Engineering Bafilomycin High-Producers by Manipulating Regulatory and Biosynthetic Genes in the Marine Bacterium Streptomyces lohii
Abstract
1. Introduction
2. Results
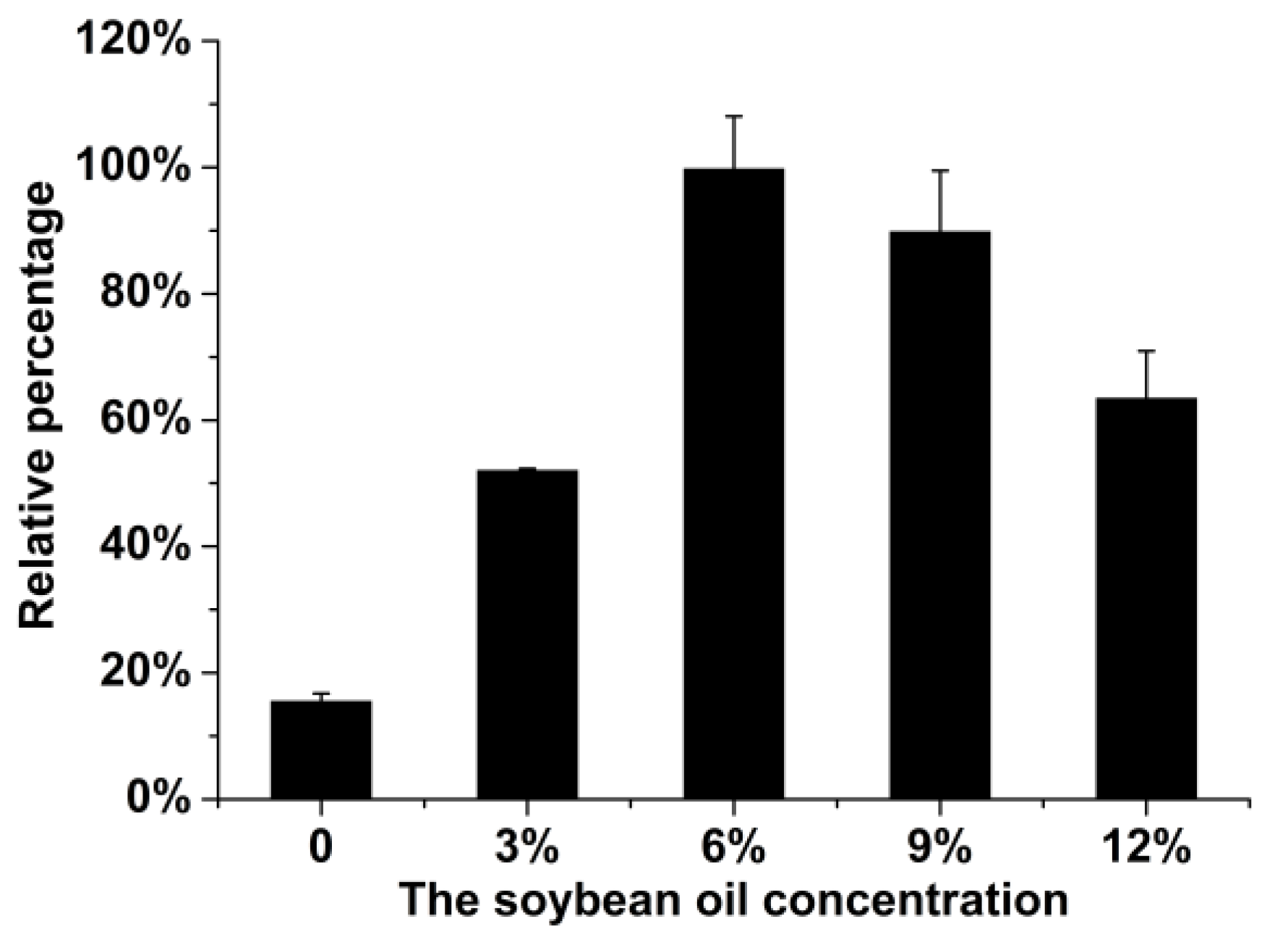
2.1. Optimization of the Fermentation Medium
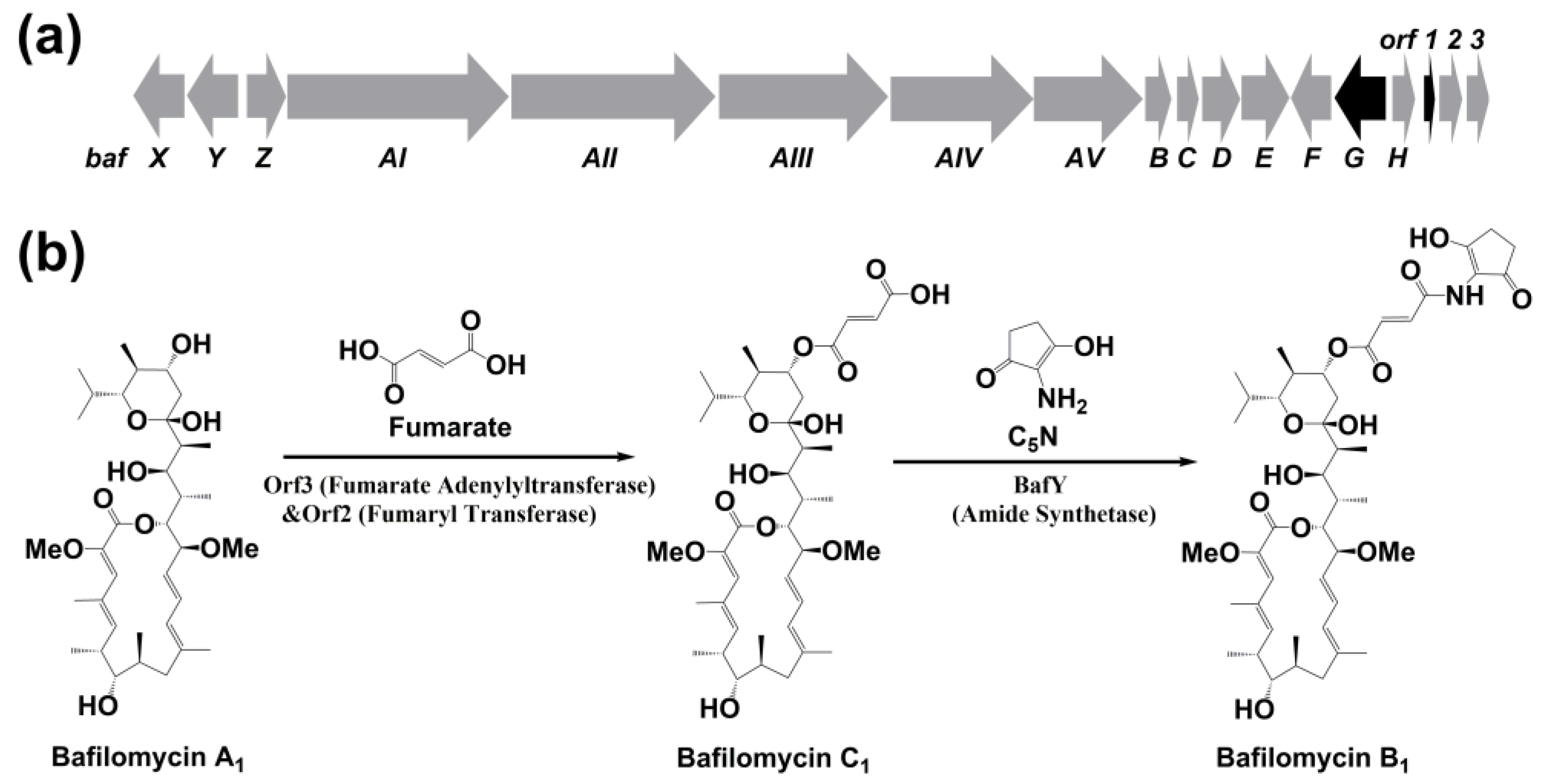
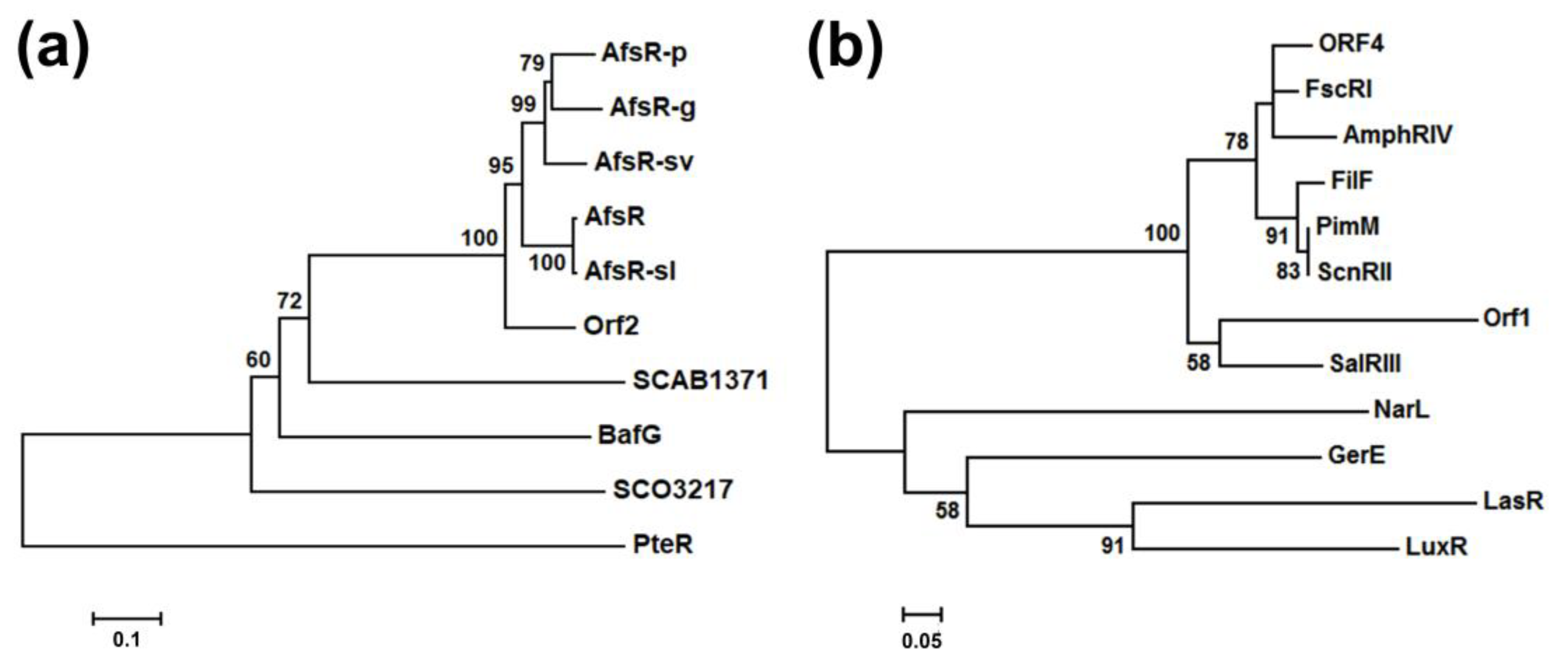
2.2. Bioinformatics Analysis of the Regulatory Genes bafG and orf1
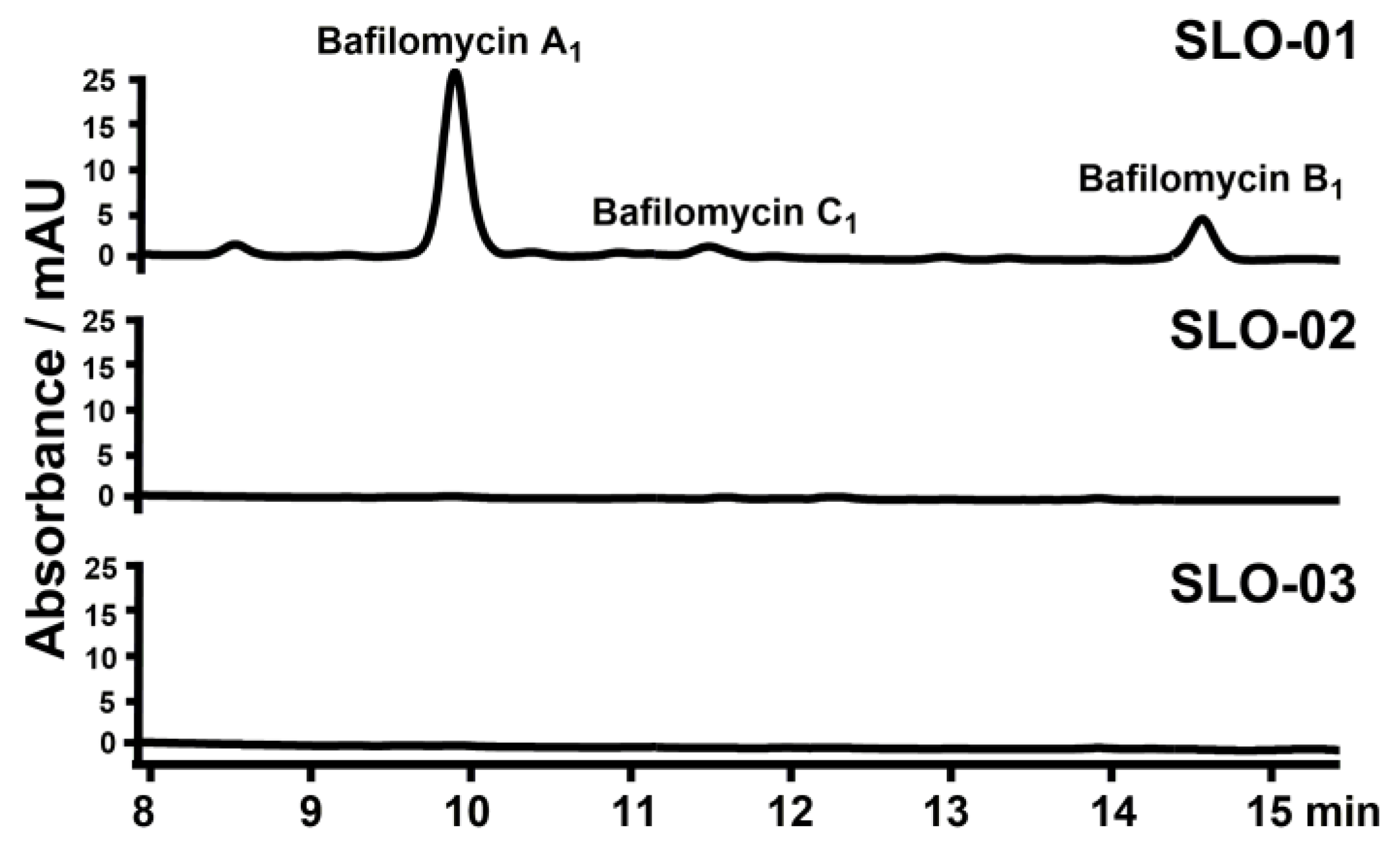
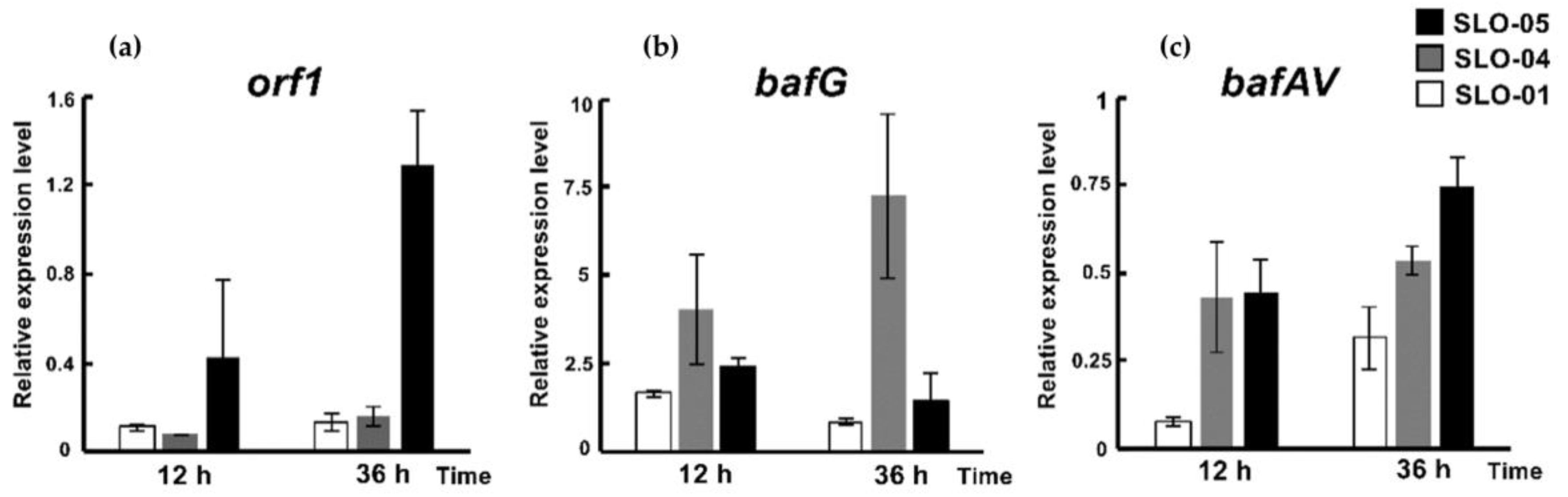
2.3. The Regulatory Roles of bafG and orf1 in Bafilomycin Production
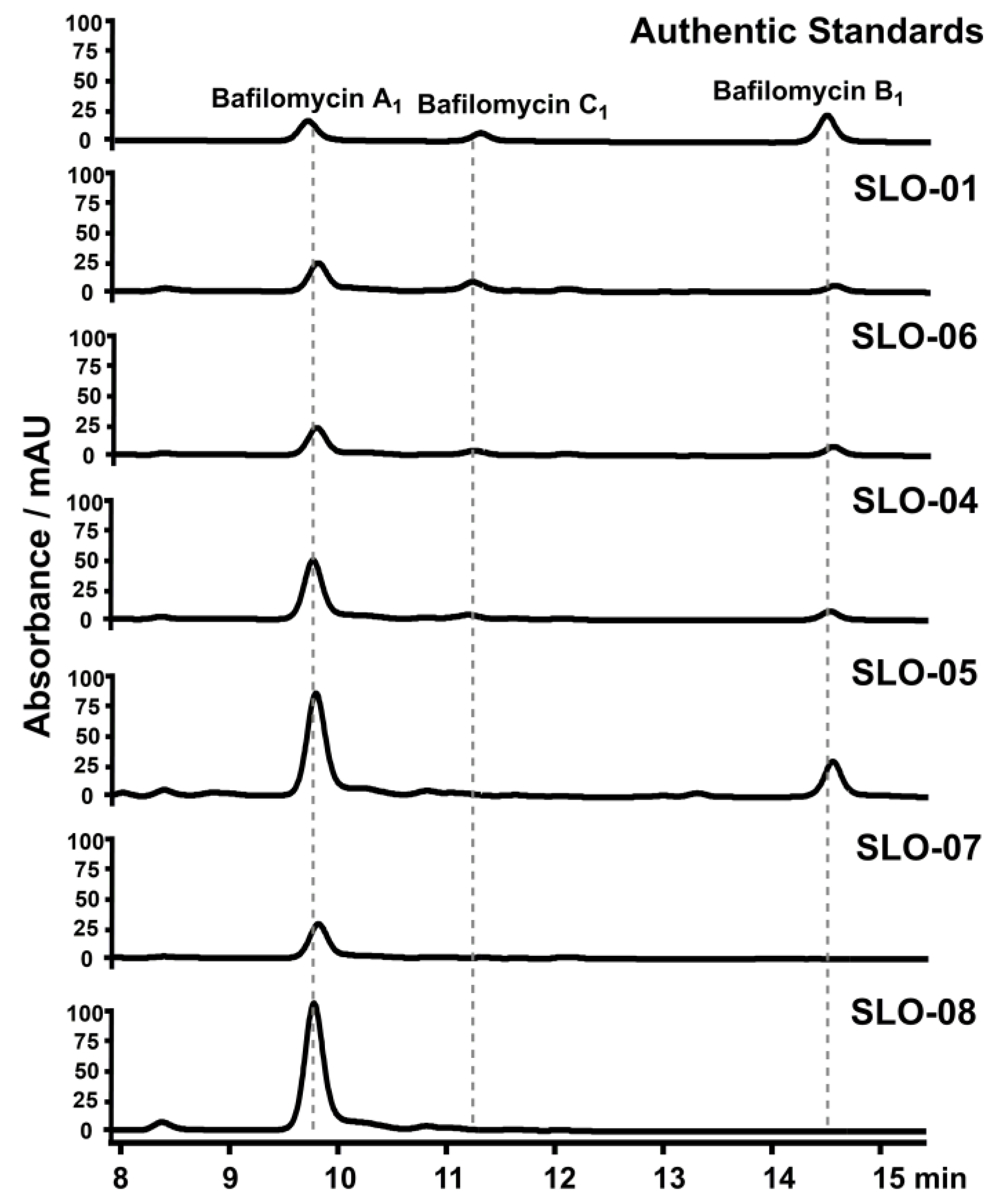
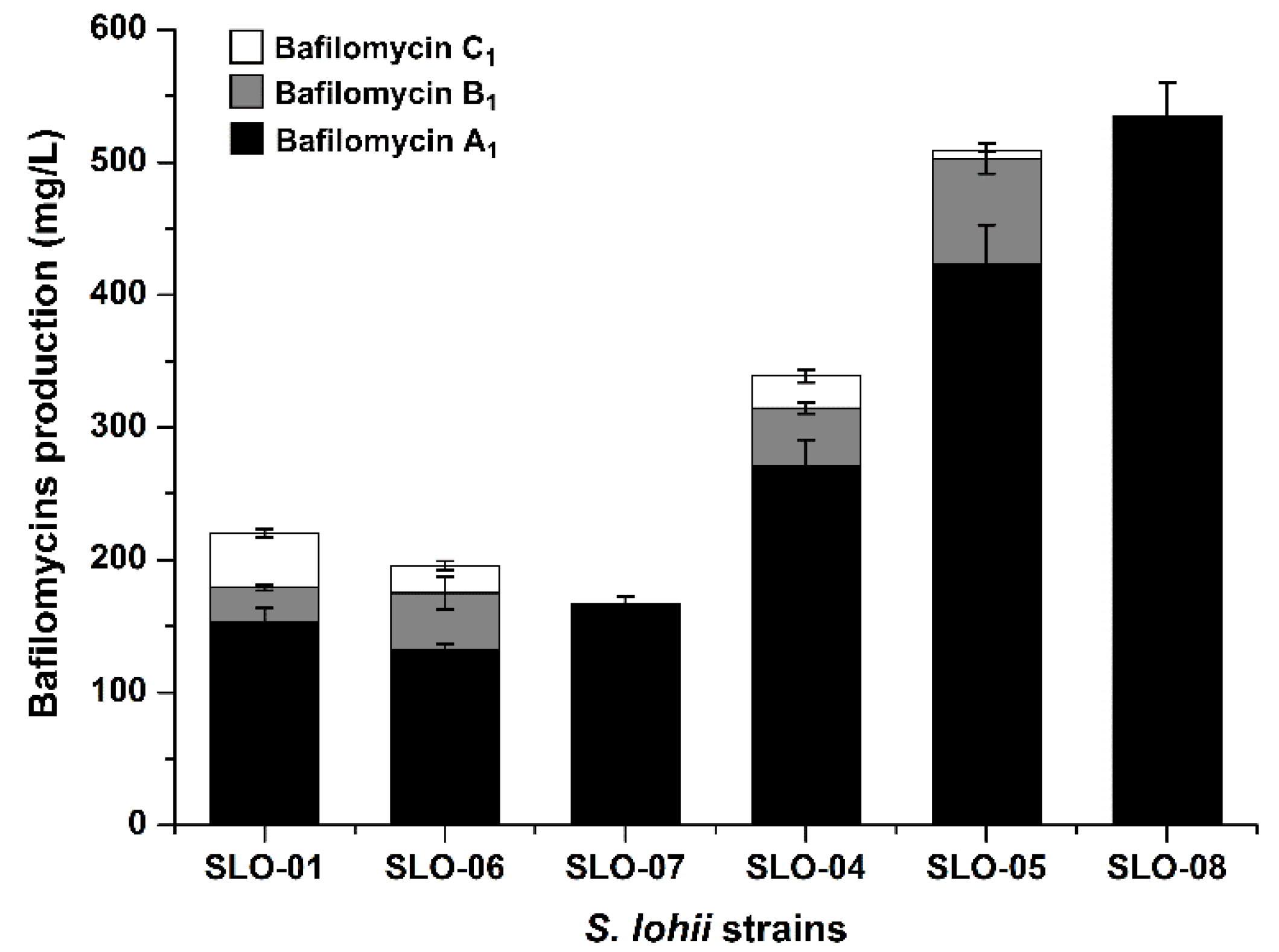
2.4. Construction of Bafilomycin A1 High-Producing Strains
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Strains, Plasmids, and Bacterial Growth Conditions
4.3. DNA Sequencing and Bioinformatics Analysis
4.4. Construction of the Suicide Knockout Vectors
4.5. Gene Inactivation in S. lohii
4.6. Construction of Integrative Plasmids for Regulatory Gene Overexpression
4.7. Overexpression of Regulatory Genes
4.8. Genotypic Confirmation of S. lohii Mutants
4.9. Fermentation and HPLC Analysis
4.10. Transcriptional Analysis of the Wild Type and Mutant S. lohii Strains by qRT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Werner, G.; Hagenmaier, H.; Dpautz, H.; Baumgartner, A.; Zähner, H. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. 1984, 37, 110–117. [Google Scholar] [CrossRef]
- Kinashi, H.; Someno, K.; Sakaguchi, K. Isolation and characterization of concanamycins A, B and C. J. Antibiot. 1984, 37, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Wilton, J.; Hokanson, G.; French, J.C. PD 118,576: A new antitumor macrolide antibiotic. J. Antibiot. 1985, 38, 1449–1452. [Google Scholar] [CrossRef] [PubMed]
