From Crystal Structures of RgIA4 in Complex with Ac-AChBP to Molecular Determinants of Its High Potency of α9α10 nAChR
Abstract
:1. Introduction
2. Results
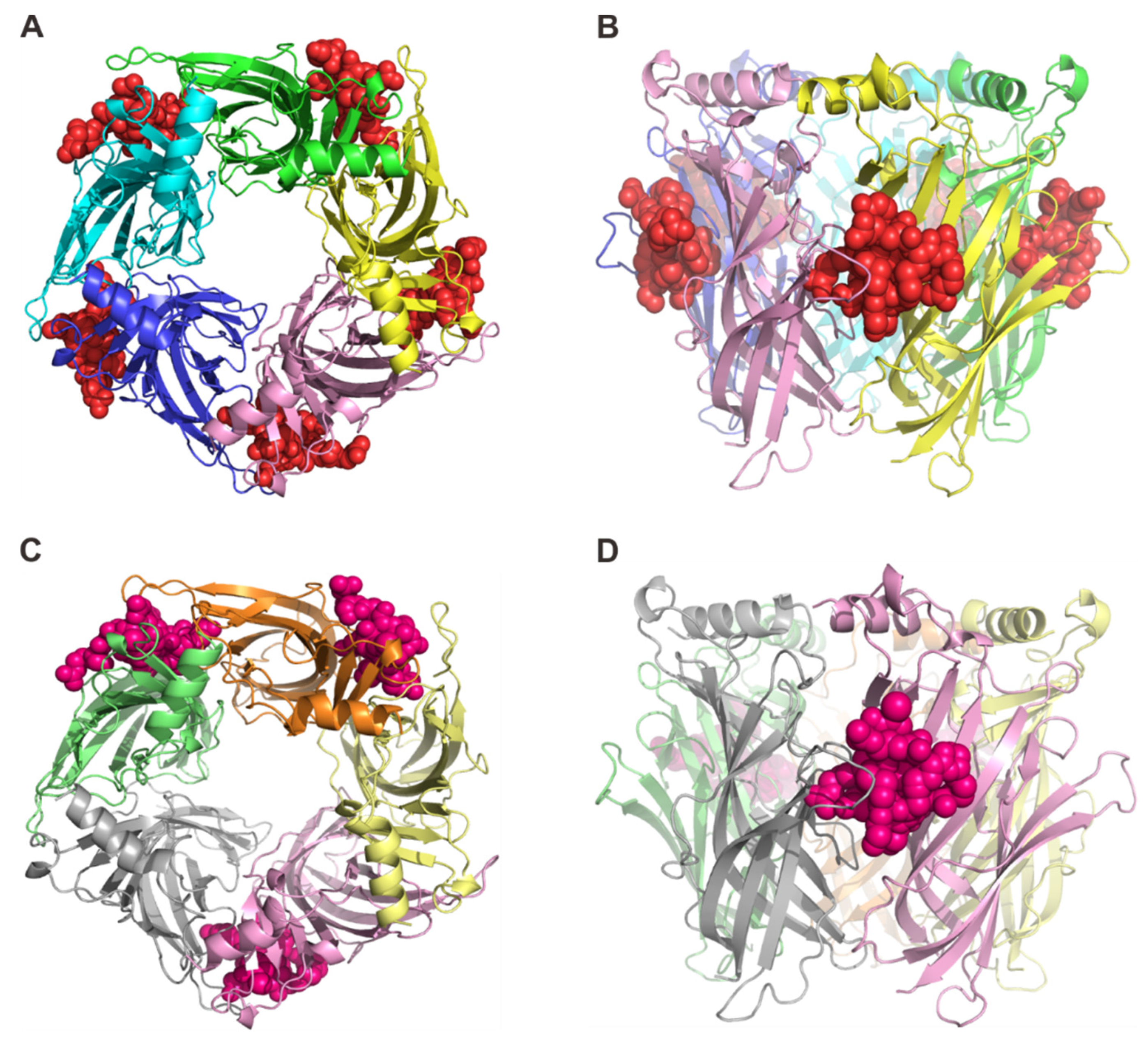
2.1. Overall Structure of RgIA and RgIA4 Bound to Ac-AChBP
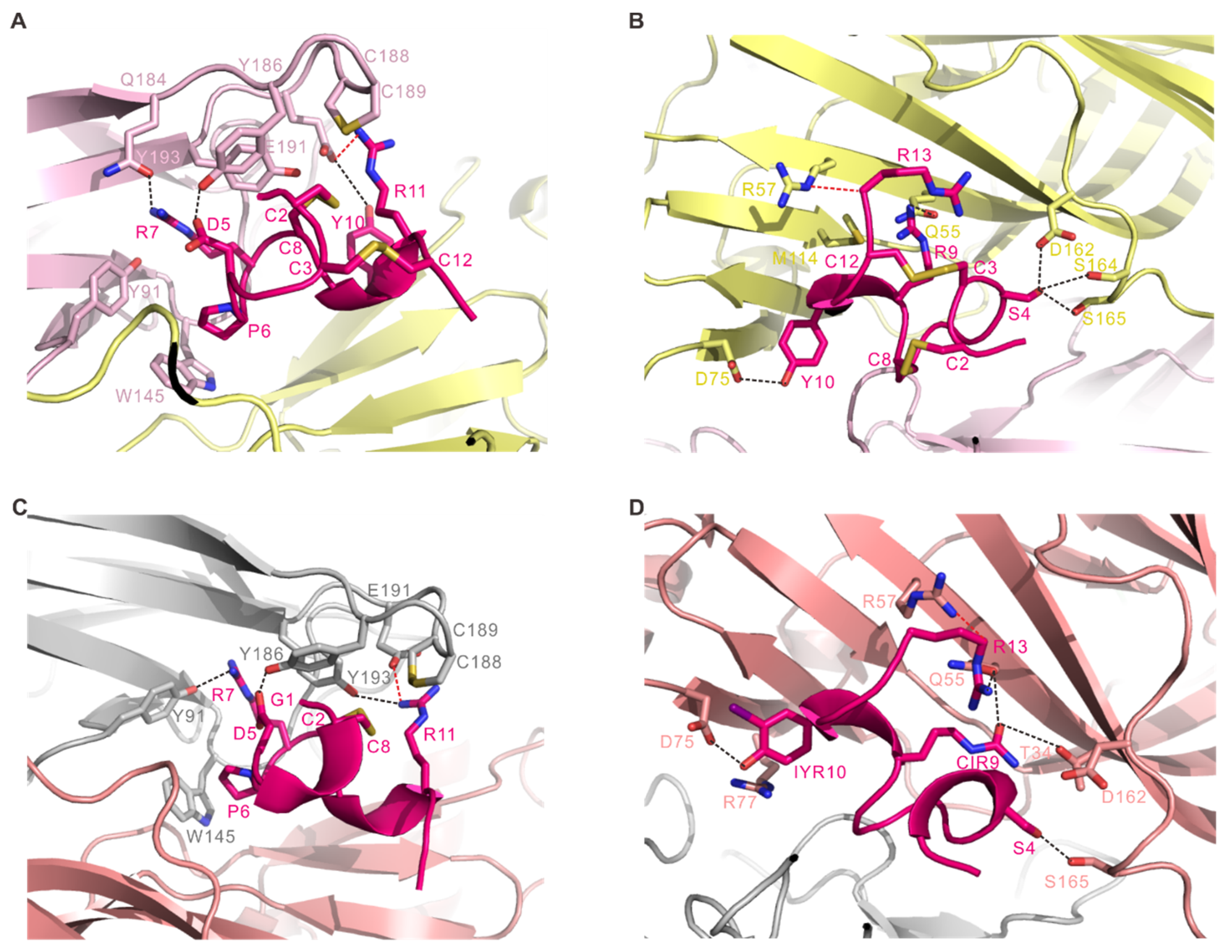
2.2. Interactions of RgIA with Ac-AChBP
2.3. Interactions of RgIA4 with Ac-AChBP
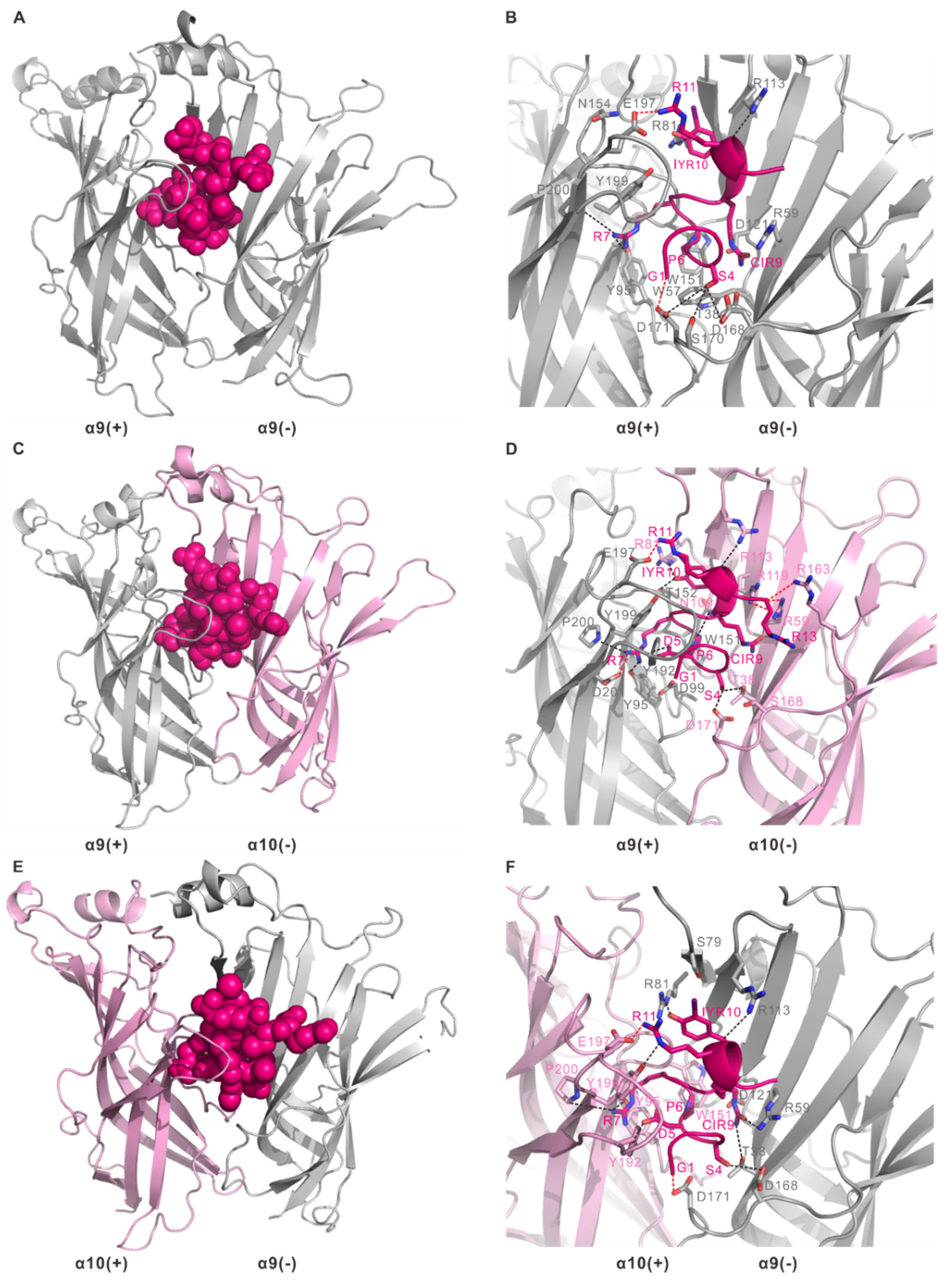
2.4. Homology Modelling of Human α9α10 nAChR and Docking with RgIA and Its Mutant
3. Discussion
4. Methods and Materials
4.1. Overall Structure of RgIA and RgIA4 Bound to Ac-AChBP
4.2. Protein Expression and Purification
4.3. Crystallization, Data Collection and Structural Determination
4.4. Homology Modeling and Model Refinement
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miller, P.S.; Smart, T.G. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 2010, 31, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.A.; Wang, W.; Hales, T.G.; Kirkness, E.F. A novel class of ligand-gated ion channel is activated by Zn2+. J. Biol. Chem. 2003, 278, 712–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotti, C.; Clementi, F. Neuronal nicotinic receptors: From structure to pathology. Prog. Neurobiol. 2004, 74, 363–396. [Google Scholar] [CrossRef] [PubMed]
- Gotti, C.; Clementi, F.; Fornari, A.; Gaimarri, A.; Guiducci, S.; Manfredi, I.; Moretti, M.; Pedrazzi, P.; Pucci, L.; Zoli, M. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem. Pharmacol. 2009, 78, 703–711. [Google Scholar] [CrossRef] [Green Version]
- Dellisanti, C.D.; Ghosh, B.; Hanson, S.M.; Raspanti, J.M.; Grant, V.A.; Diarra, G.M.; Schuh, A.M.; Satyshur, K.; Klug, C.S.; Czajkowski, C. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLoS Biol. 2013, 11, e1001714. [Google Scholar] [CrossRef]
