Briarenols W–Z: Chlorine-Containing Polyoxygenated Briaranes from Octocoral Briareum stechei (Kükenthal, 1908)
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemical Identification of Isolated Briaranes
2.2. Bioactivity of Isolated Briaranes
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Single-Crystal X-Ray Crystallography of Brianolide (8)
3.5. Molecular Mechanics Calculations
3.6. In Vitro Inflammatory Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burks, J.E.; van der Helm, D.; Chang, C.Y.; Ciereszko, L.S. The crystal and molecular structure of briarein A, a diterpenoid from the gorgonian Briareum asbestinum. Acta Cryst. 1977, B33, 704–709. [Google Scholar] [CrossRef]
- Wahlberg, I.; Eklund, A.-M. Cyclized cembranoids of natural occurrence. Fortschr. Chem. Org. Naturst. 1992, 60, 1–141. [Google Scholar]
- Chen, Y.-H.; Chin, H.-K.; Peng, B.-R.; Chen, Y.-Y.; Hu, C.-C.; Zheng, L.-G.; Huynh, T.-H.; Su, T.-P.; Zhang, Y.-L.; Wen, Z.-H.; et al. Survey of briarane-type diterpenoids–Part VII. Heterocycles 2020, 100, 857–870. [Google Scholar]
- Sung, P.-J.; Hu, W.-P.; Fang, L.-S.; Fan, T.-Y.; Wang, J.-J. Briarenol A, a new diterpenoid from a gorgonian Briareum sp. (Briareidae). Nat. Prod. Res. 2005, 19, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-J.; Su, Y.-D.; Liao, Z.-J.; Wen, Z.-H.; Su, J.-H.; Wu, Y.-C.; Sung, P.-J. Briarenol B, a new polyoxygenated briarane from the octocoral Briareum excavatum. Nat. Prod. Commun. 2017, 12, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.-F.; Su, Y.-D.; Hwnag, T.-L.; Liao, Z.-J.; Tsui, K.-H.; Wen, Z.-H.; Wu, Y.-C.; Sung, P.-J. Briarenols C–E, new polyoxygenated briaranes from the octocoral Briareum excavatum. Molecules 2017, 22, 475. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.-H.; Lee, G.-H.; Fang, L.-S.; Sheu, J.-H.; Sung, P.-J. Briarenols F–H: New polyoxygenated briarane diterpenoids produced by the octocoral Briareum excavatum. Tetrahedron Lett. 2020, 61, 151826. [Google Scholar] [CrossRef]
- Huynh, T.-H.; Fang, L.-S.; Chen, Y.-H.; Peng, B.-R.; Chen, Y.-Y.; Zheng, L.-G.; Wu, Y.-J.; Wen, Z.-H.; Chen, J.-J.; Lin, T.-C.; et al. new anti-inflammatory 8,17-epoxybriaranes from the octocoral Briareum excavatum (Briareidae). Molecules 2020, 25, 1405. [Google Scholar] [CrossRef]
- Huynh, T.-H.; Chang, Y.-M.; Yang, S.-N.; Lee, G.-H.; Wen, Z.-H.; Wu, Y.-J.; Su, T.-R.; Sung, P.-J. Briarenol L, a new chlorine-containing briaranes from Briareum excavatum (Briareidae). J. Mol. Struct. 2021, 1223, 128970. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Kuo, L.-M.; Chen, L.-Y.; Lee, G.-H.; Peng, B.-R.; Chen, Y.-Y.; Chen, Y.-H.; Hwang, T.-L.; Sheu, J.-H.; Sung, P.-J. Identification and characterization of chlorine-containing briaranes from a cultured octocoral Briareum excavatum (Briareidae). Heterocycles 2020, 100, 1633–1644. [Google Scholar]
- Chi, W.-C.; Kuo, L.-M.; Yang, S.-N.; Lee, Y.-T.; Wen, Z.-H.; Tsui, K.-H.; Hwang, T.-L.; Zhang, Y.-L.; Sung, P.-J. Briarenols O and P: Novel briaranes from a cultured octocoral Briareum excavatum (Briareidae). Phytochem. Lett. 2021, 41, 134–138. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Chiang, C.-C.; Lee, Y.-T.; Wen, Z.-H.; Wu, Y.-C.; Wu, Y.-J.; Hwang, T.-L.; Wu, T.-Y.; Chang, C.-Y.; Sung, P.-J. Briarenols Q–T: Briaranes from a cultured octocoral Briareum stechei (Kükenthal, 1908). Mar. Drugs 2020, 18, 383. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Calado, R.; Sheridan, C.; Alimonti, A.; Osinga, R. Coral aquaculture to support drug discovery. Trends Biotechnol. 2013, 31, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Samimi-Namin, K.; van Ofwegen, L.P. Overview of the genus Briareum (Cnidaria, Octocorallia, Briareidae) in the Indo-Pacific, with the description of a new species. Zookeys 2016, 557, 1–44. [Google Scholar] [CrossRef]
