Coupling Mass Spectral and Genomic Information to Improve Bacterial Natural Product Discovery Workflows
Abstract
1. Introduction
2. Concepts and Examples for Linking Genomic and Metabolomic Data
2.1. Experiment-Guided Genome Mining: Peptidogenomics and Glycogenomics
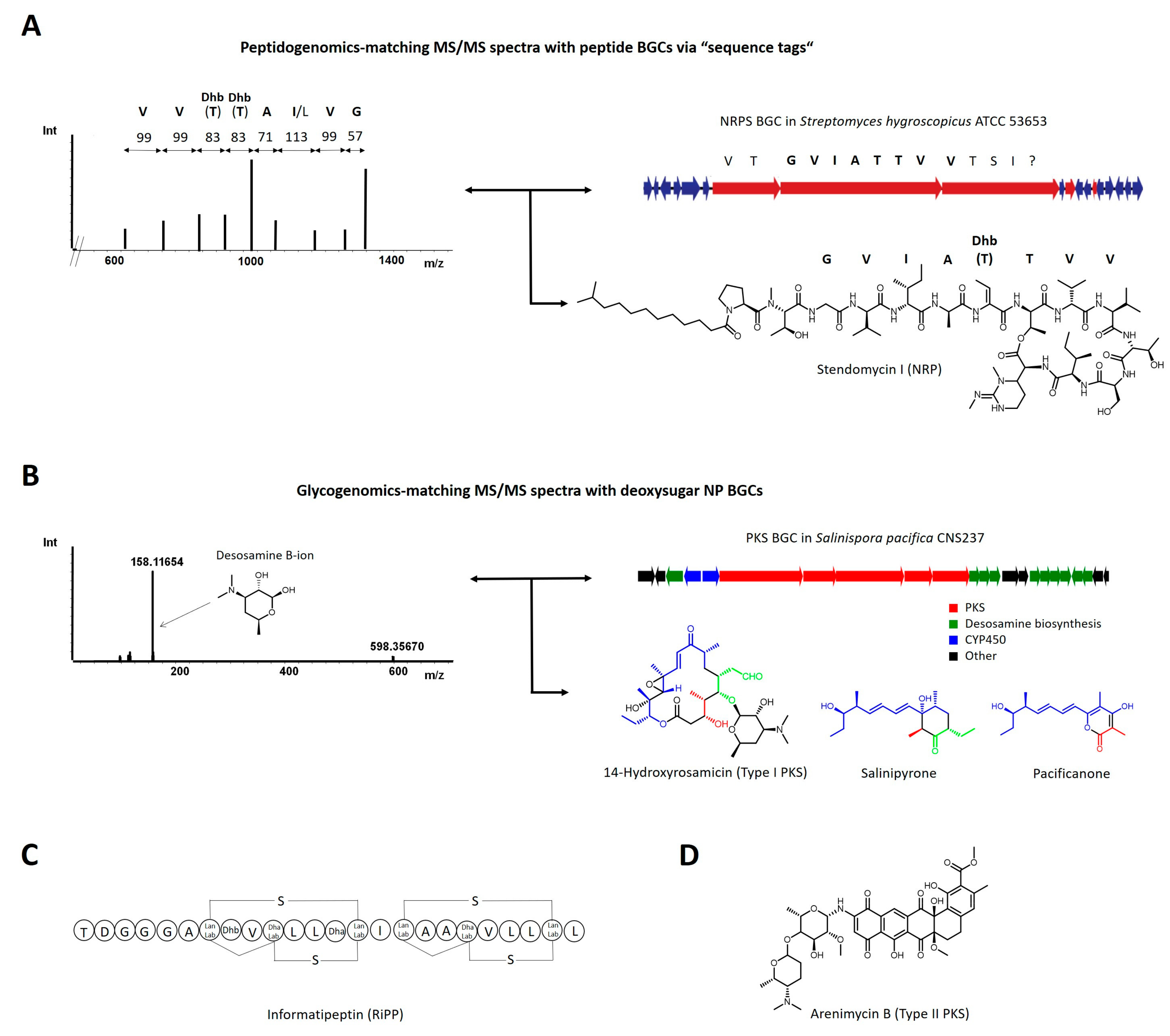
2.2. Correlation-Based Approaches on Larger Paired Datasets: Pattern-Based Genome Mining, Metabologenomics

Funding
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [PubMed]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed]
- Kenshole, E.; Herisse, M.; Michael, M.; Pidot, S.J. Natural product discovery through microbial genome mining. Curr. Opin. Chem. Biol. 2020, 60, 47–54. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Van der Hooft, J.J.J.; De Ridder, D.; Medema, M.H. BiG-SLiCE: A highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. Gigascience 2021, 10, giaa154. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Weber, T.; Medema, M.H. BiG-FAM: The biosynthetic gene cluster families database. Nucleic Acids Res. 2021, 49, D490–D497. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Wu, C.H.; Moree, W.J.; Lamsa, A.; Medema, M.H.; Zhao, X.; Gavilan, R.G.; Aparicio, M.; Atencio, L.; Jackson, C.; et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA 2013, 110, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Van der Hooft, J.J.J.; Wandy, J.; Young, F.; Padmanabhan, S.; Gerasimidis, K.; Burgess, K.E.V.; Barrett, M.P.; Rogers, S. Unsupervised Discovery and Comparison of Structural Families Across Multiple Samples in Untargeted Metabolomics. Anal. Chem. 2017, 89, 7569–7577. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.R.; Wang, M.; Nothias, L.F.; Van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Jarmusch, A.K.; Wang, M.; Aceves, C.M.; Advani, R.S.; Aguirre, S.; Aksenov, A.A.; Aleti, G.; Aron, A.T.; Bauermeister, A.; Bolleddu, S.; et al. ReDU: A framework to find and reanalyze public mass spectrometry data. Nat. Methods 2020, 17, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jarmusch, A.K.; Vargas, F.; Aksenov, A.A.; Gauglitz, J.M.; Weldon, K.; Petras, D.; Da Silva, R.R.; Quinn, R.A.; Melnik, A.V.; et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020, 38, 23–26. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Mikheenko, A.; Garg, N.; Nothias, L.F.; Ninomiya, A.; Takada, K.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 2017, 13, 30–37. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef]
- Zhang, J.J.; Tang, X.; Moore, B.S. Genetic platforms for heterologous expression of microbial natural products. Nat. Prod. Rep. 2019, 36, 1313–1332. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Weber, T.; Lee, S.Y. CRISPR/Cas-based genome engineering in natural product discovery. Nat. Prod. Rep. 2019, 36, 1262–1280. [Google Scholar] [CrossRef] [PubMed]
- Soldatou, S.; Eldjarn, G.H.; Huerta-Uribe, A.; Rogers, S.; Duncan, K.R. Linking biosynthetic and chemical space to accelerate microbial secondary metabolite discovery. FEMS Microbiol. Lett. 2019, 366, fnz142. [Google Scholar] [CrossRef]