- Dineley, K.T.; Pandya, A.A.; Yakel, J.L. Nicotinic ACh receptors as therapeutic targets in CNS disorders. Trends Pharmacol. Sci. 2015, 36, 96–108. [Google Scholar] [CrossRef] [Green Version]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, S.A.; Christie, M.J. Conotoxin interactions with α9α10-nAChRs: Is the α9α10-nicotinic acetylcholine receptor an important therapeutic target for pain management? Toxins 2015, 7, 3916–3932. [Google Scholar] [CrossRef] [Green Version]
- Castro, J.; Harrington, A.M.; Garcia-Caraballo, S.; Maddern, J.; Grundy, L.; Zhang, J.; Page, G.; Miller, P.E.; Craik, D.J.; Adams, D.J. α-Conotoxin Vc1. 1 inhibits human dorsal root ganglion neuroexcitability and mouse colonic nociception via GABAB receptors. Gut 2017, 66, 1083–1094. [Google Scholar] [CrossRef] [Green Version]
- Hone, A.J.; McIntosh, J.M. Nicotinic acetylcholine receptors in neuropathic and inflammatory pain. FEBS Lett. 2018, 592, 1045–1062. [Google Scholar] [CrossRef] [Green Version]
- Sgard, F.; Charpantier, E.; Bertrand, S.; Walker, N.; Caput, D.; Graham, D.; Bertrand, D.; Besnard, F. A novel human nicotinic receptor subunit, alpha10, that confers functionality to the alpha9-subunit. Mol. Pharmacol. 2002, 61, 150–159. [Google Scholar] [CrossRef] [Green Version]
- Elgoyhen, A.B.; Vetter, D.E.; Katz, E.; Rothlin, C.V.; Heinemann, S.F.; Boulter, J. alpha10: A determinant of nicotinic cholinergic receptor function in mammalian vestibular and cochlear mechanosensory hair cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3501–3506. [Google Scholar] [CrossRef] [Green Version]
- Ellison, M.; Feng, Z.P.; Park, A.J.; Zhang, X.; Olivera, B.M.; McIntosh, J.M.; Norton, R.S. Alpha-RgIA, a novel conotoxin that blocks the alpha9alpha10 nAChR: Structure and identification of key receptor-binding residues. J. Mol. Biol. 2008, 377, 1216–1227. [Google Scholar] [CrossRef] [Green Version]