- Groweiss, A.; Look, S.A.; Fenical, W. Solenolides, new antiinflammatory and antiviral diterpenoids from a marine octocoral of the genus Solenopodium. J. Org. Chem. 1988, 53, 2401–2406. [Google Scholar] [CrossRef]
- Su, Y.-D.; Wen, Z.-H.; Wu, Y.-C.; Fang, L.-S.; Chen, Y.-H.; Chang, Y.-C.; Sheu, J.-H.; Sung, P.-J. Briarenolides M–T, new briarane diterpenoids from a Formosan octocoral Briareum sp. Tetrahedron 2016, 72, 944–951. [Google Scholar] [CrossRef]
- Sheu, J.-H.; Sung, P.-J.; Su, J.-H.; Liu, H.-Y.; Duh, C.-Y.; Chiang, M.Y. Briaexcavatolides A–J, new diterpenes from the gorgonian Briareum excavatum. Tetrahedron 1999, 55, 14555–14564. [Google Scholar] [CrossRef]
- Kobayashi, J.; Cheng, J.-F.; Nakamura, H.; Ohizumi, Y.; Tomotake, Y.; Matsuzaki, T.; Grace, K.J.S.; Jacobs, R.S.; Kato, Y.; Brinen, L.S.; et al. Structure and stereochemistry of brianolide, a new antiinflammatory diterpenoid from the Okinawan gorgonian Briareum sp. Experientia 1991, 47, 501–502. [Google Scholar] [CrossRef]
- Allinger, N.L. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Buchholz, B.M.; Chanthaphavong, R.S.; Bauer, A.J. Nonhemopoietic cell TLR4 signaling is critical in causing early lipopolysaccharide-induced ileus. J. Immunol. 2009, 183, 6744–6753. [Google Scholar] [CrossRef]
- Jean, Y.-H.; Chen, W.-F.; Duh, C.-Y.; Huang, S.-Y.; Hsu, C.-H.; Lin, C.-S.; Sung, C.-S.; Chen, I.-M.; Wen, Z.-H. Inducible nitric oxide synthase and cyclooxygenase-2 participate in anti-inflammatory and analgesic effects of the natural marine compound lemnalol from Formosan soft coral Lemnalia cervicorni. Eur. J. Pharmacol. 2008, 578, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.-J.; Su, J.-H.; Huang, M.-S.; Wen, Z.-H.; Dai, C.-F.; Sheu, J.-H. Bioactive eunicellin-based diterpenoids from the soft coral Cladiella krempfi. Mar. Drugs 2011, 9, 2036–2045. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-H.; Wen, Z.-H. Bioactive cembrane-based diterpenoids from the soft coral Sinularia triangular. Mar. Drugs 2011, 9, 944–951. [Google Scholar] [CrossRef] [PubMed]
- Seibert, K.; Zhang, Y.; Leahy, K.; Hauser, S.; Masferrer, J.; Perkins, W.; Lee, L.; Isakson, P. Pharmacological and biochemical demonstration of the role of cyclooxygenase 2 in inflammation and pain. Proc. Natl. Acad. Sci. USA 1994, 91, 12013–12017. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Chen, S.R.; Eisenach, J.C.; Busija, D.W.; Pan, H.L. Spinal cyclooxygenase-2 is involved in development of allodynia after nerve injury in rats. Neuroscience 2000, 97, 743–748. [Google Scholar] [CrossRef]
- Wei, W.-C.; Sung, P.-J.; Duh, C.-Y.; Chen, B.-W.; Sheu, J.-H.; Yang, N.-S. Anti-inflammatory activities of natural products isolated from soft corals of Taiwan between 2008 and 2012. Mar. Drugs 2013, 11, 4083–4126. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Flack, H.D. On enantiomorph-polarity estimation. Acta Crystallogr. 1983, A39, 876–881. [Google Scholar] [CrossRef]
- Flack, H.D.; Bernardinelli, G. Absolute structure and absolute configuration. Acta Crystallogr. 1999, A55, 908–915. [Google Scholar] [CrossRef]
- CCDC Homepage. Available online: http://www.ccdc.cam.ac.uk/conts/retrieving.html (accessed on 30 January 2021).
