Aromatic Polyketides from a Symbiotic Strain Aspergillus fumigatus D and Characterization of Their Biosynthetic Gene D8.t287
Abstract
1. Introduction
2. Results
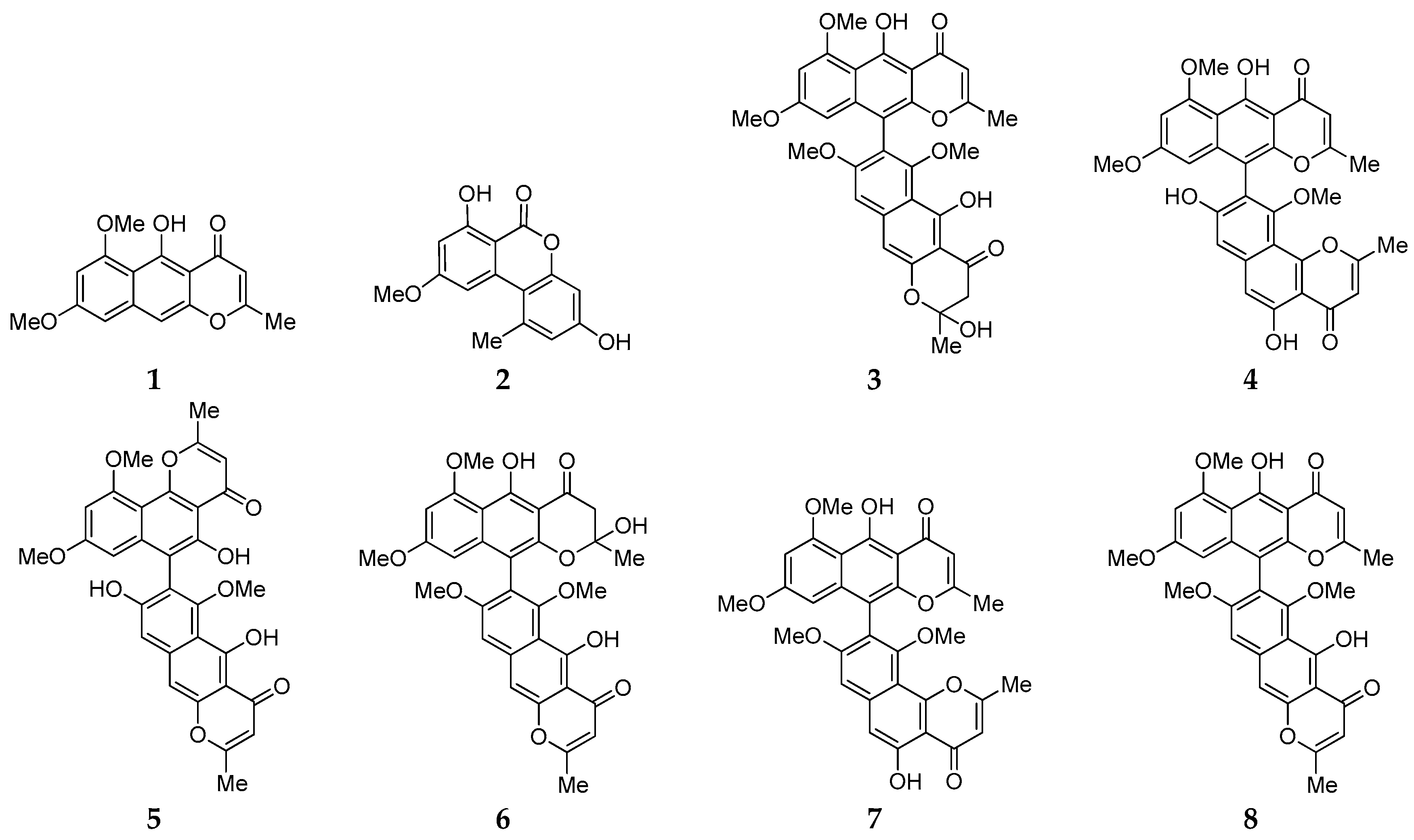
2.1. Isolation, Identification and Antimicrobial Assay of Aromatic Polyketides from Strain D
2.2. Genome Features of Strain D
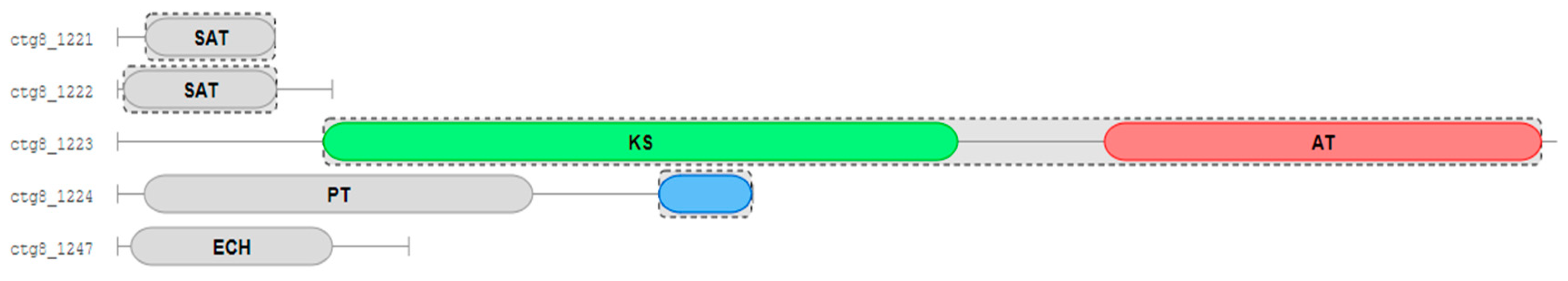
2.3. Biosynthesis Gene Cluster (BGC) Analysis of Secondary Metabolite in Strain D
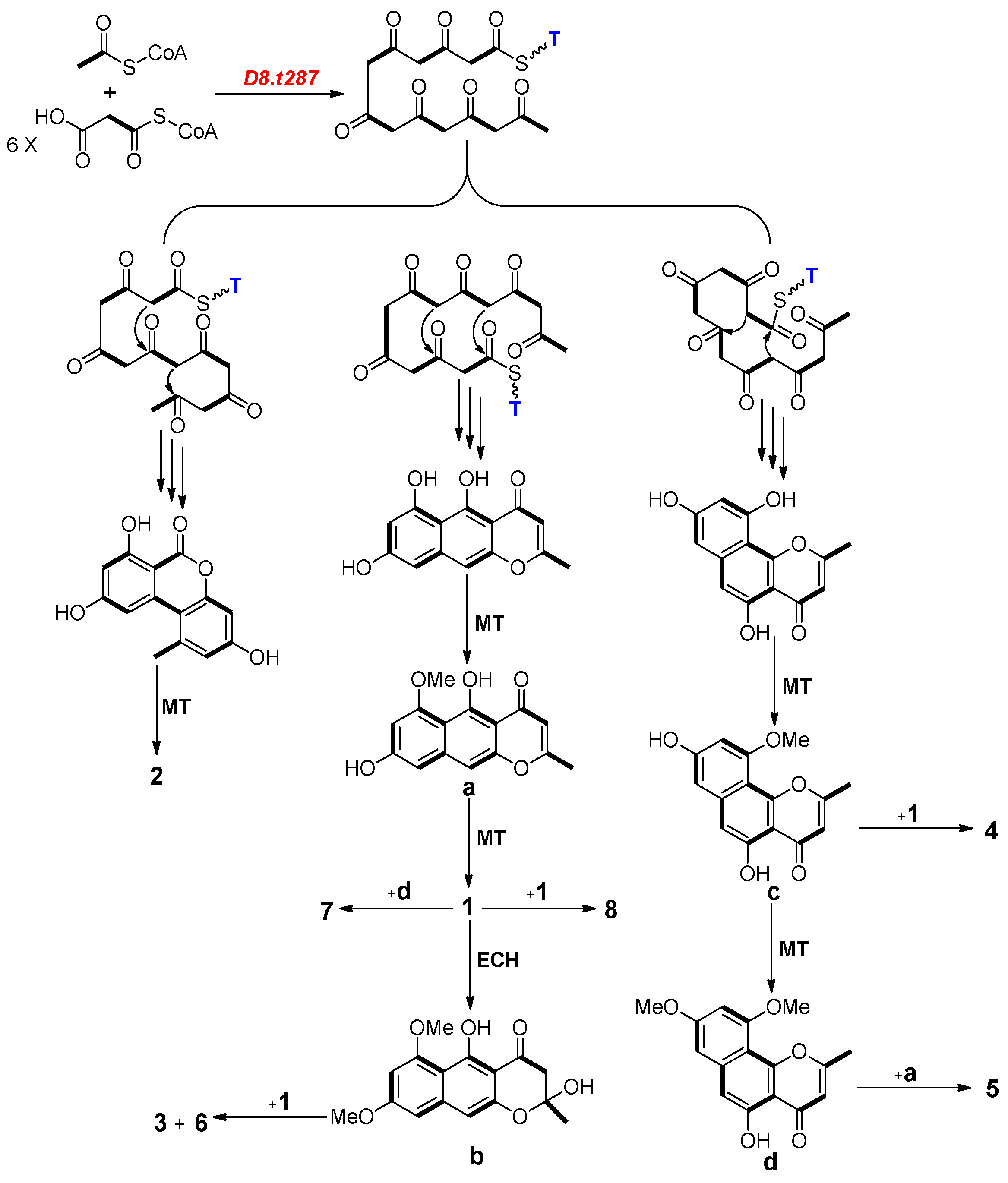
2.4. Biosynthesis Analysis of Compounds 1–8 in Strain D
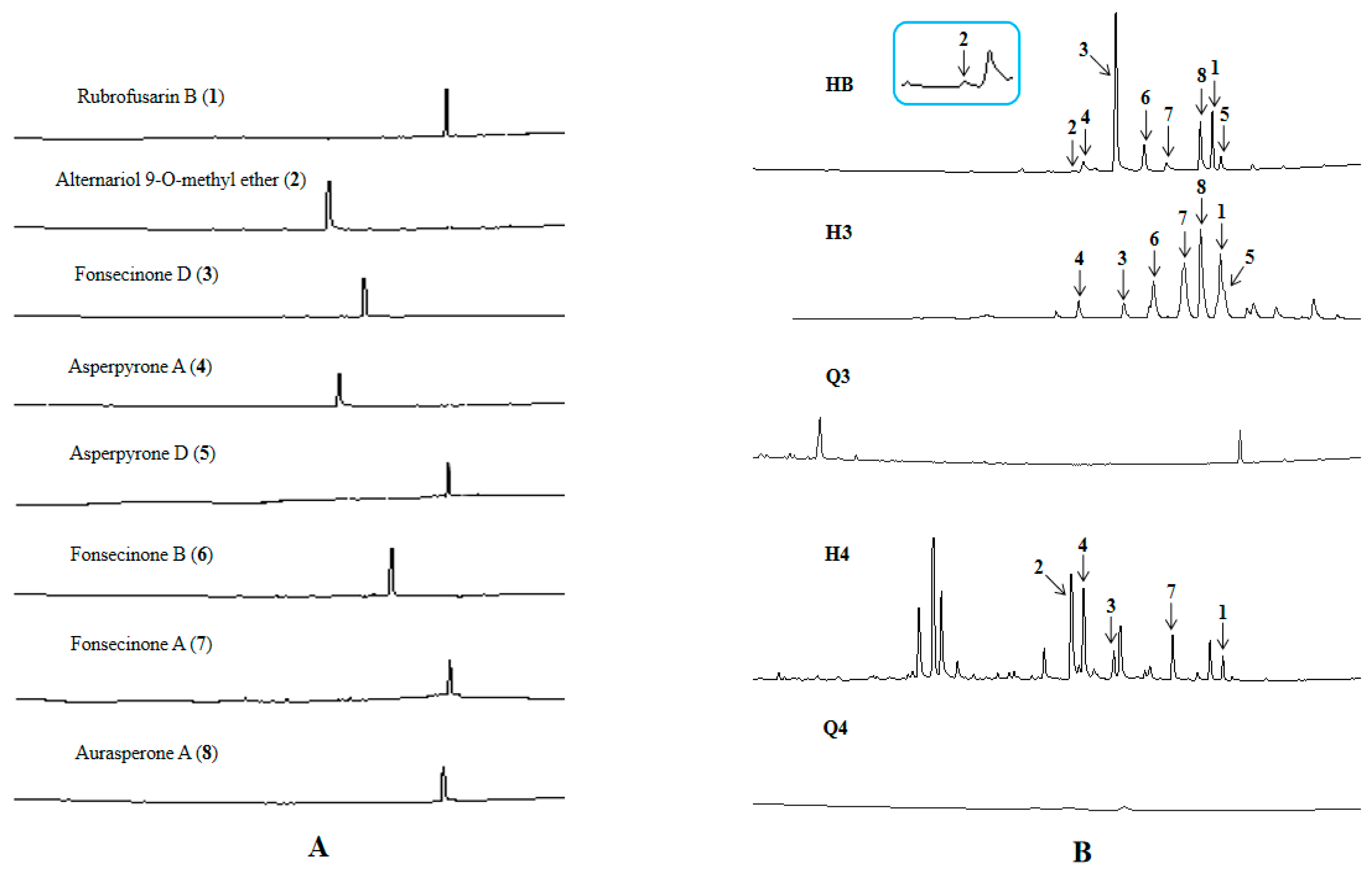
2.5. Functional Verification of the PKS Biosynthetic Gene Scaffold8.t287
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain, Medium and Cultural Conditions
3.3. Fermentation, Extraction and Isolation
3.4. Antimicrobial Test
3.5. Genome Sequencing and Analysis
3.6. Gene D8.t287 Knockout Experiment
3.6.1. Screening Resistance Markers
3.6.2. Construction of a Knockout Vector
3.6.3. Transformation and Verification of Mutant Strain
3.6.4. UPLC-MS Analysis of Strains D and ∆HY44
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, D.; Han, T.; Guan, L.-P.; Bai, J.; Zhao, N.; Li, Z.-L.; Wu, X.; Hua, H. New naphthopyrones from marine-derived fungus Aspergillus niger 2HL-M-8 and their in vitro antiproliferative activity. Nat. Prod. Res. 2015, 30, 1–7. [Google Scholar] [CrossRef]