- Azam, L.; McIntosh, J.M. Molecular basis for the differential sensitivity of rat and human alpha9alpha10 nAChRs to alpha-conotoxin RgIA. J. Neurochem. 2012, 122, 1137–1144. [Google Scholar] [CrossRef] [Green Version]
- Romero, H.K.; Christensen, S.B.; Di Cesare Mannelli, L.; Gajewiak, J.; Ramachandra, R.; Elmslie, K.S.; Vetter, D.E.; Ghelardini, C.; Iadonato, S.P.; Mercado, J.L.; et al. Inhibition of alpha9alpha10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci. USA 2017, 114, E1825–E1832. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.B.; Hone, A.J.; Roux, I.; Kniazeff, J.; Pin, J.P.; Upert, G.; Servent, D.; Glowatzki, E.; McIntosh, J.M. RgIA4 Potently Blocks Mouse alpha9alpha10 nAChRs and Provides Long Lasting Protection against Oxaliplatin-Induced Cold Allodynia. Front. Cell Neurosci. 2017, 11, 219. [Google Scholar] [CrossRef] [Green Version]
- Zouridakis, M.; Papakyriakou, A.; Ivanov, I.A.; Kasheverov, I.E.; Tsetlin, V.; Tzartos, S.; Giastas, P. Crystal Structure of the Monomeric Extracellular Domain of alpha9 Nicotinic Receptor Subunit in Complex With alpha-Conotoxin RgIA: Molecular Dynamics Insights Into RgIA Binding to alpha9alpha10 Nicotinic Receptors. Front. Pharmacol. 2019, 10, 474. [Google Scholar] [CrossRef]
- Unwin, N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 2005, 346, 967–989. [Google Scholar] [CrossRef]
- Li, S.X.; Huang, S.; Bren, N.; Noridomi, K.; Dellisanti, C.D.; Sine, S.M.; Chen, L. Ligand-binding domain of an alpha7-nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 2011, 14, 1253–1259. [Google Scholar] [CrossRef] [Green Version]
- Morales-Perez, C.L.; Noviello, C.M.; Hibbs, R.E. X-ray structure of the human α4β2 nicotinic receptor. Nature 2016, 538, 411–415. [Google Scholar] [CrossRef] [Green Version]