- Van der Hooft, J.J.J.; Mohimani, H.; Bauermeister, A.; Dorrestein, P.C.; Duncan, K.R.; Medema, M.H. Linking genomics and metabolomics to chart specialized metabolic diversity. Chem Soc. Rev. 2020, 49, 3297–3314. [Google Scholar] [CrossRef]
- Kersten, R.D.; Yang, Y.L.; Xu, Y.; Cimermancic, P.; Nam, S.J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef]
- Kersten, R.D.; Ziemert, N.; Gonzalez, D.J.; Duggan, B.M.; Nizet, V.; Dorrestein, P.C.; Moore, B.S. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc. Natl. Acad. Sci. USA 2013, 110, E4407–E4416. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Pevzner, P.A. Dereplication, sequencing and identification of peptidic natural products: From genome mining to peptidogenomics to spectral networks. Nat. Prod. Rep. 2016, 33, 73–86. [Google Scholar] [CrossRef]
- Liu, W.T.; Lamsa, A.; Wong, W.R.; Boudreau, P.D.; Kersten, R.D.; Peng, Y.; Moree, W.J.; Duggan, B.M.; Moore, B.S.; Gerwick, W.H.; et al. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. J. Antibiot. 2014, 67, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Kersten, R.D.; Yang, Y.L.; Moore, B.S.; Dorrestein, P.C. Imaging mass spectrometry and genome mining via short sequence tagging identified the anti-infective agent arylomycin in Streptomyces roseosporus. J. Am. Chem. Soc. 2011, 133, 18010–18013. [Google Scholar] [CrossRef]
- Spraker, J.E.; Sanchez, L.M.; Lowe, T.M.; Dorrestein, P.C.; Keller, N.P. Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 2016, 10, 2317–2330. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Kersten, R.D.; Liu, W.T.; Wang, M.; Purvine, S.O.; Wu, S.; Brewer, H.M.; Pasa-Tolic, L.; Bandeira, N.; Moore, B.S.; et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 2014, 9, 1545–1551. [Google Scholar] [CrossRef]
- Mohimani, H.; Liu, W.T.; Kersten, R.D.; Moore, B.S.; Dorrestein, P.C.; Pevzner, P.A. NRPquest: Coupling Mass Spectrometry and Genome Mining for Nonribosomal Peptide Discovery. J. Nat. Prod. 2014, 77, 1902–1909. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Paalvast, Y.; Nguyen, D.D.; Melnik, A.; Dorrestein, P.C.; Takano, E.; Breitling, R. Pep2Path: Automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol. 2014, 10, e1003822. [Google Scholar] [CrossRef]
- Cao, L.; Gurevich, A.; Alexander, K.L.; Naman, C.B.; Leão, T.; Glukhov, E.; Luzzatto-Knaan, T.; Vargas, F.; Quinn, R.A.; Bouslimani, A.; et al. MetaMiner: A Scalable Peptidogenomics Approach for Discovery of Ribosomal Peptide Natural Products with Blind Modifications from Microbial Communities. Cell Syst. 2019, 9, 600–608.e4. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. 2010, 63, 37–39. [Google Scholar] [CrossRef]
- Awakawa, T.; Crüsemann, M.; Munguia, J.; Ziemert, N.; Nizet, V.; Fenical, W.; Moore, B.S. Salinipyrone and Pacificanone Are Biosynthetic By-products of the Rosamicin Polyketide Synthase. Chembiochem 2015, 16, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; Da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing Natural Product Diversity in a Collection of 146 Bacterial Strains Based on Growth and Extraction Protocols. J. Nat. Prod. 2017, 80, 588–597. [Google Scholar] [CrossRef]