- Yodsing, N.; Lekphrom, R.; Sangsopha, W.; Aimi, T.; Boonlue, S. Secondary Metabolites and Their Biological Activity from Aspergillus aculeatus KKU-CT2. Curr. Microbiol. 2017, 75, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-M.; Zhang, Y.-H.; Hai, Y.; Zheng, J.-Y.; Gu, Y.; Wang, C.; Shao, C.-L. Aspersymmetide A, a new centrosymmetric cyclohexapeptide from the marine-derived fungus Aspergillus versicolor. Mar. Drugs 2017, 15, 363. [Google Scholar] [CrossRef]
- Xu, L.L.; Zhang, C.C.; Zhu, X.Y.; Cao, F.; Zhu, H.J. Bioactive phenyl ether derivatives from the marine-derived fungus Aspergillus carneus. Nat. Prod. Res. 2016, 31, 1875–1879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ruan, C.; Bai, X. Isolation and antimicrobial effects of endophytic fungi from Edgeworthia chrysantha. Bangladesh J. Pharmacol. 2015, 10, 529. [Google Scholar] [CrossRef]
- Zhang, H.; Ruan, C.; Bai, X.; Chen, J.; Wang, H. Heterocyclic alkaloids as antimicrobial agents of Aspergillus fumigatus D endophytic on Edgeworthia chrysantha. Chem. Nat. Compd. 2018, 54, 411–414. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Z.; Chen, J.; Bai, X.; Wang, H. Tricycloalternarene analogs from a symbiotic fungus Aspergillus sp. D and their antimicrobial and cytotoxic effects. Molecules 2018, 23, 855. [Google Scholar] [CrossRef]
- Zhang, H.; Bai, X.; Wang, H. O1 Isolation and identification of an antimicrobial and cytotoxic chlorated perylenequinone from the symbiotic fungus Aspergillus fumigatus D. Biochem. Pharmacol. 2017, 139, 110. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619. [Google Scholar] [CrossRef]