- Bowman, E.J.; Siebers, A.; Altendorf, K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 1988, 85, 7972–7976. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.; Hagenmaier, H.; Albert, K.; Kohlshorn, H. The structure of the bafilomycins, a new group of macrolide antibiotics. Tetrahedron Lett. 1983, 24, 5193–5196. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, M.; Chung, H.; Weng, C.; Su, J.; Yang, Y.; Su, Y.; Chang, Y.; Kuo, J.; Wu, Y. Bafilomycin M, a new cytotoxic bafilomycin produced by a Streptomyces sp. isolated from a marine sponge Theonella sp. Tetrahedron Lett. 2016, 57, 4863–4865. [Google Scholar] [CrossRef]
- Li, J.-Q.; Zhao, H.-W.; Ma, Z.-J. Cytotoxic bafilomycin analogues 6/5/5 with tricyclic ring system from a marine-derived Streptomyces sp. Tetrahedron Lett. 2020, 61, 151874. [Google Scholar] [CrossRef]
- Nara, A.; Hashimoto, T.; Komatsu, M.; Nishiyama, M.; Kuzuyama, T.; Ikeda, H. Characterization of bafilomycin biosynthesis in Kitasatospora setae KM-6054 and comparative analysis of gene clusters in Actinomycetales microorganisms. J. Antibiot. 2017, 70, 616–624. [Google Scholar] [CrossRef]
- Li, Z.; Du, L.; Zhang, W.; Zhang, X.; Jiang, Y.; Liu, K.; Men, P.; Xu, H.; Fortman, J.L.; Sherman, D.H. Complete elucidation of the late steps of bafilomycin biosynthesis in Streptomyces lohii. J. Biol. Chem. 2017, 292, 7095–7104. [Google Scholar] [CrossRef]
- Wu, C.; Medema, M.H.; Läkamp, R.M.; Zhang, L.; Dorrestein, P.C.; Choi, Y.H.; van Wezel, G.P. Leucanicidin and endophenasides result from methyl-rhamnosylation by the same tailoring enzymes in Kitasatospora sp. MBT66. ACS Chem. Biol. 2015, 11, 478–490. [Google Scholar] [CrossRef]
- Mauvezin, C.; Nagy, P.; Juhász, G.; Neufeld, T.P. Autophagosome–lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 2015, 6, 7007. [Google Scholar] [CrossRef] [PubMed]
