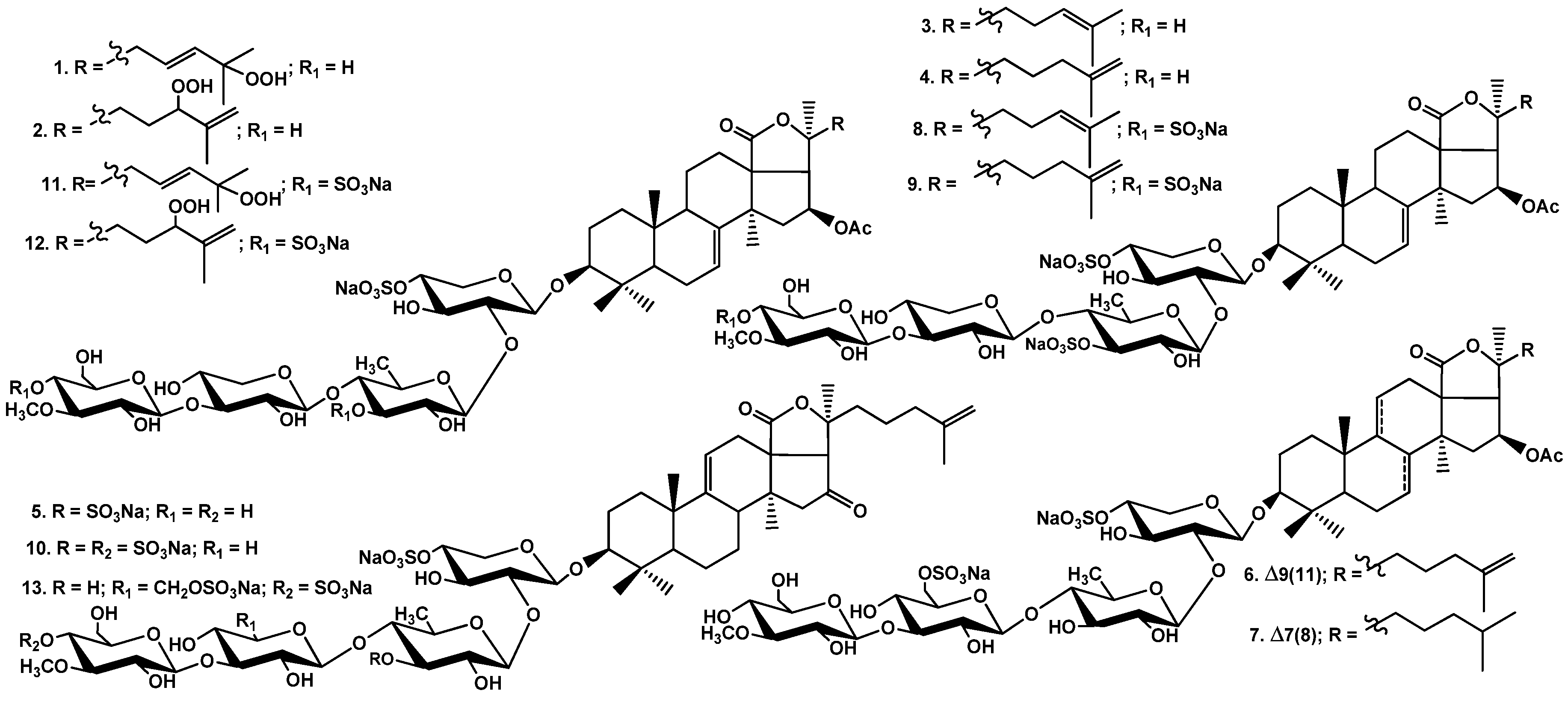
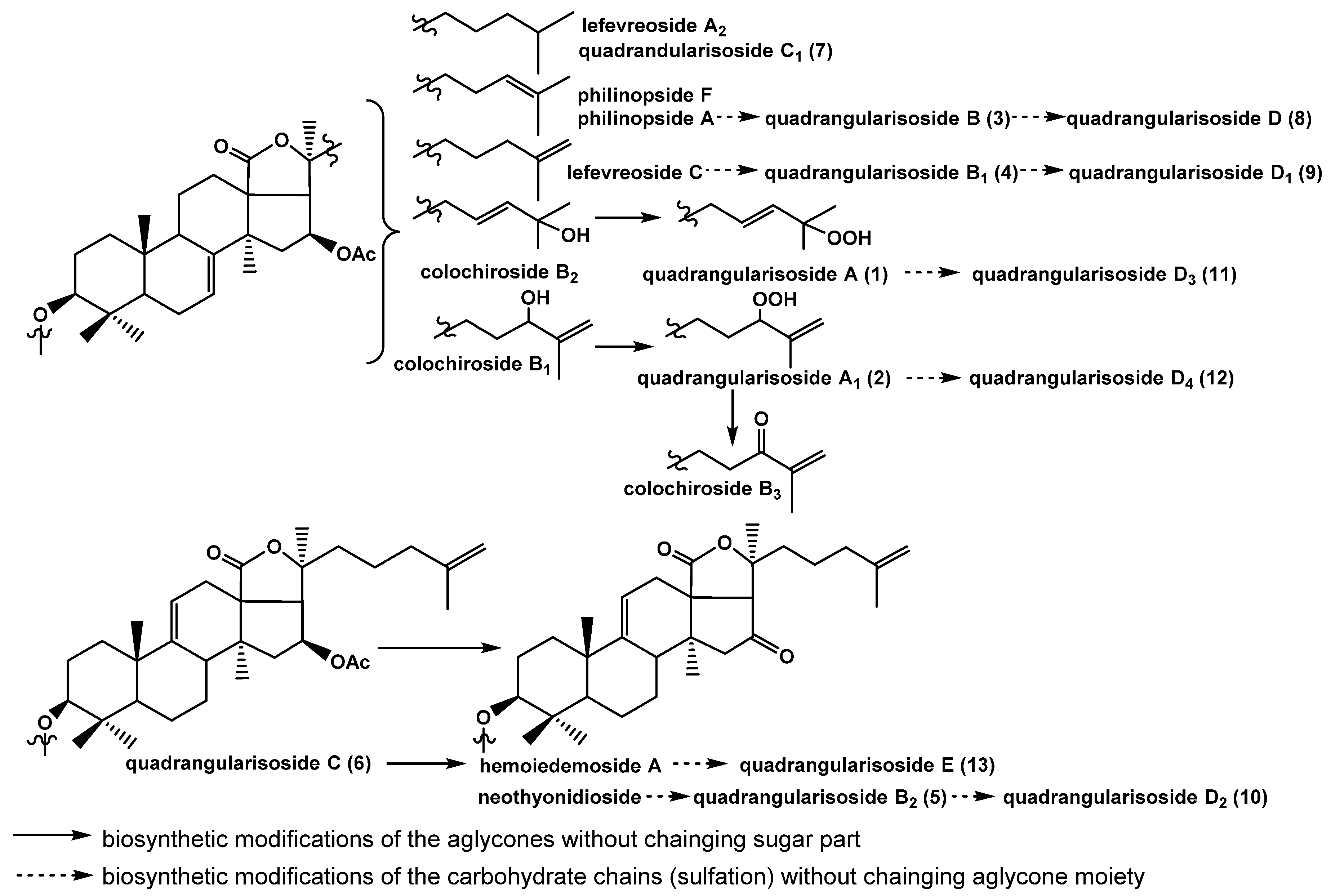
2.2. Structural Elucidation of the Glycosides
The concentrated ethanolic extract of the sea cucumber
Colochirus quadrangularis was chromatographed on a Polychrom-1 column (powdered Teflon, Biolar, Latvia). The glycosides were eluted with 50% EtOH and separated by chromatography on a Si gel column using CHCl
3/EtOH/H
2O (100:100:17) and (100:125:25) as mobile phases. The obtained fractions were subsequently subjected to HPLC on a silica-based Supelcosil LC-Si (4.6 × 150 mm) column and on a reversed-phase semipreparative Supelco Discovery HS F5-5 (10 × 250 mm) column to yield compounds
1–
13 (
Figure 1) along with nine known earlier glycosides: colochirosides B
1, B
2, and B
3 [
20], lefevreosides A
2 and C [
21], neothyonidioside [
22], hemoiedemoside A [
23], and philinopsides A [
12] and F [
13], which were isolated earlier from different species of sea cucumbers. The known compounds were identified by the comparison of their
1H and
13C NMR spectra with those reported in the literature.