- Jiang, W.; Jiang, H.C.; Wei, X.; Xu, J.J.; Shan, T.Z.; Pan, Y. Structures and activities of naphthopyrones from marine fungus Aspergillus niger XJJ-3. Microbiol. China 2018, 45, 1897–1903. (In Chinese) [Google Scholar] [CrossRef]
- Andreas, A.H.; Beate, N.K.; Erika, P. Conjugation of the mycotoxins alternariol and alternariol monomethyl ether in tobacco suspension cells. J. Agric. Food Chem. 2015, 63, 4728–4736. [Google Scholar] [CrossRef]
- Priestap, H. New naphthopyrones from Aspergillus fonsecaeus. Tetrahedron 1984, 40, 3617–3624. [Google Scholar] [CrossRef]
- Siriwardane, A.; Kumar, N.S.; Jayasinghe, U.; Fujimoto, Y. Chemical investigation of metabolites produced by an endophytic Aspergillus sp. isolated from Limonia acidissima. Nat. Prod. Res. 2015, 29, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhang, Q.; Gao, Y.-Q.; Shi, X.; Gao, J. Antifungal and antibacterial metabolites from an endophytic Aspergillus sp. associated with Melia azedarach. Nat. Prod. Res. 2014, 28, 1388–1392. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Gunaherath, G.M.K.B.; Wijeratne, E.M.K.; Gunatilaka, A.A.L. Asperpyrone D and other metabolites of the plant-associated fungal strain Aspergillus tubingensis. Phytochemistry 2007, 68, 368–372. [Google Scholar] [CrossRef]