- Del Poeta, M.; Cruz, M.C.; Cardenas, M.E.; Perfect, J.R.; Heitman, J. Synergistic antifungal activities of bafilomycin A1, fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob. Agents Chemother. 2000, 44, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Miyazawa, K.; Moriya, S.; Ohtomo, T.; Che, X.; Naito, M.; Itoh, M.; Tomoda, A. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: Crosstalk among proteasome, autophagy-lysosome and ER stress. Int. J. Oncol. 2011, 38, 643. [Google Scholar]
- Li, L.Q.; Xie, W.J.; Pan, D.; Chen, H.; Zhang, L. Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity of gastric cancer cells. Tumor Biol. 2016, 37, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Shen, H.M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S.; Altendorf, K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 1997, 200, 1–8. [Google Scholar]
- Gagliardi, S.; Gatti, P.A.; Belfiore, P.; Zocchetti, A.; Clarke, G.D.; Farina, C. Synthesis and structure-activity relationships of bafilomycin A1 derivatives as inhibitors of vacuolar H+-ATPase. J. Med. Chem. 1998, 41, 1883–1893. [Google Scholar] [CrossRef] [PubMed]
- Kleinbeck, F.; Carreira, E.M. Total synthesis of bafilomycin A1. Angew. Chem. Int. Edit. 2009, 48, 578–581. [Google Scholar] [CrossRef]
- Scheidt, K.A.; Bannister, T.D.; Tasaka, A.; Wendt, M.D.; Savall, B.M.; Fegley, G.J.; Roush, W.R. Total synthesis of (-)-bafilomycin A1. J. Am. Chem. Soc. 2002, 124, 6981–6990. [Google Scholar] [CrossRef]
- Zhang, W.; Fortman, J.L.; Carlson, J.C.; Yan, J.; Liu, Y.; Bai, F.; Guan, W.; Jia, J.; Matainaho, T.; Sherman, D.H.; et al. Characterization of the bafilomycin biosynthetic gene cluster from Streptomyces lohii. ChemBioChem 2013, 14, 301–306. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Kim, H.S.; Kim, S.H.; Oh, H.R.; Nam, D.H. Organization and characterization of a biosynthetic gene cluster for bafilomycin from Streptomyces griseus DSM 2608. AMB Express 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Kim, S.H.; Oh, H.R.; Kwon, E.; Nam, D.H. Analysis of a draft genome sequence of Kitasatospora cheerisanensis KCTC 2395 producing bafilomycin antibiotics. J. Microbiol. 2015, 53, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Li, G.; Jiang, Y.; Han, L.; Huang, X.; Lu, T.; Jiang, C. The complete genome sequence of Streptomyces albolongus YIM 101047, the producer of novel bafilomycins and odoriferous sesquiterpenoids. J. Biotechnol. 2017, 262, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, H.; Huang, D.; Jin, L.; Wang, C.; Wen, J. Comparative proteomic and metabolomic analysis of Streptomyces tsukubaensis reveals the metabolic mechanism of FK506 overproduction by feeding soybean oil. Appl. Microbiol. Biotechnol. 2017, 101, 2447–2465. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Kieser, T. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Bierman, M.I.; Logan, R.; Obrien, K.; Seno, E.T.; Rao, R.N.; Schoner, B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef]
- Li, P.; Li, J.; Guo, Z.; Tang, W.; Han, J.; Meng, X.; Hao, T.; Zhu, Y.; Zhang, L.; Chen, Y. An efficient blue-white screening based gene inactivation system for Streptomyces. Appl. Microbiol. Biotechnol. 2015, 99, 1923–1933. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tanaka, A.; Takano, Y.; Ohnishi, Y.; Horinouchi, S. AfsR recruits RNA polymerase to the afsS promoter: A model for transcriptional activation by SARPs. J. Mol. Biol. 2007, 369, 322–333. [Google Scholar] [CrossRef]
- Chater, K.F.; Chandra, G. The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. J. Microbiol. 2008, 46, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Leskiw, B.K.; Lawlor, E.J.; Fernandez-Abalos, J.M.; Chater, K.F. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc. Natl. Acad. Sci. USA 1991, 88, 2461–2465. [Google Scholar] [CrossRef] [PubMed]