The
1H and
13C NMR spectra corresponding to the carbohydrate chains of quadrangularisosides A (
1) and A
1 (
2) (
Table 1) were identical to each other and to those of known compounds isolated from this species: colochirosides B
1, B
2, and B
3, lefevreosides A
2 and C, neothyonidioside and philinopside A, having linear tetrasaccharide monosulfated carbohydrate moieties with the xylose residue as the third unit. Such a sugar chain is common in the glycosides of sea cucumbers of different taxa [
2].
The molecular formula of quadrangularisoside A (
1) was determined to be C
55H
85O
27SNa from the [M
Na − Na]
− ion peak at
m/
z 1209.5004 (calc. 1209.5004) in the (−)HR-ESI-MS and [M
Na + Na]
+ ion peak at
m/
z 1255.4779 (calc. 1255.4789) in the (+)HR-ESI-MS. The analysis of the
13C and
1H NMR spectra of the aglycone part of
1 suggested the presence of an 18(20)-lactone [from the signals of C(18) at δ
C 180.0 and C(20) at δ
C 84.8], a 7(8)-double bond [from the signal of secondary carbon C(7) at δ
C 120.3 and the corresponding proton signal at δ
H 5.61 (m, H(7)), the signal of quaternary carbon C(8) at δ
C 145.5] as well as the
β-
OAc group at C(16) [from the signal of carbon at δ
C 74.8 (C(16)) and the corresponding proton signal at δ
H 5.92 (H(16), q,
J = 8.5 Hz) along with the signals of two carbons of the
O-acetic group at δ
C 170.7 and 21.1 and a signal of protons of the methyl group at δ
H 1.97(s)] (
Table 2). All these data indicated the presence of a holostane-type nucleus in
1. The signals of olefinic carbons at δ
C 124.2 (C(23)) and 139.5 (C(24)) indicated the presence of a double bond in the side chain of quadrangularisoside A (
1). The δ
C values of the olefinic carbons were close to those in the
13C NMR spectrum of psolusoside D
3 [
24] that allowed supposing the 23(24)-position of the double bond. The HMBC correlations from methyl groups H(26) and H(27) to the olefinic carbons C(23) and C(24) corroborated the position of the double bond. The coupling pattern of olefinic protons H(23) (δ
H 5.71, dt,
J = 6.6; 15.5 Hz) and H(24) (δ
H 5.97, d,
J = 15.8 Hz) indicated a 23
E-configuration of the double bond in
1. The signal of the tertiary bearing oxygen carbon C(25) was deduced from the HMBC correlations between the signals of methyl groups H(26) (δ
H 1.46, s) and H(27) (δ
H 1.48, s) and the signal at δ
C 81.3. This chemical shift was also close to that of the corresponding carbon (δ
C 80.8, C(25)) in the spectrum of psolusoside D
3 [
24]. So, the NMR and HR-ESI-MS data indicated the presence of a 25-hydroperoxy-23
E-ene fragment in the side chain of
1. This is the third finding of a hydroperoxyl group in the triterpene glycosides aglycones of sea cucumbers [
24,
25].
The (+)ESI-MS/MS of 1 demonstrated the fragmentation of the [MNa + Na]+ ion at m/z 1255.5. The peaks of fragment ions were observed at m/z 1223.5 [MNa + Na − OOH + H]+, 1103.5 [MNa + Na − OOH − NaSO4 + H]+, 927.5 [MNa + Na − OOH − NaSO4 − C7H12O5 (MeGlc) + H]+, 795.4 [MNa + Na − OOH − NaSO4 − C7H12O5 (MeGlc) − C5H8O4 (Xyl) + H]+, 729.2 [MNa + Na − C32H47O6 (Agl) + H]+, 649.3 [MNa + Na− OOH − NaSO4 − C7H12O5 (MeGlc) − C5H8O4 (Xyl) − C6H10O4 (Qui) + H]+, 609.2 [MNa + Na − C32H47O6 (Agl) − NaHSO4]+, 477.1 [MNa + Na − C32H47O6 (Agl) − NaHSO4 − C5H8O4 (Xyl) + H]+, corroborating the structure of quadrangularisoside A (1).
All these data indicate that quadrangularisoside A (1) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-25-peroxy-16β-acetoxyholosta-7,23E-diene.
