Fungi Are Genetic Resources for Chronic Disease Drug Discovery" Marine Drugs 19, no. 7: 390. https://doi.org/10.3390/md19070390
APA StyleMarchese, P., Young, R., O’Connell, E., Afoullouss, S., Baker, B. J., Allcock, A. L., Barry, F., & Murphy, J. M. (2021). Deep-Sea Coral Garden Invertebrates and Their Associated Fungi Are Genetic Resources for Chronic Disease Drug Discovery. Marine Drugs, 19(7), 390. https://doi.org/10.3390/md19070390