- Chen, C.-H.; Chen, N.-F.; Feng, C.-W.; Cheng, S.-Y.; Hung, H.-C.; Tsui, K.-H.; Hsu, C.-H.; Sung, P.-J.; Chen, W.-F.; Wen, Z.-H. A coral-derived compound improves functional recovery after spinal cord injury through its antiapoptotic and anti-inflammatory effects. Mar. Drugs 2016, 14, 160. [Google Scholar] [CrossRef] [PubMed]
 ), heteronuclear multiple bond coherence (HMBC) (
), heteronuclear multiple bond coherence (HMBC) ( ), and protons with nuclear Overhauser effect spectroscopy (NOESY) (
), and protons with nuclear Overhauser effect spectroscopy (NOESY) ( ) correlations for 1.
) correlations for 1.
 ), heteronuclear multiple bond coherence (HMBC) (
), heteronuclear multiple bond coherence (HMBC) ( ), and protons with nuclear Overhauser effect spectroscopy (NOESY) (
), and protons with nuclear Overhauser effect spectroscopy (NOESY) (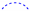 ) correlations for 1.
) correlations for 1.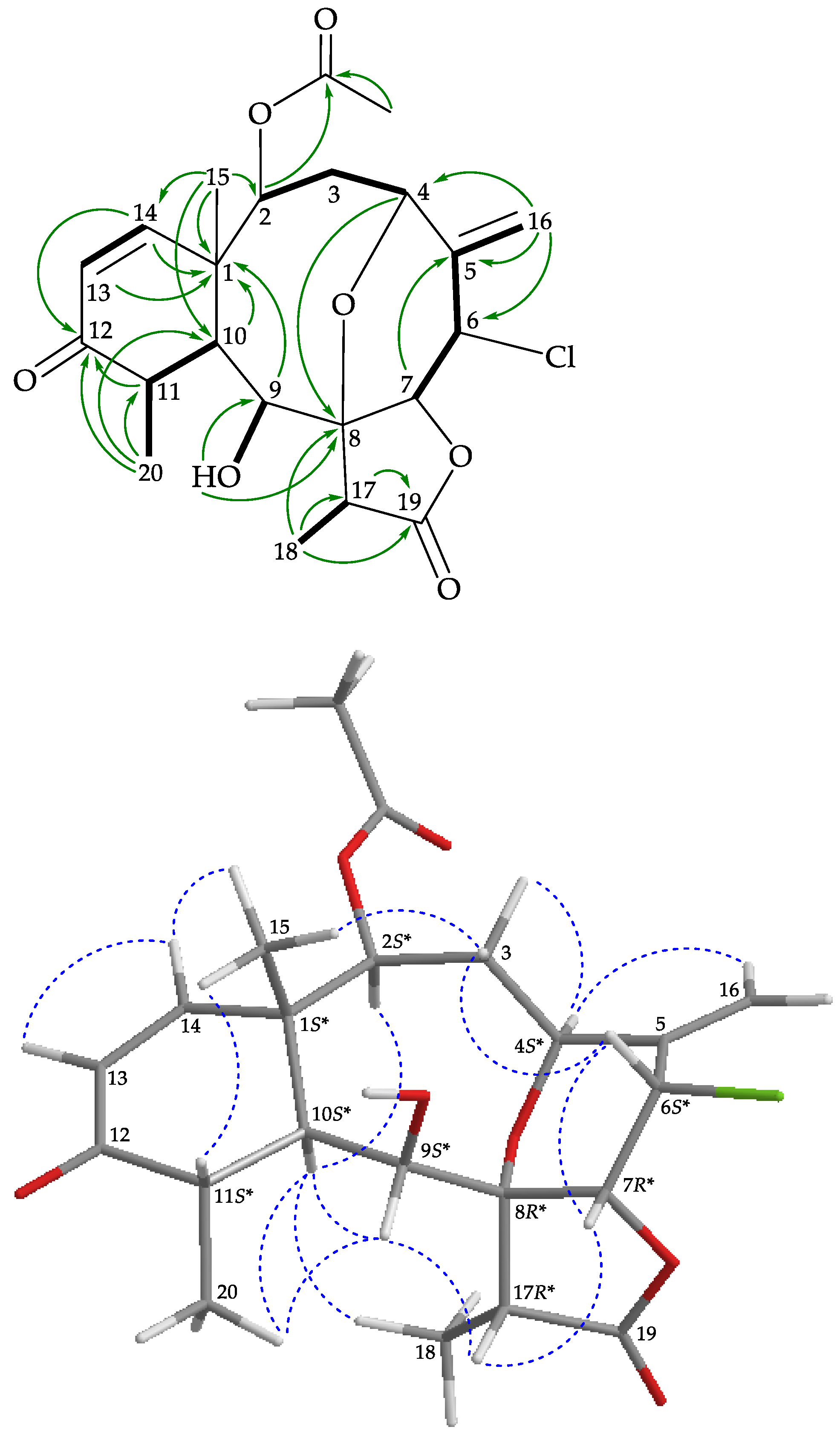
 ), HMBC (
), HMBC ( ), and protons with NOESY (
), and protons with NOESY ( ) correlations for 2.