The molecular formula of quadrangularisoside A
1 (
2) was determined to be the same as that of
1 (C
55H
85O
27SNa) from the [M
Na − Na]
− ion peak at
m/
z 1209.5006 (calc. 1209.5004) in the (−)HR-ESI-MS and [M
Na + Na]
+ ion peak at
m/
z 1255.4772 (calc. 1255.4789) in the (+)HR-ESI-MS. The signals in the
13C NMR spectrum of
2 assigning to triterpene nucleus (C(1)–C(20), C(30)–C(32)) coincided with the corresponding signals in the spectrum of
1 indicating the difference of these glycoside only in the side chains structures (
Table 3). An isolated spin system formed by the protons H(22)–H(24) was deduced from the
1H,
1H-COSY spectrum of
2. The signal of H(24) was deshielded and observed at δ
H 4.51 (brt,
J = 6.2 Hz) due to the attachment of a hydroperoxyl group to C(24) in the glycoside
2, which was corroborated by the deshielding of the signal of C(24) to δ
C 89.2. The signals of olefinic carbons at δ
C 144.9 (C(25)) and 113.6 (C(26) as well as olefinic protons at δ
H 5.14 (brs, H(26)) and 5.02 (brs, H(26′)) indicated the presence of a terminal double bond in the side chain of
2. The positions of a double bond and hydroperoxyl group were confirmed by the HMBC correlations H(24)/C(26); H(26) and H(26′)/C(24), C(27); H(27)/C(24), C(25), C(26). Hence, quadrangularisosides A (
1) and A
1 (
2) are the isomers by the position of the hydroperoxy-ene fragment in their side chains.
The 24(
S)-configuration was assigned to the 24(
S)-hydroxy-25-dehydroechinoside A isolated earlier from the sea cucumber
Actinopyga flammea [
26]. The same configuration of C(24)-stereocenter was established by Mosher′s method in the aglycone of cucumarioside A
7 from the sea cucumber
Eupentacta fraudatrix [
27], which differs from the aglycone of
2 only by a hydroxyl substituent at C(24) instead of a hydroperoxyl group. Thus, 24(
S)-configuration can be attributed to the aglycone of quadrangularisoside A
1 (
2) based on its biogenetic background.
The (+) ESI-MS/MS of 2 demonstrated the fragmentation of the [MNa + Na − OOH]+ ion at m/z 1237.5 corresponding to a dehydrated molecule, whose formation was caused by the presence of a hydroperoxyl group in the aglycone. The peaks of fragment ions were observed at m/z 1177.4 [MNa + Na − OOH − CH3COOH]+, 1117.5 [MNa + Na − OOH − NaHSO4]+. The fragmentation of the [MNa + Na]+ ion at m/z 1237.5 resulted in the formation of fragment ions at the same m/z as in 1: 729.2 [MNa + Na− C32H47O6 (Agl) + H]+, 609.2 [MNa + Na − C32H47O6 (Agl) − NaHSO4]+, 477.1 [MNa + Na − C32H47O6 (Agl) − NaHSO4 − C5H8O4 (Xyl) + H]+, corroborating their isomerism.
All these data indicate that quadrangularisosides A1 (2) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-24S-peroxy-16β-acetoxyholosta-7,25-diene.
The
1H and
13C NMR spectra corresponding to the carbohydrate chains of quadrangularisosides B (
3), B
1 (
4), and B
2 (
5) (
Table 4) were identical to each other and coincided with the spectra of the carbohydrate part of pseudostichoposide B isolated earlier from the sea cucumber
Pseudostichopus trachus (=
P. mollis) (family Pseudostichopodidae, order Persiculida) [
28] and violaceusoside E from
Pseudocolochirus violaceus [
29]. This linear disulfated tetrasaccharide chain was characterized by the attachment of the second sulfate group to C(3) of the quinovose residue. The structure of the carbohydrate chain of glycosides
3–
5 was established by the thorough analysis of the
1H,
1H-COSY, HSQC, and 1D TOCSY spectra for each monosaccharide unit, and the positions of interglycosidic linkages were elucidated based on the ROESY and HMBC correlations (
Table 4). The finding of sugar moieties of the glycosides with the sulfate group at C(3) of the quinovose is very rare. Only three glycosides having such structural features are found so far [
28,
29] among more than 150 known triterpene glycosides from the sea cucumbers.
It is necessary to note that pentactasides B and C were reported earlier to have the disulfated tetrasaccharide chains with the same monosacaccharide composition as in
3–
5 but differing by the position of a second sulfate group that was positioned at C(2) of the quinovose residue [
14]. The analysis of the NMR data provided in the paper [
14] allowed supposing the erroneous interpretation of these data by the authors. When the sulfate group is bonded to C(2) of the monosaccharide residue in the pyranose form, it resulted in the up-field shifting of the signal of anomeric carbon to δ
C approximately 100 [
30]. Whenever the signals of C(1) of a quinovose unit in pentactasides B and C were observed, δ
C 102.5 and 102.9, correspondingly, indicating the absence of a sulfate group at C(2) of this residue. So, the signals of C(2) at δ
C 81.1 and 81.4 and C(3) at δ
C 74.5 and 75.0 of the quinovose units in pentactasides B and C, correspondingly, probably were displaced with each other, and the second sulfate group is located at C(3)Qui2 in these glycosides. Therefore, the structures of pentactasides B and C most likely are identical to those of quadrangularisosides B (
3) and B
1 (
4), correspondingly.
The molecular formula of quadrangularisoside B (
3) was determined to be C
55H
84O
28S
2Na
2 from the [M
2Na − Na]
− ion peak at
m/
z 1279.4489 (calc. 1279.4494) and [M
2Na − 2Na]
2− ion peak at
m/
z 628.2311 (calc. 628.2301) in the (−)HR-ESI-MS as well as from the [M
2Na + Na]
+ ion peak at
m/
z 1325.4272 (calc. 1325.4278) in the (+)HR-ESI-MS. The signals in the
13C NMR spectrum of the aglycone part of
3 were very close to those in the spectrum of cucumarioside A
1 from
E. fraudatrix [
28] and pentactaside B [
14], indicating the identity of their aglycones (
Table 5). This holostane-type aglycone with 7(8)- and 24(25)-double bonds and a 16
β-acetoxy-group is quite common for the glycosides of sea cucumbers of the order Dendrochirotida [
2,
4,
5].
The (−)ESI-MS/MS of 3 showed the fragmentation of the [M2Na − Na]− ion at m/z 1279.5. The peaks of fragment ions were observed at m/z 1219.4 [M2Na − Na − CH3COOH]−, 1177.5 [M2Na − Na − NaSO3 + H]−. The (+)ESI-MS/MS of 3 showed that the fragmentation of the [M2Na + Na]+ ion at m/z 1325.4 resulted in the formation of fragment ions whose peaks were observed at m/z 1223.5 [M2Na + Na − NaSO3 + H]+, 915.4 [M2Na + Na − NaSO3 − C7H12O5 (MeGlc) − C5H8O4 (Xyl) + H]+, 813.4 [M2Na + Na − C32H47O5 (Agl) − H]+, 711.1 [M2Na + Na − C32H47O5 (Agl) − SO3Na]+, 579.1 [M2Na + Na − C32H47O5 (Agl) − C5H7O7SNa (XylSO3Na) − H]+, 535.3 [M2Na + Na − C32H47O5 (Agl) − SO3Na − C7H12O5 (MeGlc)]+, 403.0 [M2Na + Na − C32H47O5 (Agl) − SO3Na − C7H12O5 (MeGlc) − C5H8O4 (Xyl)]+, and 331.1 [M2Na + Na − C32H47O5 (Agl) − C5H7O7SNa (XylSO3Na) − C6H9O7SNa (QuiSO3Na) − H]+, which corroborate the monosaccharide sequence in the carbohydrate chain of 3.