- Devine, J.H.; Shadel, G.S.; Baldwin, T.O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC 7744. Proc. Natl. Acad. Sci. USA 1989, 86, 5688–5692. [Google Scholar] [CrossRef]
- Ducros, V.M.; Lewis, R.J.; Verma, C.S.; Dodson, E.J.; Leonard, G.; Turkenburg, J.P.; Murshudov, G.N.; Wilkinson, A.J.; Brannigan, J.A. Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis. J. Mol. Biol. 2001, 306, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Gambello, M.J.; Iglewski, B.H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 1991, 173, 3000. [Google Scholar] [CrossRef]
- Stewart, V.; Parales, J., Jr.; Merkel, S.M. Structure of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 1988, 170, 1589–1597. [Google Scholar] [CrossRef]
- Henikoff, S.; Wallace, J.C.; Brown, J.P. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 1989, 183, 111–132. [Google Scholar]
- Fuqua, W.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef]
- Knirschová, R.; Nováková, R.; Fecková, L.; Timko, J.; Turna, J.; Bistáková, J.; Kormanec, J. Multiple regulatory genes in the salinomycin biosynthetic gene cluster of Streptomyces albus CCM 4719. Folia Microbiol. 2007, 52, 359–365. [Google Scholar] [CrossRef]
- Santos-Aberturas, J.; Vicente, C.M.; Guerra, S.M.; Payero, T.D.; Martín, J.F.; Aparicio, J.F. Molecular control of polyene macrolide biosynthesis: Direct binding of the regulator PimM to eight promoters of pimaricin genes and identification of binding boxes. J. Biol. Chem. 2011, 286, 9150–9161. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, Z.; Li, H.; Chen, X.L.; Deng, Z.; Bai, L.; Pang, X. Production of the antibiotic FR-008/Candicidin in Streptomyces sp. FR-008 is coregulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology 2015, 161, 539. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Greenberg, E.P. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. USA 1991, 88, 11115–11119. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Gao, S. The effect of surfactant on fermentation of kitasamycin in Streptomyces kitasatoensis. Biotechnol. Appl. Biochem. 2016, 63, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Kirst, H.A.; Wild, G.; Baltz, R.; Seno, E.; Hamill, R.; Paschal, J.; Dorman, D. Elucidation of structure of novel macrolide antibiotics produced by mutant strains of Streptomyces fradiae. J. Antibiot. 1983, 36, 376–382. [Google Scholar] [CrossRef]
- Horinouchi, S.; Kito, M.; Nishiyama, M.; Furuya, K.; Hong, S.-K.; Miyake, K.; Beppu, T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3 (2). Gene 1990, 95, 49–56. [Google Scholar] [CrossRef]
- Horinouchi, S. AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3 (2). J. Ind. Microbiol. Biotechnol. 2003, 30, 462–467. [Google Scholar] [CrossRef]
- Maharjan, S.; Oh, T.-J.; Lee, H.C.; Sohng, J.K. Identification and functional characterization of an afsR homolog regulatory gene from Streptomyces venezuelae ATCC 15439. J. Microbiol. Biotechnol. 2009, 19, 121–127. [Google Scholar]
- Seipke, R.F.; Song, L.; Bicz, J.; Laskaris, P.; Yaxley, A.M.; Challis, G.L.; Loria, R. The plant pathogen Streptomyces scabies 87-22 has a functional pyochelin biosynthetic pathway that is regulated by TetR-and AfsR-family proteins. Microbiology 2011, 157, 2681–2693. [Google Scholar] [CrossRef]
- Wang, W.; Wang, H.; Hu, H.; Peng, H.; Zhang, X. Overexpression of afsR and optimization of metal chloride to improve lomofungin production in Streptomyces lomondensis S015. J. Microbiol. Biotechnol. 2015, 25, 672–680. [Google Scholar] [CrossRef]