- Kouvatsos, N.; Giastas, P.; Chroni-Tzartou, D.; Poulopoulou, C.; Tzartos, S.J. Crystal structure of a human neuronal nAChR extracellular domain in pentameric assembly: Ligand-bound α2 homopentamer. Proc. Natl. Acad. Sci. USA 2016, 113, 9635–9640. [Google Scholar] [CrossRef] [Green Version]
- Brejc, K.; van Dijk, W.J.; Klaassen, R.V.; Schuurmans, M.; van Der Oost, J.; Smit, A.B.; Sixma, T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 2001, 411, 269–276. [Google Scholar] [CrossRef]
- Smit, A.B.; Syed, N.I.; Schaap, D.; van Minnen, J.; Klumperman, J.; Kits, K.S.; Lodder, H.; van der Schors, R.C.; van Elk, R.; Sorgedrager, B.; et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature 2001, 411, 261–268. [Google Scholar] [CrossRef]
- Vazquez, A.E. α9α10 Acetylcholine receptors: Structure and functions. Neurotransmitter 2016, 3, e1298. [Google Scholar]
- Vincler, M.; Wittenauer, S.; Parker, R.; Ellison, M.; Olivera, B.M.; McIntosh, J.M. Molecular mechanism for analgesia involving specific antagonism of alpha9alpha10 nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. USA 2006, 103, 17880–17884. [Google Scholar] [CrossRef] [Green Version]
- Ellison, M.; Haberlandt, C.; Gomez-Casati, M.E.; Watkins, M.; Elgoyhen, A.B.; McIntosh, J.M.; Olivera, B.M. Alpha-RgIA: A novel conotoxin that specifically and potently blocks the alpha9alpha10 nAChR. Biochemistry 2006, 45, 1511–1517. [Google Scholar] [CrossRef]
- Perez, E.G.; Cassels, B.K.; Zapata-Torres, G. Molecular modeling of the alpha9alpha10 nicotinic acetylcholine receptor subtype. Bioorg. Med. Chem. Lett. 2009, 19, 251–254. [Google Scholar] [CrossRef]