All these data indicate that quadrangularisoside B (3) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16β-acetoxyholosta-7,24-diene.
The molecular formula of quadrangularisoside B
1 (
4) was determined to be C
55H
84O
28S
2Na
2 from the [M
2Na − Na]
− ion peak at
m/
z 1279.4502 (calc. 1279.4494) and [M
2Na − 2Na]
2− ion peak at
m/
z 628.2320 (calc. 628.2301) in the (−)HR-ESI-MS as well as from the [M
2Na + Na]
+ ion peak at
m/
z 1325.4272 (calc. 1325.4278) in the (+)HR-ESI-MS, which was coincident with the formula of quadrangularisoside B (
3) and indicated their isomerism. In the
1H and
13C NMR spectra of the aglycone part of
4, the signals of holostane-type aglycone having 7(8)- and 25(26)-double bonds as well as a 16
β-acetoxy-group were observed (
Table 6). This aglycone is identical to that of pentactasides C [
14] and II [
15] isolated earlier from the same species and is frequently occurred in the glycosides of the sea cucumbers of the order Dendrochirotida [
2,
4,
5,
21,
30].
The (+)ESI-MS/MS of 4 showed the fragmentation of the [M2Na + Na]+ ion at m/z 1325.4. The peaks of fragment ions were observed at m/z 1223.5 [M2Na + Na − NaSO3 + H]+, 1205.5 [M2Na + Na − NaHSO4]+, 1085.5 [M2Na + Na − 2NaHSO4]+, 897.4 [M2Na + Na − NaHSO4 − C7H12O5 (MeGlc) − C5H8O4 (Xyl)]+, 711.1 [M2Na + Na − C32H47O5 (Agl) − SO3Na]+, 579.1 [M2Na + Na − C32H47O5 (Agl) − C5H7O7SNa (XylSO3Na) − H]+, 403.0 [M2Na + Na − C32H47O5 (Agl) − SO3Na − C7H12O5 (MeGlc) − C5H8O4 (Xyl)]+, and 331.1 [M2Na + Na − C32H47O5 (Agl) − C5H7O7SNa (XylSO3Na) − C6H9O7SNa (QuiSO3Na) − H]+, corroborating the structure of quadrangularisoside B1 (4).
All these data indicate that quadrangularisoside B1 (4) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16β-acetoxyholosta-7,25-diene.
The molecular formula of quadrangularisoside B
2 (
5) was determined to be C
53H
80O
27S
2Na
2 from the [M
2Na − Na]
− ion peak at
m/
z 1235.4243 (calc. 1235.4232) and [M
2Na − 2Na]
2− ion peak at
m/
z 606.2189 (calc. 606.2170) in the (−)HR-ESI-MS as well as the [M
2Na + Na]
+ ion peak at
m/
z 1281.4004 (calc. 1281.4016) in the (+)HR-ESI-MS. The signals in the
1H and
13C NMR spectra of the aglycone part of
5 were characteristic of holostane-type aglycone having 9(11)- and 25(26)-double bonds and a 16-keto-group (
Table 7). This aglycone is known as holotoxinogenin, and it was found first in the glycosides named as holotoxins A
1 and B
1, which were isolated from the sea cucumber
Apostichopus japonicus (Stichopodidae, Synallactida) [
31] and are rather common for the glycosides of the sea cucumbers of different orders [
2,
4,
5].
The (+)ESI-MS/MS of 5 demonstrated the fragmentation of the [M2Na + Na]+ ion at m/z 1281.4. The peaks of fragment ions were observed at m/z 1179.4 [M2Na + Na − NaSO3 + H]+, 1059.5 [M2Na + Na − NaSO3 − NaHSO4 + H]+, confirming the presence of two sulfate groups. The peaks of fragment ions at m/z 711.1, 579.1, 403.0, and 331.1 were the same as in the mass spectra of the glycosides 3 and 4 due to the identity of their carbohydrate chains.
All these data indicate that quadrangularisoside B2 (5) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16-ketoholosta-7,25-diene.
The
13C NMR spectra of the carbohydrate moieties of quadrangularisosides C (
6) and C
1 (
7) were identical to each other (
Table 8) and to those of hemoiedemoside A isolated first from the sea cucumber
Hemoiedema spectabilis [
23] and identified by us in the glycosidic sum of
C. quadrangularis. The glycosides
6 and
7 have a tetrasaccharide linear carbohydrate chain differing from that of
1 and
2 by the sulfated by C(6) glucose residue as the third unit in the chain instead of a xylose residue. The carbohydrate chain structure of quadrangularisosides C (
6) and C
1 (
7) was elucidated based on a thorough analysis of 1D and 2D NMR spectra (
Table 8). Besides compounds
6,
7, and hemoiedemoside A, such a sugar moiety is a part of another six glycosides [
32,
33,
34,
35,
36,
37] isolated from the sea cucumbers of the order Dendrochirotida.
The molecular formula of quadrangularisoside C (
6) was determined to be C
56H
86O
29S
2Na
2 from the [M
2Na − Na]
− ion peak at
m/
z 1309.4608 (calc. 1309.4599) and [M
2Na − 2Na]
2− ion peak at
m/
z 643.2369 (calc. 643.2354) in the (−)HR-ESI-MS. In the (+)HR-ESI-MS, the [M
2Na + Na]
+ ion peak observed at
m/
z 1355.4373 (calc. 1355.4384) corresponded to the same molecular formula. The signals of C(1)–C(4) and C(15)–C(32) in the
13C NMR spectrum of
6 (
Table 9) were close to those in the spectrum of quadrangularisoside B
1 (
4), but the signals characteristic for the 9(11)-double bond in the triterpene nucleus [at δ
C 150.6 (C(9)) and 110.9 (C(11)) as well as δ
H 5.16 (m, H(11))] were observed instead of the signals assigned to the 7(8)-double bond in
4. So, these compounds were the isomers by the double bond position in the aglycone nuclei. The aglycone identical to that of quadrangularisoside C (
6) was earlier found only in cladoloside A
3 from the sea cucumber
Cladolabes schmeltzii [
36].