) correlations for 2.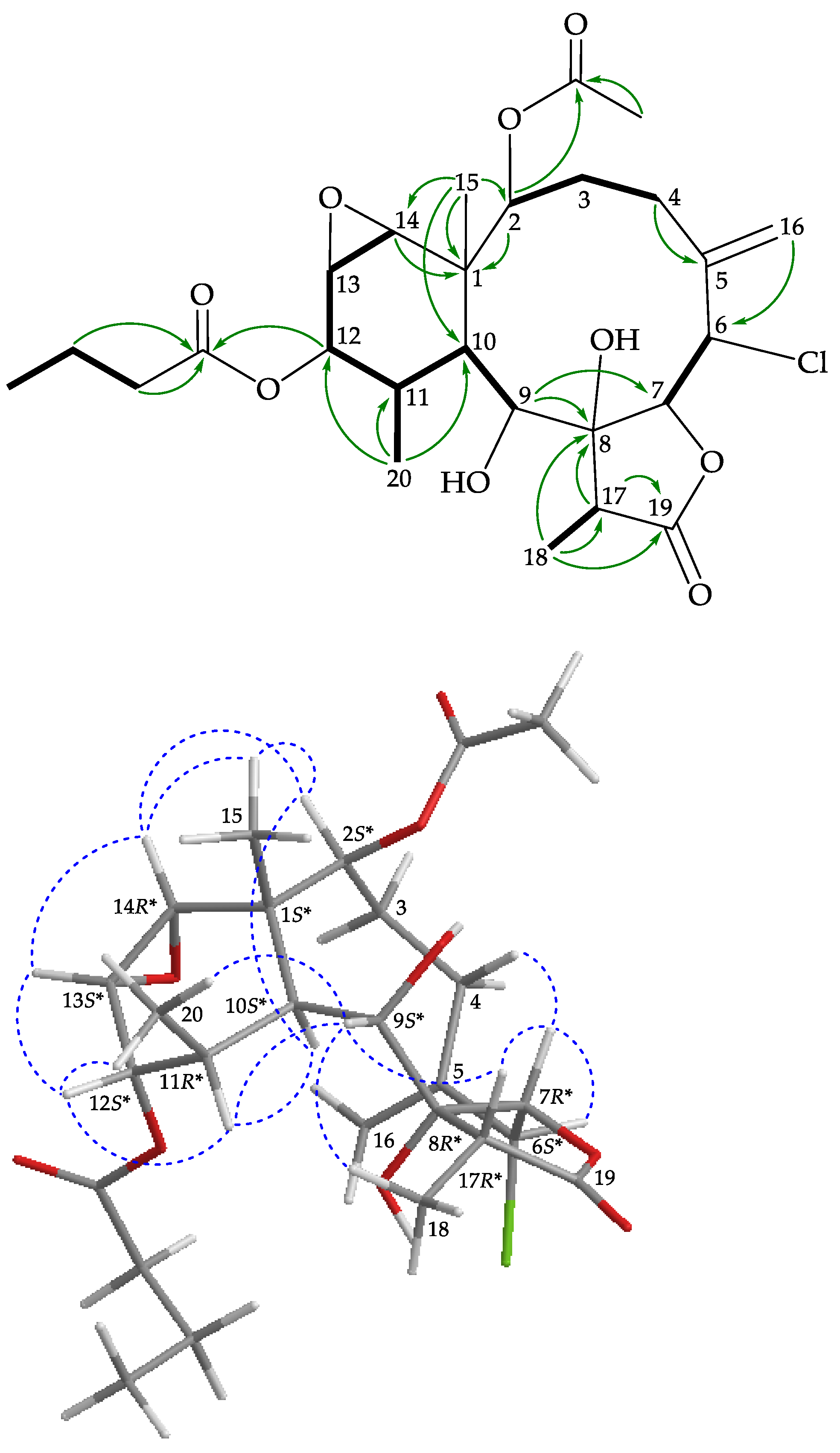
 ), HMBC (
), HMBC ( ), and protons with NOESY (
), and protons with NOESY ( ) correlations for 3.