- Lin, B.; Xu, M.; Zhu, X.; Wu, Y.; Liu, X.; Zhangsun, D.; Hu, Y.; Xiang, S.-H.; Kasheverov, I.E.; Tsetlin, V.I.; et al. From crystal structure of α-conotoxin GIC in complex with Ac-AChBP to molecular determinants of its high selectivity for α3β2 nAChR. Sci. Rep. 2016, 6, 22349. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Zhu, X.; Yu, J.; Yu, J.; Luo, S.; Wang, X. The crystal structure of Ac-AChBP in complex with α-conotoxin LvIA reveals the mechanism of its selectivity towards different nAChR subtypes. Protein Cell 2017, 8, 675–685. [Google Scholar] [CrossRef]
- Hansen, S.B.; Sulzenbacher, G.; Huxford, T.; Marchot, P.; Taylor, P.; Bourne, Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005, 24, 3635–3646. [Google Scholar] [CrossRef]
- Celie, P.H.; Kasheverov, I.E.; Mordvintsev, D.Y.; Hogg, R.C.; van Nierop, P.; van Elk, R.; van Rossum-Fikkert, S.E.; Zhmak, M.N.; Bertrand, D.; Tsetlin, V.; et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 2005, 12, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhangsun, D.; Zhu, X.; Kaas, Q.; Zhangsun, M.; Harvey, P.J.; Craik, D.J.; McIntosh, J.M.; Luo, S. alpha-Conotoxin [S9A]TxID Potently Discriminates between alpha3beta4 and alpha6/alpha3beta4 Nicotinic Acetylcholine Receptors. J. Med. Chem. 2017, 60, 5826–5833. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhu, X.; Harvey, P.J.; Kaas, Q.; Zhangsun, D.; Craik, D.J.; Luo, S. Single Amino Acid Substitution in alpha-Conotoxin TxID Reveals a Specific alpha3beta4 Nicotinic Acetylcholine Receptor Antagonist. J. Med. Chem. 2018, 61, 9256–9265. [Google Scholar] [CrossRef] [PubMed]
- Azam, L.; Yoshikami, D.; McIntosh, J.M. Amino acid residues that confer high selectivity of the alpha6 nicotinic acetylcholine receptor subunit to alpha-conotoxin MII[S4A,E11A,L15A]. J. Biol. Chem. 2008, 283, 11625–11632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halai, R.; Clark, R.J.; Nevin, S.T.; Jensen, J.E.; Adams, D.J.; Craik, D.J. Scanning mutagenesis of alpha-conotoxin Vc1.1 reveals residues crucial for activity at the alpha9alpha10 nicotinic acetylcholine receptor. J. Biol. Chem. 