The (−)ESI-MS/MS of 6 demonstrated the fragmentation of the [M2Na − Na]− ion at m/z 1309.5. The peaks of fragment ions were observed at m/z 1249.4 [M2Na − Na − CH3COOH]−, 1189.5 [M2Na − Na − NaHSO4]−, 1129.5 [M2Na − Na − CH3COOH − NaHSO4]−, 677.2 [M2Na − Na − NaHSO4 − C32H47O5 (Agl) − H]−, 563.1 [M2Na − Na − NaHSO4 − C32H47O5 (Agl) − C5H7O3 (Xyl)]−, 417.1 [M2Na − Na − NaHSO4 − C32H47O5 (Agl) − C5H7O3 (Xyl) − C6H10O4 (Qui)]−, and 241.0 [M2Na − Na − NaHSO4 − C32H47O5 (Agl) − C5H7O3 (Xyl) − C6H10O4 (Qui) − C7H12O5 (MeGlc)]−.
All these data indicate that quadrangularisoside C (6) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-6-O-sodium sulfate-β-d-glucopyranosyl-(1→4)-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16β-acetoxyholosta-9(11),25-diene.
The molecular formula of quadrangularisoside C
1 (
7) was determined to be C
56H
88O
29S
2Na
2 from the [M
2Na − Na]
− ion peak at
m/
z 1311.4763 (calc. 1311.4756), [M
2Na − 2Na]
2− ion peak at
m/
z 644.2447 (calc. 644.2432), and [M
2Na + Na]
+ ion peak at
m/
z 1357.4520 (calc. 1357.4540) in the (−) and (+)HR-ESI-MS, correspondingly. The signals of carbons corresponding to the holostane-type nucleus in the
13C NMR spectrum of
7 (
Table 10) were coincident with those in the spectra of quadrangularisosides A (
1), A
1 (
2), B (
3), and B
1 (
4), indicating the presence of the aglycone having a 7(8)-double bond and 16
β-acetoxy-group. The signals of H(22)–H(27) form the isolated spin system in the
1H,
1H-COSY spectrum of
7. So, the presence of an unsubstituted saturated side chain was supposed for this compound. Actually, the
13C NMR spectrum of the aglycone part of
7 was coincident with that of lefevreoside A
2, isolated first from
A. lefevrei [
21] and identified in the glycosidic sum of
C. quadrangularis. The aglycone identical to that of quadrangularisoside C
1 (
7) is frequently occurred in the glycosides of representatives of the order Dendrochirotida [
2,
4,
5].
The (−)ESI-MS/MS of 7 demonstrated the fragmentation of the [M2Na − Na]− ion at m/z 1311.5. The peaks of fragment ions were observed at m/z 797.1 [M2Na − Na − C32H49O5 (Agl) − H]−, 677.2 [M2Na − Na − NaHSO4 − C32H49O5 (Agl) − H]−, 563.1 [M2Na − Na − NaHSO4 − C32H49O5 (Agl) − C5H7O3 (Xyl)]−, 417.1 [M2Na − Na − NaHSO4 − C32H49O5 (Agl) − C5H7O3 (Xyl) − C6H10O4 (Qui)]−, and 241.0 [M2Na − Na − NaHSO4 − C32H49O5 (Agl) − C5H7O3 (Xyl) − C6H10O4 (Qui) − C7H12O5 (MeGlc)]−, which were coincident with those in the MS/MS spectrum of 6, confirming the identity of their carbohydrate chains.
All these data indicate that quadrangularisoside C1 (7) is 3β-O-[3-O-methyl-β-d-glucopyranosyl-(1→3)-6-O-sodium sulfate-β-d-glucopyranosyl-(1→4)-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16β-acetoxyholost-7-ene.
The
13C NMR spectra of the carbohydrate moieties of quadrangularisosides D–D
4 (
8–
12) were coincident to each other (
Table 11), indicating the presence and identity of sugar moieties in these glycosides. In the
1H and
13C NMR spectra of the carbohydrate part of
8–
12, four characteristic doublets at δ
H 4.63–5.12 (
J = 7.1–8.0 Hz), and corresponding to them, four signals of anomeric carbons at δ
C 102.8–104.6 were indicative of a tetrasaccharide chain and
β-configurations of glycosidic bonds. The
1H,
1H-COSY, HSQC, and 1D TOCSY spectra of
8–
12 showed the signals of the isolated spin systems assigned to two xylose residues, one quinovose residue, and one 3-
O-methylglucose residue. These data indicated the same monosaccharide composition of the sugar chain of
8–
12 as in quadrangularisosides of the groups A (
1,
2) and B (
3–
5). The comparison of the
13C NMR spectra of
8–
12 and
3–
5 showed the coincidence of the signals corresponding to the monosaccharide residues from the first to the third. The signals, assigning to the terminal 3-
O-methylglucose residue in
8–
12, were deduced by the analysis of
1H,
1H-COSY and 1D TOCSY spectra. The signal of C(4) MeGlc4 was observed at δ
C 76.2, the signal of C(3) was observed at δ
C 85.2, and the signal of C(5) was observed at δ
C 76.3 in the spectra of
8–
12. Hence, the α- and β-shifting effects due to the attachment of a sulfate group to C(4) MeGlc4 of quadrangularisosides of the group D (
8–
12) were observed in comparison with the δ
C values of the corresponding signals in the spectra of quadrangularisosides of the group B (
3–
5) (δ
C 70.3 for C(4) MeGlc4, δ
C 86.8 for C(3) MeGlc4, and δ
C 77.4 for C(5) MeGlc4) having non-sulfated 3-
O-methylglucose residue in the same position of the carbohydrate chain.
Hence, three sulfate groups are present in the carbohydrate chain of quadrangularisosides of the group D (
8–
12): at C(4) Xyl1—the common position of this functionality for the sea cucumber glycosides, at C(3) Qui2—the rare position of the group, and finally, at C(4) MeGlc4—the unique for the glycosides position of a sulfate group. The similar structural feature was found earlier in the carbohydrate chains of some glycosides from
Psolus fabricii, but the sulfate group was attached to C(4) of the terminal disulfated glucose residue having an additional sulfate group at the C(2) or C(6) position [
37] and in the carbohydrate chain of stichorrenoside B isolated from
Stichopus horrens [
38], where the sulfate group was attached to C(4) of the glucose residue terminal in its disaccharide chain. The majority of known sulfated glycosides contain terminal 3-
O-methylglucose residue with the sulfate group attached to C(6); in contrast, the sulfation of C(4) of the 3-
O-methylglucose residue in the linear tetrasaccharidechain as in compounds
8–
12 has never been discovered earlier in the sea cucumbers glycosides.