) correlations for 3.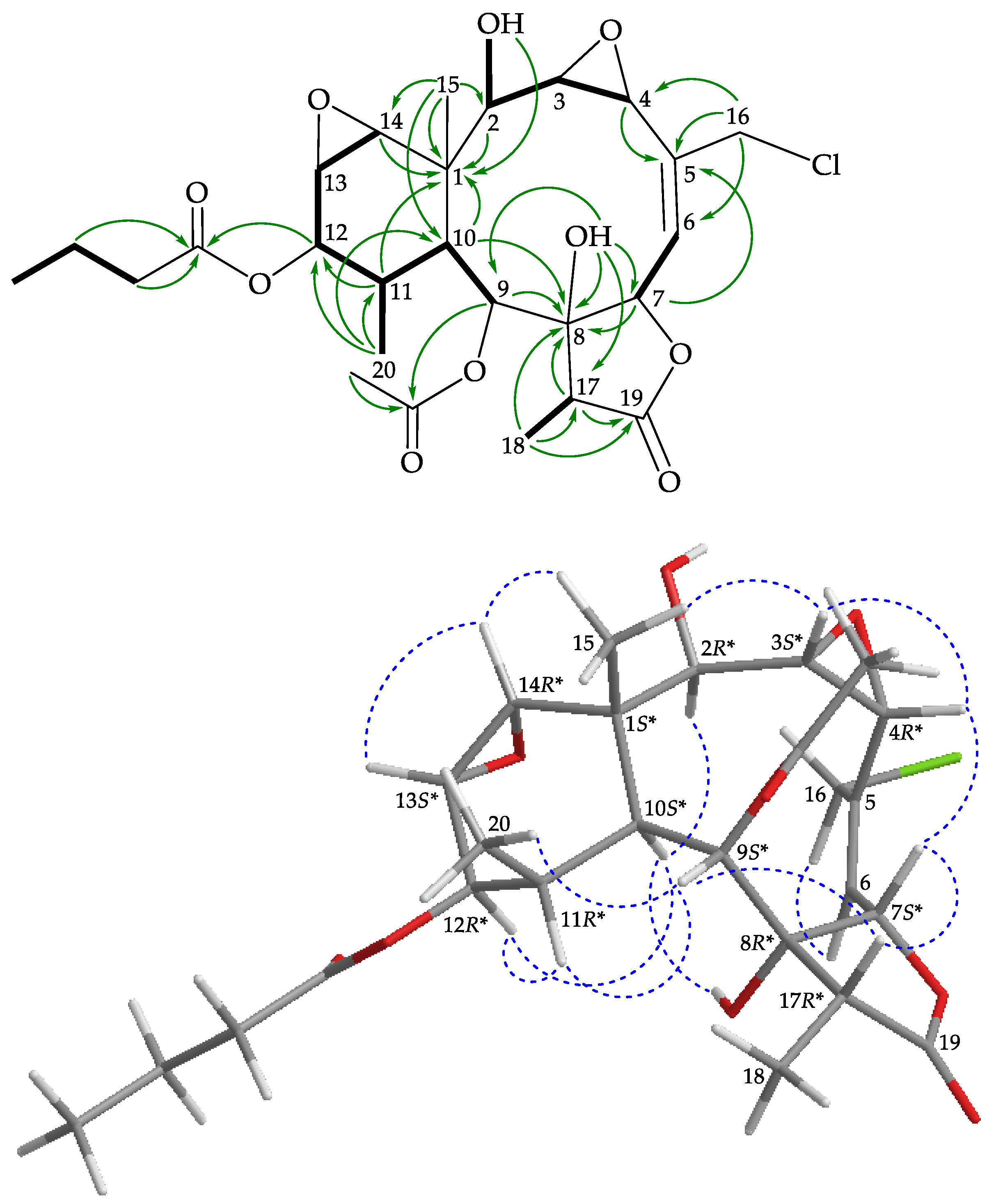
 ), HMBC (
), HMBC ( ), and protons with NOESY (
), and protons with NOESY ( ) correlations for 4.