2009, 284, 20275–20284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azam, L.; Papakyriakou, A.; Zouridakis, M.; Giastas, P.; Tzartos, S.J.; McIntosh, J.M. Molecular interaction of alpha-conotoxin RgIA with the rat alpha9alpha10 nicotinic acetylcholine receptor. Mol. Pharmacol. 2015, 87, 855–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuny, H.; Kompella, S.N.; Tae, H.S.; Yu, R.; Adams, D.J. Key Structural Determinants in the Agonist Binding Loops of Human beta2 and beta4 Nicotinic Acetylcholine Receptor Subunits Contribute to alpha3beta4 Subtype Selectivity of alpha-Conotoxins. J. Biol. Chem. 2016, 291, 23779–23792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kompella, S.N.; Hung, A.; Clark, R.J.; Mari, F.; Adams, D.J. Alanine scan of alpha-conotoxin RegIIA reveals a selective alpha3beta4 nicotinic acetylcholine receptor antagonist. J. Biol. Chem. 2015, 290, 1039–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.-S.; Zhang, K.-H.; Cui, Y.; Wang, Z.-J.; Pan, Q.-Y.; Liu, K.; Sun, B.; Zhou, H.; Li, M.-J.; Xu, Q.; et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018, 29, 68. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997, 276, 307–326. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Grosse-Kunstleve, R.W.; Hung, L.W.; Ioerger, T.R.; McCoy, A.J.; Moriarty, N.W.; Read, R.J.; Sacchettini, J.C.; Sauter, N.K.; Terwilliger, T.C. PHENIX: Building new software for automated crystallographic structure determination. Acta Cryst. D Biol. Cryst. 2002, 58, 1948–1954. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Cryst. D Biol. Cryst. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D Biol. Cryst. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.Y. RED Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [Green Version]