The positions of interglycosidic linkages were established by the ROESY and HMBC spectra of
8–
12 (
Table 11) where the cross-peaks between H(1) of the xylose and H(3) (C(3)) of an aglycone, H(1) of the second residue (quinovose) and H(2) (C(2)) of the xylose, H(1) of the third residue (xylose) and H(4) (C(4)) of the second residue (quinovose), H(1) of the fourth residue (3-O-methylglucose) and H-3 (C(3)) of the third residue (xylose) were observed.
The molecular formula of quadrangularisoside D (
8) was determined to be C
55H
83O
31S
3Na
3 from the [M
3Na − Na]
− ion peak at
m/
z 1381.3888 (calc. 1381.1881), [M
3Na − 2Na]
2− ion peak at
m/
z 679.2012 (calc. 679.1995), [M
3Na − 3Na]
3− ion peak at
m/
z 445.1387 (calc. 445.1366), and [M
3Na + Na]
+ ion peak at
m/
z 1427.3663 (calc. 1427.3666) in the (−) and (+) HR-ESI-MS, respectively. The
13C NMR spectrum of the aglycone part of
8 (
Table 5) coincided with that of quadrangularisoside B (
3), indicating the identity of their aglycones.
The (−)ESI-MS/MS of quadrangularisoside D (8) demonstrated the fragmentation of the [M3Na − Na]− ion at m/z 1381.4. The peaks of fragment ions were observed at m/z 1261.4 [M3Na − Na − NaHSO4]−, 1103.4 [M3Na − Na − C7H11O8SNa (MeGlcSO3Na)]−, 751.3 [M3Na − Na −NaHSO4− C32H47O5 (Agl) + H]−, 533.1 [M3Na − Na − 2NaSO3 − C33H47O5 (Agl) − C5H8O4 (Xyl) + H]−, 387.1 [M3Na − Na − 2NaSO3 − C33H47O5 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) + H]−, and 255.0 [M3Na − Na − 2NaSO3 − C33H47O5 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) − C5H8O4 (Xyl) + 2H]−.
All these data indicate that quadrangularisoside D (8) is 3β-O-[4-O-sodium sulfate-3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16β-acetoxyholosta-7,24-diene.
The molecular formula of quadrangularisoside D
1 (
9) was determined to be the same as for quadrangularisoside D (
8) (C
55H
83O
31S
3Na
3) from the [M
3Na − Na]
− ion peak at
m/
z 1381.3891 (calc. 1381.3881), [M
3Na − 2Na]
2− ion peak at
m/
z 679.2015 (calc. 679.1995), and [M
3Na − 3Na]
3− ion peak at
m/
z 445.1390 (calc. 445.1366) in the (−)HR-ESI-MS as well as from the [M
3Na + Na]
+ ion peak at
m/
z 1427.3653 (calc. 1427.3666) in the (+)HR-ESI-MS. The aglycone of quadrangularisoside D
1 (
9) was identical to that of quadrangularisoside B
1 (
4), which was deduced from the coincidence of their
13C NMR spectra (
Table 6).
The (−)ESI-MS/MS of 9 demonstrated the fragmentation of the [M3Na − Na]− ion at m/z 1381.4. The peaks of fragment ions were observed at m/z 1279.5 [M3Na − Na − NaSO3 + H]−, 1177.5 [M3Na − Na − 2NaSO3 + 2H]−, 1001.4 [M3Na − Na − 2NaSO3−C7H12O5 (MeGlc) + 2H]−, 941.4 [M3Na − Na − 2NaSO3−C7H12O5 (MeGlc) − CH3COO + H]−, 751.3 [M3Na − Na − NaHSO4 − C32H47O5 (Agl) + H]−, and 255.0 [M3Na − Na − 2NaSO3 − C33H47O5 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) − C5H8O4 (Xyl) + 2H]−. The (+)ESI-MS/MS of 9 demonstrated the sequential loss by the [M3Na + Na]+ ion at m/z 1427.4 of three sulfate groups (ion peaks at m/z 1325.4 [M3Na + Na − NaSO3 + H]+, 1223.5 [M3Na + Na − 2NaSO3 + 2H]+, and 1121.5 [M3Na + Na − 3NaSO3 + 3H]+).
All these data indicate that quadrangularisoside D1 (9) is 3β-O-[4-O-sodium sulfate-3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16β-acetoxyholosta-7,25-diene.
The molecular formula of quadrangularisoside D
2 (
10) was determined to be C
53H
79O
30S
3Na
3 from the [M
3Na − Na]
− ion peak at
m/
z 1337.3625 (calc. 1337.3619), [M
3Na − 2Na]
2− ion peak at
m/
z 657.1881 (calc. 657.1863), and [M
3Na − 3Na]
3− ion peak at
m/
z 430.4633 (calc. 430.4612) in the (−)HR-ESI-MS as well as from the [M
3Na + Na]
+ ion peak at
m/
z 1383.3390 (calc. 1383.3404) in the (+)HR-ESI-MS. The aglycone of quadrangularisoside D
2 (
10) was identical to that of quadrangularisoside B
2 (
5) (
Table 7), having holotoxinogenin as a triterpenoid nucleus [
31].
The (−)ESI-MS/MS of 10 demonstrated the fragmentation of the [M3Na − Na]− ion at m/z 1381.4. The peaks of fragment ions were observed at m/z 1217.4 [M3Na − Na − NaHSO4]−, 1059.4 [M3Na − Na − C7H11O8SNa (MeGlcSO3Na)]−, 939.4 [M3Na − Na − NaHSO4 − C7H11O8SNa (MeGlcSO3Na)]−, 533.1 [M3Na − Na − 2NaSO3 − C30H43O4 (Agl) − C5H8O4 (Xyl) + H]−, 387.1 [M3Na − Na − 2NaSO3 − C30H43O4 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) + H]−, and 255.0 [M3Na − Na − 2NaSO3 − C30H43O4 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) − C5H8O4 (Xyl) + 2H]−, corroborating the identity of the carbohydrate chains of compounds 8−10. The (+)ESI-MS/MS of 10 demonstrated the loss of two sulfate groups: ion peaks at m/z 1264.4 [M3Na + Na − NaSO4]+ and 1143.4 [M3Na + Na − 2NaSO4]+.
All these data indicate that quadrangularisoside D2 (10) is 3β-O-[4-O-sodium sulfate-3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16-ketoholosta-9(11),25-diene.