) correlations for 4.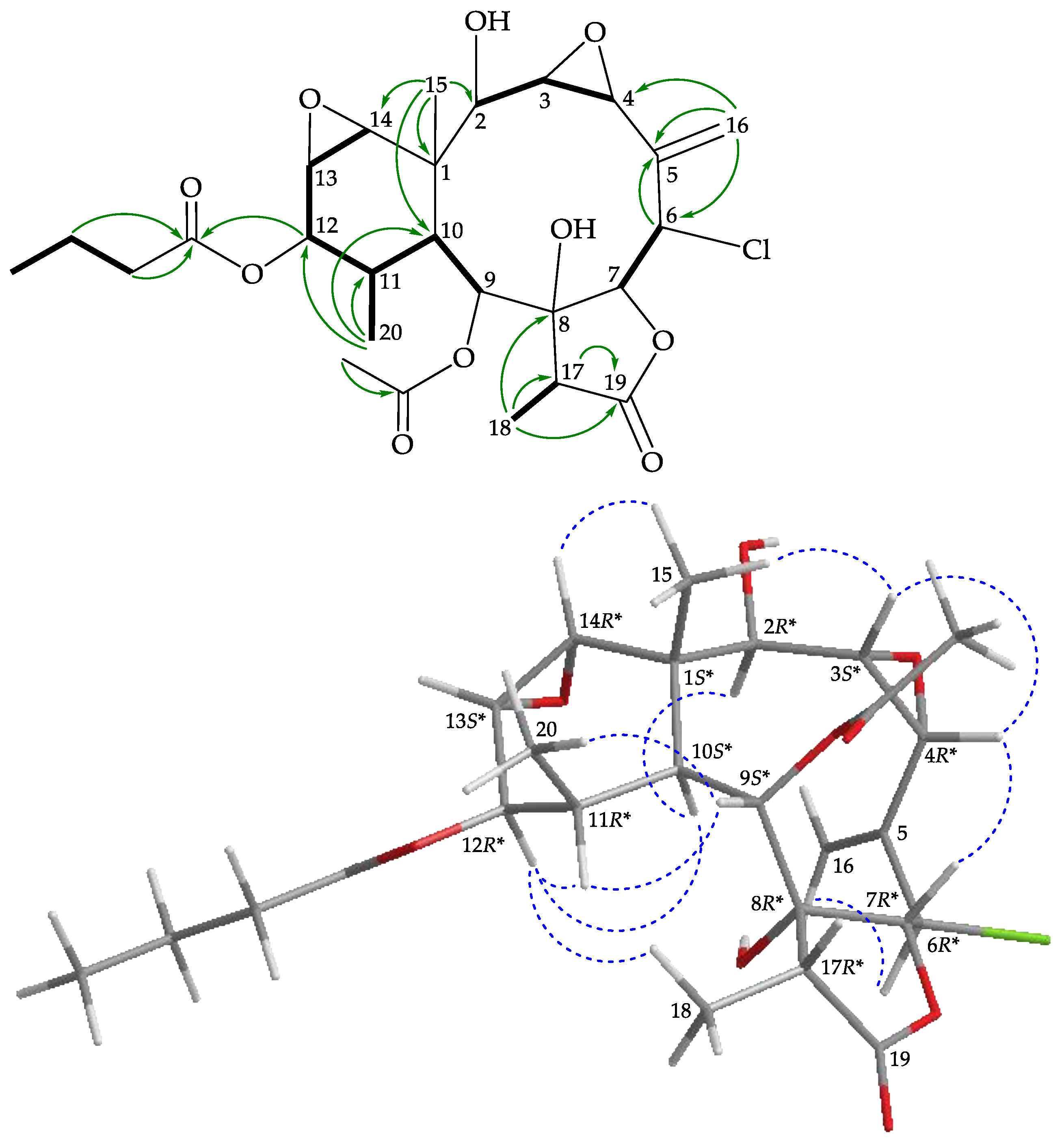

| Position | 1 a | 2 a | 3 b | 4 b |
|---|---|---|---|---|
| 1 | 44.2, C c | 40.4, C | 39.1, C | 38.4, C |
| 2 | 76.3, CH | 82.0, CH | 76.2, CH | 74.1, CH |
| 3 | 35.4, CH2 | 25.3, CH2 | 60.5, CH | 62.4, CH |
| 4 | 76.6, CH | 21.2, CH2 | 58.1, CH | 56.1, CH |
| 5 | 137.5, C | 140.3, C | 137.9, C | 137.4, C |
| 6 | 54.9, CH | 64.9, CH | 126.1, CH | 63.2, CH |
| 7 | 80.4, CH | 76.3, CH | 76.7, CH | 83.0, CH |
| 8 | 82.4, C | 86.1, C | 81.7, C | 80.9, C |
| 9 | 76.9, CH | 71.8, CH | 69.1, CH | 69.5, CH |
| 10 | 45.8, CH | 33.9, CH | 36.5, CH | 35.9, CH |
| 11 | 42.1, CH | 36.8, CH | 36.5, CH | 36.0, CH |
| 12 | 205.6, C | 70.3, CH | 71.6, CH | 72.2, CH |
| 13 | 126.0, CH | 49.7, CH | 57.2, CH | 57.2, CH |
| 14 | 156.3, CH | 62.1, CH | 62.1, CH | 62.0, CH |
| 15 | 16.2, CH3 | 20.2, CH3 | 15.2, CH3 | 15.3, CH3 |
| 16 | 116.1, CH2 | 118.6, CH2 | 44.1, CH2 | 121.2 CH2 |
| 17 | 50.2, CH | 44.4, CH | 43.6, CH | 46.1, CH |
| 18 | 8.2, CH3 | 7.2, CH3 | 6.3, CH3 | 6.0, CH3 |
| 19 | 175.3, C | 176.4, C | 175.5, C | 173.8, C |
| 20 | 15.4, CH3 | 14.5, CH3 | 9.7, CH3 | 9.4, CH3 |
| OAc-2 | 170.6, C | 168.3, C | ||
| 21.1, CH3 | 20.9, CH3 | |||
| OAc-9 | 169.5, C | 169.9, C | ||
| 21.8, CH3 | 21.8, CH3 | |||
| n-butyrate-12 | 173.3, C | 173.1, C | 173.1, C | |
| 35.8, CH2 | 36.2, CH2 | 36.2, CH2 | ||
| 18.4, CH2 | 18.4, CH2 | 18.4, CH2 | ||
| 13.7, CH3 | 13.6, CH3 | 13.6, CH3 |
| Position | 1 a | 2 a | 3 b | 4 b |
|---|---|---|---|---|
| 2 | 4.86 d (7.2) | 5.17 d (7.2) | 3.11 dd (9.6, 2.8) | 3.58 dd (8.8, 4.0) |
| 3α | 1.46 dd (15.6, 4.2) | 1.66 m | ||