- Oh, D.C.; Gontang, E.A.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinipyrones and pacificanones, mixed-precursor polyketides from the marine actinomycete Salinispora pacifica. J. Nat. Prod. 2008, 71, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Lechner, A.; Wietz, M.; Millán-Aguiñaga, N.; Chavarria, K.L.; Jensen, P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA 2014, 111, E1130–E1139. [Google Scholar] [CrossRef] [PubMed]
- Letzel, A.C.; Li, J.; Amos, G.C.A.; Millán-Aguiñaga, N.; Ginigini, J.; Abdelmohsen, U.R.; Gaudêncio, S.P.; Ziemert, N.; Moore, B.S.; Jensen, P.R. Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environ. Microbiol. 2017, 19, 3660–3673. [Google Scholar] [CrossRef]
- Duncan, K.R.; Crüsemann, M.; Lechner, A.; Sarkar, A.; Li, J.; Ziemert, N.; Wang, M.; Bandeira, N.; Moore, B.S.; Dorrestein, P.C.; et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 2015, 22, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Tobias, N.J.; Wolff, H.; Djahanschiri, B.; Grundmann, F.; Kronenwerth, M.; Shi, Y.M.; Simonyi, S.; Grün, P.; Shapiro-Ilan, D.; Pidot, S.J.; et al. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2017, 2, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Albright, J.C.; Goering, A.W.; Ju, K.S.; Haines, R.R.; Tchalukov, K.A.; Labeda, D.P.; Kelleher, N.L.; Metcalf, W.W. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 2014, 10, 963–968. [Google Scholar] [CrossRef]
- Goering, A.W.; McClure, R.A.; Doroghazi, J.R.; Albright, J.C.; Haverland, N.A.; Zhang, Y.; Ju, K.S.; Thomson, R.J.; Metcalf, W.W.; Kelleher, N.L. Metabologenomics: Correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2016, 2, 99–108. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.A.; Goering, A.W.; Ju, K.S.; Baccile, J.A.; Schroeder, F.C.; Metcalf, W.W.; Thomson, R.J.; Kelleher, N.L. Elucidating the Rimosamide-Detoxin Natural Product Families and Their Biosynthesis Using Metabolite/Gene Cluster Correlations. ACS Chem. Biol. 2016, 11, 3452–3460. [Google Scholar] [CrossRef]
- Parkinson, E.I.; Tryon, J.H.; Goering, A.W.; Ju, K.S.; McClure, R.A.; Kemball, J.D.; Zhukovsky, S.; Labeda, D.P.; Thomson, R.J.; Kelleher, N.L.; et al. Discovery of the Tyrobetaine Natural Products and Their Biosynthetic Gene Cluster via Metabologenomics. ACS Chem. Biol. 2018, 13, 1029–1037. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Iorio, M.; Maffioli, S.I.; Crüsemann, M.; Donadio, S.; Sosio, M. Planomonospora: A Metabolomics Perspective on an Underexplored Actinobacteria Genus. J. Nat. Prod. 2021, 84, 204–219. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Alanjary, M.M.; Zarazúa, G.S.; Maffioli, S.I.; Crüsemann, M.; Medema, M.H.; Donadio, S.; Sosio, M. A biaryl-linked tripeptide from Planomonospora reveals a widespread class of minimal RiPP gene clusters. Cell Chem. Biol. 2020. [Google Scholar] [CrossRef]