The molecular formula of quadrangularisoside D
3 (
11) was determined to be C
55H
83O
33S
3Na
3 from the [M
3Na − Na]
− ion peak at
m/
z 1413.3785 (calc. 1413.3780), [M
3Na − 2Na]
2− ion peak at
m/
z 695.1963 (calc. 695.1944), and [M
3Na − 3Na]
3− ion peak at
m/
z 455.8006 (calc. 455.7998) in the (−)HR-ESI-MS as well as from the [M
3Na + Na]
+ ion peak at
m/
z 1459.3534 (calc. 1459.3564) in the (+)HR-ESI-MS. The aglycone of quadrangularisoside D
3 (
11) was established to be 25-peroxy-16
β-acetoxyholosta-7,23
E-diene-3
β-ol and was identical to the aglycone of quadrangularisoside A (
1) (
Table 2), which was deduced from the comparison of their
13C NMR spectra.
The (−)ESI-MS/MS of 11 demonstrated the fragmentation of the [M3Na − Na]− ion at m/z 1413.4. The peaks of fragment ions were observed at m/z 1261.4 [MNa − Na − OOH − NaSO4]−, 1103.3 [MNa − Na − OOH − C7H11O8SNa (MeGlcSO3Na) + H]−, 723.3 [M3Na − Na − OOH −C7H11O8SNa (MeGlcSO3Na) − C5H8O4 (Xyl) − C6H9O7SNa (QuiSO3Na) + H]−, as well as the ion peaks at the same m/z as in the mass spectra of the other quadrangularisosides of the group D (8−10): 751.3 [M3Na − Na − NaHSO4 − C32H47O6 (Agl)+ H]−, 387.1 [M3Na − Na − 2NaSO3 − C32H47O6 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) + H]−, and 255.0 [M3Na − Na − 2NaSO3 − C32H47O6 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) − C5H8O4 (Xyl) + H]−.
All these data indicate that quadrangularisoside D3 (11) is 3β-O-[4-O-sodium sulfate-3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-25-peroxy-16β-acetoxyholosta-7,23E-diene.
The molecular formula of quadrangularisoside D
4 (
12) was determined to be the same as that for
11 (C
55H
83O
33S
3Na
3) from the [M
3Na − Na]
− ion peak at
m/
z 1413.3762 (calc. 1413.3780), [M
3Na − 2Na]
2− ion peak at
m/
z 695.1954 (calc. 695.1944), and [M
3Na − 3Na]
3− ion peak at
m/
z 455.8011 (calc. 455.7998) in the (−)HR-ESI-MS as well as from the [M
3Na + Na]
+ ion peak at
m/
z 1459.3512 (calc. 1459.3564) in the (+)HR-ESI-MS, indicating the isomerism of these glycosides. The comparison of the
13C NMR spectra of the aglycone parts of quadrangularisosides D
4 (
12) and A
1 (
2) showed their identity (
Table 3). Thus, the aglycone of quadrangularisoside D
4 (
12) was established to be 24
S-peroxy-16
β-acetoxyholosta-7,25-diene-3
β-ol.
The (−)ESI-MS/MS of 12 demonstrated the fragmentation of the [M3Na − Na]− ion at m/z 1413.4. The peaks of fragment ions were observed at m/z 999.4 [MNa − Na − OOH−NaSO3 − C7H11O8SNa (MeGlcSO3Na)]−, 867.4 [MNa − Na − OOH − NaSO3 − C7H11O8SNa (MeGlcSO3Na) − C5H8O4 (Xyl)]−, and 255.0 [M3Na − Na − 2NaSO3 − C32H47O6 (Agl) − C5H8O4 (Xyl) − C6H10O4 (Qui) − C5H8O4 (Xyl) + H]−.
All these data indicate that quadrangularisoside D4 (12) is 3β-O-[4-O-sodium sulfate-3-O-methyl-β-d-glucopyranosyl-(1→3)-β-d-xylopyranosyl-(1→4)-3-O-sodium sulfate-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-24S-peroxy-16β-acetoxyholosta-7,25-diene.
In the
1H and
13C NMR spectra of the carbohydrate moiety of quadrangularisoside E (
13), four characteristic doublets at δ
H 4.67–5.19 (
J = 7.3–7.7 Hz) and corresponding to them four signals of anomeric carbons at δ
C 104.2–104.7 were indicative of a tetrasaccharide chain and
β-configurations of glycosidic bonds (
Table 12). The
1H,
1H-COSY and 1D TOCSY spectra of
13 showed the signals of the isolated spin systems assigned to the xylose, quinovose, glucose, and 3-
O-methylglucose residues. So, the monosaccharide composition of
13 was identical to that in the quadrangularisosides of group C (
6,
7). The comparison of their
13C NMR spectra showed the coincidence of the signals corresponding to the monosaccharide residues from the first to the third. The signals of terminal 3-
O-methylglucose residue in the
13C NMR spectrum of
13 differed from the corresponding signals in the spectrum of
6,
7 due to the attachment of the third sulfate group to C(4) MeGlc4, causing α- and β-shifting effects (δ
C 76.1 (C(4) MeGlc4); δ
C 85.2 (C(3) MeGlc4); δ
C 76.4 (C(5) MeGlc4)) in quadrangularisoside E (
13). Actually, the signals of terminal sugar moieties in the spectra of quadrangularisosides E (
13) and D (
8) were coincident, corroborating the sulfation of C(4) MeGlc4 in glycoside
13. Hence, compound
13 has a novel carbohydrate chain with an unusual position of the third sulfate group in the terminal sugar unit.
The molecular formula of quadrangularisoside E (
13) was determined to be C
54H
81O
31S
3Na
3 from the [M
3Na − Na]
− ion peak at
m/
z 1367.3739 (calc. 1367.3725), [M
3Na − 2Na]
2− ion peak at
m/
z 672.1941 (calc. 672.1916), and [M
3Na − 3Na]
3− ion peak at
m/
z 440.4674 (calc. 440.4647) in the (−)HR-ESI-MS as well as from the [M
3Na + Na]
+ ion peak at
m/
z 1413.3488 (calc. 1413.3509) in the (+)HR-ESI-MS. The aglycone of quadrangularisoside E (
13) was identical to the aglycone of quadrangularisosides B
2 (
5) and D
2 (
10)—holotoxinogenin (
Table 7).