| β | 3.32 ddd (15.6, 13.2, 7.2) | 2.41 m | 3.38 dd (9.2, 4.0) | 3.33 dd (8.8, 4.4) |
| 4α | 2.08 m | |||
| β | 4.87 dd (13.2, 4.2) | 2.01 m | 4.14 dd (4.0, 2.0) | 3.91 d (4.4) |
| 6 | 5.52 m | 5.02 br s | 6.03 ddd (9.2, 1.2, 1.2) | 4.92 d (10.4) |
| 7 | 4.65 d (3.0) | 5.35 d (1.8) | 5.28 d (9.2) | 4.71 d (10.4) |
| 9 | 4.53 d (6.0) | 3.57 dd (7.2, 7.2) | 5.29 d (8.0) | 5.33 d (8.4) |
| 10 | 2.08 d (12.0) | 2.63 br s | 1.77 dd (8.0, 2.8) | 1.82 dd (8.4, 2.4) |
| 11 | 2.72 dq (12.0, 7.2) | 2.15 m | 2.14 m | 1.95 m |
| 12 | 4.74 dd (5.4, 1.8) | 4.71 d (4.8) | 4.58 d (4.8) | |
| 13 | 5.95 d (10.8) | 3.47 dd (5.4, 3.6) | 3.19 dd (3.6, 0.8) | 3.24 d (4.0) |
| 14 | 6.95 d (10.8) | 2.88 d (3.6) | 3.29 d (3.6) | 3.27 d (4.0) |
| 15 | 1.39 s | 1.30 s | 1.19 s | 1.16 s |
| 16a/b | 5.34 d (1.8); 5.48 d (1.8) | 5.72 s; 5.89 s | 4.19 d (12.4); 4.09 d (12.4) | 5.40 s; 5.64 s |
| 17 | 2.53 q (7.2) | 3.11 q (7.2) | 2.42 q (7.2) | 2.76 q (7.2) |
| 18 | 1.22 d (7.2) | 1.14 d (7.2) | 1.19 d (7.2) | 1.18 d (7.2) |
| 20 | 1.30 d (7.2) | 1.05 d (7.8) | 1.03 d (7.2) | 1.01 d (7.2) |
| OH-2 | 2.43 s | 2.35 d (4.0) | ||
| OH-8 | 2.86 br s | 3.12 s | 3.06 s | |
| OH-9 | 2.84 d (6.0) | |||
| OAc-2 | 2.15 s | 2.19 s | ||
| OAc-9 | 2.22 s | 2.23 s | ||
| n-butyrate-12 | 2.34 t (7.2) | 2.35 t (7.2) | 2.35 t (7.2) | |
| 1.69 sext (7.2) | 1.67 sext (7.2) | 1.67 sext (7.2) | ||
| 0.99 t (7.2) | 0.96 t (7.2) | 0.96 t (7.2) |
| Compound | iNOS | COX-2 | β-Actin |
|---|---|---|---|
| 10 µM | Expression (% of LPS) | ||
| Control | 2.88 ± 0.86 | 0.94 ± 0.10 | 107.01 ± 2.73 |
| Vehicle | 100.00 ± 1.84 | 100.00 ± 3.98 | 100.00 ± 1.66 |
| 1 | 88.18 ± 12.38 | 103.23 ± 5.20 | 101.63 ± 4.23 |