- Soldatou, S.; Eldjárn, G.H.; Ramsey, A.; Van der Hooft, J.J.J.; Hughes, A.H.; Rogers, S.; Duncan, K.R. Comparative Metabologenomics Analysis of Polar Actinomycetes. Marine Drugs 2021, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Eldjárn, G.H.; Ramsay, A.; Van der Hooft, J.J.J.; Duncan, K.R.; Soldatou, S.; Rousu, R.; Daly, R.; Wandy, J.; Rogers, S. Ranking microbial metabolomic and genomic links in the NPLinker framework using complementary scoring functions. bioRxiv 2020. [Google Scholar] [CrossRef]
- Männle, D.; McKinnie, S.M.K.; Mantri, S.S.; Steinke, K.; Lu, Z.; Moore, B.S.; Ziemert, N.; Kaysser, L. Comparative Genomics and Metabolomics in the Genus Nocardia. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; Van der Hooft, J.J.J.; Van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef] [PubMed]
- Schorn, M.A.; Verhoeven, S.; Ridder, L.; Huber, F.; Acharya, D.D.; Aksenov, A.A.; Aleti, G.; Amiri Moghaddam, J.; Aron, A.T.; Aziz, S.; et al. A community resource for paired genomic and metabolomic data mining. Nat. Chem. Biol. 2021. [Google Scholar] [CrossRef]
- Dejong, C.A.; Chen, G.M.; Li, H.; Johnston, C.W.; Edwards, M.R.; Rees, P.N.; Skinnider, M.A.; Webster, A.L.; Magarvey, N.A. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol. 2016, 12, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.W.; Skinnider, M.A.; Wyatt, M.A.; Li, X.; Ranieri, M.R.; Yang, L.; Zechel, D.L.; Ma, B.; Magarvey, N.A. An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat. Commun. 2015, 6, 8421. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Nothias, L.F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Vázquez-Baeza, Y.; Gauglitz, J.M.; Wang, M.; Dührkop, K.; Nothias-Esposito, M.; Acharya, D.D.; Ernst, M.; Van der Hooft, J.J.J.; Zhu, Q.; et al. Chemically informed analyses of metabolomics mass spectrometry data with Qemistree. Nat. Chem. Biol. 2021, 17, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Reher, R.; Kim, H.W.; Zhang, C.; Mao, H.H.; Wang, M.; Nothias, L.F.; Caraballo-Rodriguez, A.M.; Glukhov, E.; Teke, B.; Leao, T.; et al. A Convolutional Neural Network-Based Approach for the Rapid Annotation of Molecularly Diverse Natural Products. J. Am. Chem. Soc. 2020, 142, 4114–4120. [Google Scholar] [CrossRef] [PubMed]
- Danelius, E.; Halaby, S.; Van der Donk, W.A.; Gonen, T. MicroED in natural product and small molecule research. Nat. Prod. Rep. 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crüsemann, M. Coupling Mass Spectral and Genomic Information to Improve Bacterial Natural Product Discovery Workflows. Mar. Drugs 2021, 19, 142. https://doi.org/10.3390/md19030142
Crüsemann M. Coupling Mass Spectral and Genomic Information to Improve Bacterial Natural Product Discovery Workflows. Marine Drugs. 2021; 19(3):142. https://doi.org/10.3390/md19030142
Chicago/Turabian StyleCrüsemann, Max. 2021. "Coupling Mass Spectral and Genomic Information to Improve Bacterial Natural Product Discovery Workflows" Marine Drugs 19, no. 3: 142. https://doi.org/10.3390/md19030142
APA StyleCrüsemann, M. (2021). Coupling Mass Spectral and Genomic Information to Improve Bacterial Natural Product Discovery Workflows. Marine Drugs, 19(3), 142. https://doi.org/10.3390/md19030142