- de Lazaro, S.; Ramos Campos, F.; Rodrigues-Fo, E.; Barison, A.; Daolio, C.; Ferreira, A.G.; Polo, V.; Longo, E.; Andrés, J. Combined 13C NMR and DFT/GIAO studies of the polyketides aurasperone A and fonsecinone A. Int. J. Quantum Chem. 2008, 108, 2408–2416. [Google Scholar] [CrossRef]
- Campos, F.R.; Barison, A.; Daolio, C.; Ferreira, A.G.; Rodrigues-Fo, E. Spectral assignments and reference data-Complete H-1 and C-13 NMR assignments of aurasperone A and fonsecinone A, two bis-naphthopyrones produced by Aspergillus aculeatus. Magn. Reson. Chem. 2005, 43, 962–965. [Google Scholar] [CrossRef]
- Yan, H.; Yu, R.; Li, D.; Shi, L.; Schwarz, S.; Yao, H.; Li, X.S.; Du, X.D. A novel multiresistance gene cluster located on a plasmid-borne transposon in Listeria monocytogenes. J. Antimicrob. Chemother. 2020, 75, 868–872. [Google Scholar] [CrossRef]
- Jiang, L.; Lim, C.J.; Jeong, J.C.; Kim, C.Y.; Kim, D.-H.; Kim, S.W.; Lee, J. Whole-genome sequence data and analysis of Saccharibacillus sp. ATSA2 isolated from Kimchi cabbage seeds. Data Brief 2019, 26, 104465. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, W43, W237–W243. [Google Scholar] [CrossRef]
- Zhang, A.; Lu, P.; Dahl-Roshak, A.M.; Paress, P.S.; Kennedy, S.; Tkacz, J.S.; An, Z. Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyensis. Mol. Genet. Genom. 2003, 268, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.G.; Townsend, C.A. Molecular characterization of the cercosporin biosynthetic pathway in the fungal plant pathogen Cercospora nicotianae. J. Am. Chem. Soc. 2016, 138, 4219–4228. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Fujii, I.; Tsai, H.; Chang, Y.C.; Kwon-Chung, K.J.; Ebizuka, Y. Aspergillus fumigatus alb1 encodes naphthopyrone synthase when expressed in Aspergillus oryzae. FEMS Microbiol. Lett. 2000, 192, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Praseuth, A.P.; Wang, C.C.C. A comprehensive and engaging overview of the type III family of polyketide synthases. Curr. Opin. Chem. Boil. 2007, 11, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, M.; Feng, C.; Hu, C. Progress in fungal polyketide biosynthesis. Chin. J. Biotechnol. 2018, 34, 151–164. (In Chinese) [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Xiang, B.; Li, X.; Qian, J.; Wang, L.; Ma, L.; Tian, X.; Wang, Y. The complete chloroplast genome sequence of the medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules 2016, 21, 1029. [Google Scholar] [CrossRef]
- Sayari, M.; Van Der Nest, M.; Steenkamp, E.; Adegeye, O.; Marincowitz, S.; Wingfield, B.D. Agrobacterium-mediated transformation of Ceratocystis albifundus. Microbiol. Res. 2019, 226, 55–64. [Google Scholar] [CrossRef]
- Lander, R.J.; Winters, M.A.; Meacle, F.J.; Buckland, B.C.; Lee, A. Fractional precipitation of plasmid DNA from lysate by CTAB. Biotechnol. Bioeng. 2002, 79, 776–784. [Google Scholar] [CrossRef]