- Wilson, D.J.; Xue, Y.; Reynolds, K.A.; Sherman, D.H. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 2001, 183, 3468–3475. [Google Scholar] [CrossRef]
- Hur, Y.-A.; Choi, S.-S.; Sherman, D.H.; Kim, E.-S. Identification of TmcN as a pathway-specific positive regulator of tautomycetin biosynthesis in Streptomyces sp. CK4412. Microbiology 2008, 154, 2912–2919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuščer, E.; Coates, N.; Challis, I.; Gregory, M.; Wilkinson, B.; Sheridan, R.; Petković, H. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J. Bacteriol. 2007, 189, 4756–4763. [Google Scholar] [CrossRef]
- Intra, B.; Euanorasetr, J.; Nihira, T.; Panbangred, W. Characterization of a gamma-butyrolactone synthetase gene homologue (stcA) involved in bafilomycin production and aerial mycelium formation in Streptomyces sp. SBI034. Appl. Microbiol. Biotechnol. 2016, 100, 2749–2760. [Google Scholar] [CrossRef] [PubMed]
- Aroonsri, A.; Kitani, S.; Hashimoto, J.; Kosone, I.; Izumikawa, M.; Komatsu, M.; Fujita, N.; Takahashi, Y.; Shinya, K.; Ikeda, H. Pleiotropic control of secondary metabolism and morphological development by KsbC, a butyrolactone autoregulator receptor homologue in Kitasatospora setae. Appl. Environ. Microb. 2012, 78, 8015–8024. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-U.; Lee, C.-K.; Hwang, Y.-I.; Kinoshita, H.; Nihira, T. Cloning and functional analysis by gene disruption of a gene encoding a γ-butyrolactone autoregulator receptor from Kitasatospora setae. J. Bacteriol. 2004, 186, 3423–3430. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Strain or Plasmid | Characteristics | Reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5a | Cloning host | [25] |
| ET12567/pUZ8002 | Interspecies conjugation | [26] |
| Streptomyces strains | ||