- Dupradeau, F.Y.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, P. The R.E.D. tools: Advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010, 12, 7821–7839. [Google Scholar] [CrossRef] [Green Version]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]

| Ac-AChBP | RgIA | RgIA4 |
|---|---|---|
| Principal Side | ||
| Tyr91 | Arg7 | Arg7 |
| Trp145 | Pro6, Arg7 | Pro6 |
| Val146 | Arg7 | |
| Gln184 | Asp5, Arg7 | |
| Tyr186 | Asp5 | Gly1, Asp5 |
| Cys188 | Cys2 | |
| Cys189 | Cys8, Arg11 | Cys2, Cys8 |
| Glu191 | Tyr10, Arg11 | Arg11 |
| Tyr193 | Cys8 | Cys8, Arg11 |
| Ile194 | Arg7 | |
| Complementary Side | ||
| Thr34 | CIR9 | |
| Gln55 | Arg9 | CIR9, Arg13 |
| Arg57 | Arg13 | Arg13 |
| Asp75 | Tyr10 | IYR10 |
| Arg77 | IYR10 | |
| Met114 | Arg9 | |
| Asp162 | Ser4 | Ser4 |
| Ser164 | Ser4 | |
| Ser165 | Ser4 | Ser4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, S.; Fan, Y.; Zhu, X.; Xue, Y.; Luo, S.; Wang, X. From Crystal Structures of RgIA4 in Complex with Ac-AChBP to Molecular Determinants of Its High Potency of α9α10 nAChR. Mar. Drugs 2021, 19, 709. https://doi.org/10.3390/md19120709
Pan S, Fan Y, Zhu X, Xue Y, Luo S, Wang X. From Crystal Structures of RgIA4 in Complex with Ac-AChBP to Molecular Determinants of Its High Potency of α9α10 nAChR. Marine Drugs. 2021; 19(12):709. https://doi.org/10.3390/md19120709
Chicago/Turabian StylePan, Si, Yingxu Fan, Xiaopeng Zhu, Yi Xue, Sulan Luo, and Xinquan Wang. 2021. "From Crystal Structures of RgIA4 in Complex with Ac-AChBP to Molecular Determinants of Its High Potency of α9α10 nAChR" Marine Drugs 19, no. 12: 709. https://doi.org/10.3390/md19120709
APA StylePan, S., Fan, Y., Zhu, X., Xue, Y., Luo, S., & Wang, X. (2021). From Crystal Structures of RgIA4 in Complex with Ac-AChBP to Molecular Determinants of Its High Potency of α9α10 nAChR. Marine Drugs, 19(12), 709. https://doi.org/10.3390/md19120709