The (−)ESI-MS/MS of 13 demonstrated the fragmentation of the [M3Na − Na]− ion at m/z 1367.4. The peaks of fragment ions were observed at m/z 1247.4 [MNa − Na − NaHSO4]−, 1089.4 [MNa − Na − C7H11O8SNa (MeGlcSO3Na)]−, 969.4 [M3Na − Na − NaHSO4−C7H11O8SNa (MeGlcSO3Na)]−, 825.4 [M3Na − Na − C7H11O8SNa(MeGlcSO3Na) − C6H10O8SNa(GlcSO3Na) + H]−, and 255.0 [M3Na − Na − C30H43O4 (Agl) − C5H7O7SNa(XylSO3Na) − C6H10O4 (Qui) − C6H9O8SNa(GlcSO3Na) − H]−. As result of the fragmentation of the [M3Na + Na]+ ion at m/z 1413.4 in the (+)ESI-MS/MS of 13, the peaks of fragment ions were observed at m/z 963.5 [M3Na + Na − C30H43O3 (Agl) + H]+ and 685.4 [M3Na + Na − C30H43O3 (Agl) − C7H11O8SNa (MeGlcSO3Na) + H]+.
All these data indicate that quadrangularisoside E (13) is 3β-O-[4-O-sodium sulfate-3-O-methyl-β-d-glucopyranosyl-(1→3)-6-O-sodium sulfate-β-d-glucopyranosyl-(1→4)-β-d-quinovopyranosyl-(1→2)-4-O-sodium sulfate-β-d-xylopyranosyl]-16-ketoholosta-9(11),25-diene.
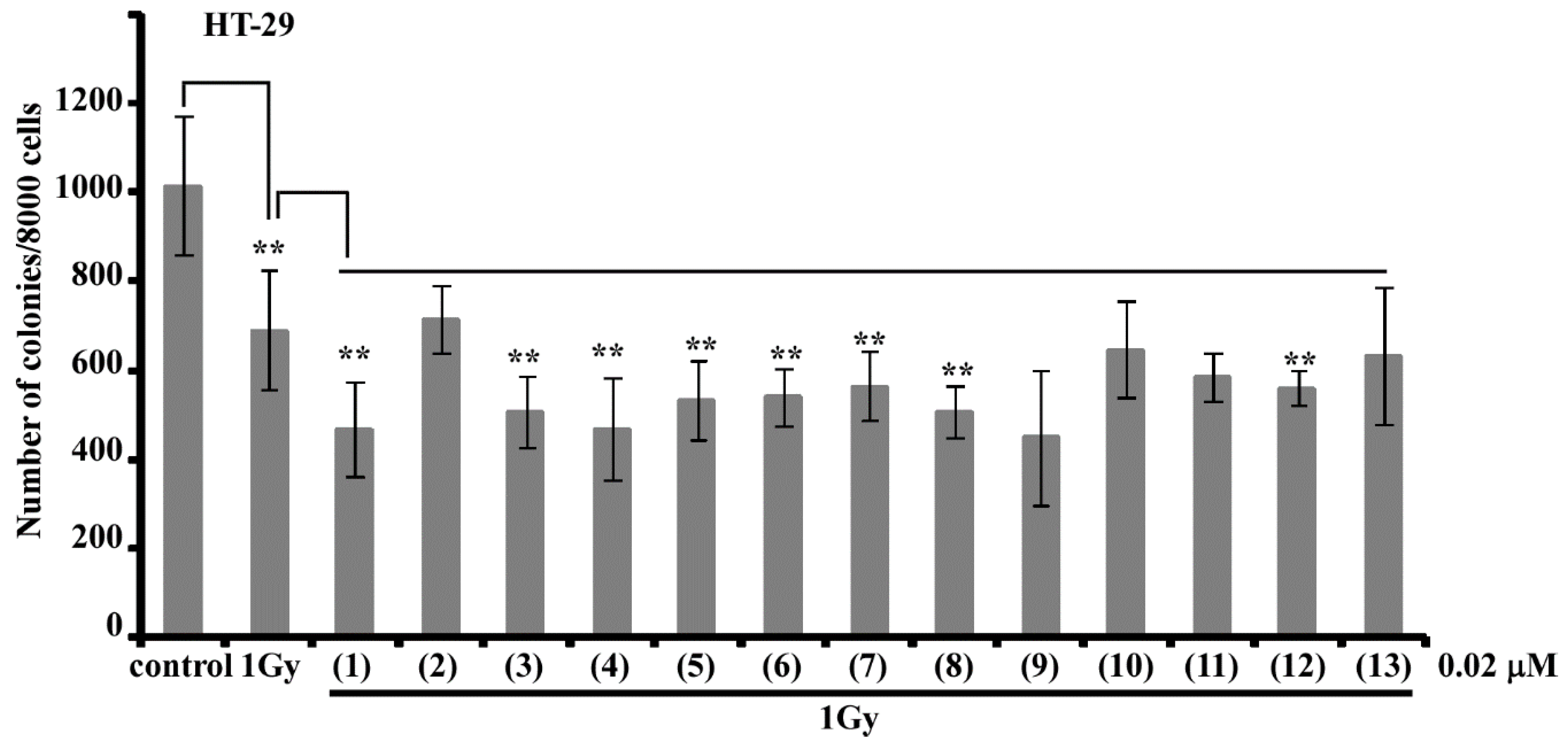
Thus, 13 unknown earlier triterpene glycosides were isolated from the Vietnamese sea cucumber
Colochirus quadrangularis. The glycosides include five different carbohydrate chains (quadrangularisosides of the groups A–E) and seven holostane aglycones. The trisulfated carbohydrate chains of quadrangularisosides of the groups D and E are novel and characterized by the position of one of the sulfate groups at C(4) of the terminal 3-
O-methylglucose residue, which is unusual for the glycosides from sea cucumbers. Two novel aglycones having hydroperoxyl groups in the side chains were discovered in quadrangularisosides A (
1) and D
3 (
11) (hydroperoxyl group at C(25)) as well as in quadrangularisosides A
1 (
2) and D
4 (
12) (hydroperoxyl group at C(24)). The finding of such functionalities in the aglycones of triterpene glycosides from the sea cucumbers is rare, and two other cases were reported only last year [
24,
25].
As to the structures of the glycosides isolated earlier by the Chinese researchers from the same species [
12,
13,
14,
15], only two compounds—philinopsides A and F—were identified by us in the glycosidic fraction of
C. quadrangularis. Other two compounds, isolated earlier—pentactasides B and C—are presumably the glycosides identical to quadrangularisosides B (
4) and B
1 (
5), because their carbohydrate chain structures were established incorrectly due to inaccuracies made by the authors in the NMR data interpretation. The glycosides with di- (pentactaside III) and trisaccharide chains (pentactasides I and II) as well as pentactaside E, having 16-ketoholosta-7,25-diene-3
β-ol as the aglycone, have not been found in the glycosidic sum obtained by us from
C. quadrangularis.