| SLO-01 | Streptomyces lohii ATCC BAA-1276 (wild-type strain) | [20] |
| SLO-02 | S. lohii ∆bafG | This study |
| SLO-03 | S. lohii ∆orf1 | This study |
| SLO-04 | S. lohii/pSET152-ermE*-bafG | This study |
| SLO-05 | S. lohii/pSET152-ermE*-orf1 | This study |
| SLO-06 | S. lohii/pSET152-ermE* | This study |
| SLO-07 | S. lohii ∆orf2&orf3 | [9] |
| SLO-08 | S. lohii ∆orf2&orf3/pSET152s-ermE*-orf1 | This study |
| Plasmids | ||
| pSET152-ermE* | Apramycin resistance | [27] |
| pSET152s-ermE* | Spectinomycin resistance | This study |
| pIJ778 | Spectinomycin resistance | [28] |
| pCIMt002 | Ampicillin and Apramycin resistance | [29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Li, S.; Du, L.; Zhang, X.; Jiang, Y.; Liu, W.; Zhang, W.; Li, S. Engineering Bafilomycin High-Producers by Manipulating Regulatory and Biosynthetic Genes in the Marine Bacterium Streptomyces lohii. Mar. Drugs 2021, 19, 29. https://doi.org/10.3390/md19010029
Li Z, Li S, Du L, Zhang X, Jiang Y, Liu W, Zhang W, Li S. Engineering Bafilomycin High-Producers by Manipulating Regulatory and Biosynthetic Genes in the Marine Bacterium Streptomyces lohii. Marine Drugs. 2021; 19(1):29. https://doi.org/10.3390/md19010029
Chicago/Turabian StyleLi, Zhong, Shuai Li, Lei Du, Xingwang Zhang, Yuanyuan Jiang, Wenhua Liu, Wei Zhang, and Shengying Li. 2021. "Engineering Bafilomycin High-Producers by Manipulating Regulatory and Biosynthetic Genes in the Marine Bacterium Streptomyces lohii" Marine Drugs 19, no. 1: 29. https://doi.org/10.3390/md19010029
APA StyleLi, Z., Li, S., Du, L., Zhang, X., Jiang, Y., Liu, W., Zhang, W., & Li, S. (2021). Engineering Bafilomycin High-Producers by Manipulating Regulatory and Biosynthetic Genes in the Marine Bacterium Streptomyces lohii. Marine Drugs, 19(1), 29. https://doi.org/10.3390/md19010029