| 2 | 142.03 ± 18.44 | 159.21 ± 13.41 | 97.81 ± 3.15 |
| 3 | 99.71 ± 13.77 | 89.20 ± 1.40 | 98.31 ± 5.33 |
| 4 | 103.25 ± 16.72 | 96.92 ± 4.72 | 99.46 ± 3.75 |
| 5 | 134.11 ± 14.70 | 196.03 ± 12.35 | 106.56 ± 1.98 |
| 6 | 86.20 ± 11.20 | 85.98 ± 2.47 | 104.50 ± 2.01 |
| 7 | 92.55 ± 10.52 | 91.71 ± 1.90 | 104.80 ± 2.53 |
| Dexamethasone | 61.24 ± 11.09 | 18.17 ± 2.65 | 104.70 ± 3.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-Y.; Zhang, Y.-L.; Lee, G.-H.; Tsou, L.K.; Zhang, M.M.; Hsieh, H.-P.; Chen, J.-J.; Ko, C.-Y.; Wen, Z.-H.; Sung, P.-J. Briarenols W–Z: Chlorine-Containing Polyoxygenated Briaranes from Octocoral Briareum stechei (Kükenthal, 1908). Mar. Drugs 2021, 19, 77. https://doi.org/10.3390/md19020077
Chen Y-Y, Zhang Y-L, Lee G-H, Tsou LK, Zhang MM, Hsieh H-P, Chen J-J, Ko C-Y, Wen Z-H, Sung P-J. Briarenols W–Z: Chlorine-Containing Polyoxygenated Briaranes from Octocoral Briareum stechei (Kükenthal, 1908). Marine Drugs. 2021; 19(2):77. https://doi.org/10.3390/md19020077
Chicago/Turabian StyleChen, You-Ying, Yi-Lin Zhang, Gene-Hsiang Lee, Lun Kelvin Tsou, Mingzi M. Zhang, Hsing-Pang Hsieh, Jih-Jung Chen, Chou-Yuan Ko, Zhi-Hong Wen, and Ping-Jyun Sung. 2021. "Briarenols W–Z: Chlorine-Containing Polyoxygenated Briaranes from Octocoral Briareum stechei (Kükenthal, 1908)" Marine Drugs 19, no. 2: 77. https://doi.org/10.3390/md19020077
APA StyleChen, Y.-Y., Zhang, Y.-L., Lee, G.-H., Tsou, L. K., Zhang, M. M., Hsieh, H.-P., Chen, J.-J., Ko, C.-Y., Wen, Z.-H., & Sung, P.-J. (2021). Briarenols W–Z: Chlorine-Containing Polyoxygenated Briaranes from Octocoral Briareum stechei (Kükenthal, 1908). Marine Drugs, 19(2), 77. https://doi.org/10.3390/md19020077