- Han, G.; Shao, Q.; Li, C.; Zhao, K.; Jiang, L.; Fan, J.; Jiang, H.; Tao, F. An efficient Agrobacterium-mediated transformation method for aflatoxin generation fungus Aspergillus flavus. J. Microbiol. 2018, 56, 356–364. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed]
| Strain | Genome Size (Mb) | G + C% | Gene | tRNA | Hypothetical Protein | Assembly |
|---|---|---|---|---|---|---|
| A1163 | 29.21 | 49.5 | 10,124 | 180 | 9929 | GCA_000150145.1 |
| Af10 | 28.76 | 49.5 | - | - | - | GCA_000225625.2 |
| Af293 | 29.39 | 49.8 | 9630 | 229 | 9916 | GCA_000002655.1 |
| CNM-CM8057 | 28.32 | 49.5 | 8910 | - | 8910 | GCA_012656215.1 |
| D | 33.40 | 51.4 | 9789 | 108 | 4417 | GCA_003069565.1 |
| HMR AF 270 | 29.48 | 49.2 | 9730 | 179 | 9549 | GCA_002234955.1 |
| HMR AF 706 | 28.23 | 49.5 | 9466 | 180 | 9284 | GCA_002234985.1 |
| ISSFT-021 | 28.24 | 49.4 | - | - | - | GCA_001643655.1 |
| LMB-35Aa | 27.52 | 50.0 | - | - | - | GCA_001715275.2 |
| SGAir0713 | 29.02 | 49.6 | - | - | - | GCA_005768625.2 |
| Z5 | 29.36 | 49.2 | - | - | - | GCA_001029325.1 |
| Region | Type | Gene Cluster Position | Most Similar Gene Cluster | Similarity (%) | |
|---|---|---|---|---|---|
| Scaffold1 | 1.1 | NRPS | 3,405,290–3,461,377 | / | / |
| Scaffold2 | 2.1 | terpene | 1,004,114–1,024,548 | / | / |
| Scaffold3 | 3.1 | T3PKS | 985,697–1,026,775 | / | / |
| Scaffold4 | 4.1 | terpene | 388,980–408,695 | terpene | 40 |
| Scaffold4 | 4.2 | terpene | 2,345,955–2,367,289 | / | / |
| Scaffold5 | 5.1 | T1PKS | 1,087,051–1,131,054 | polyketide | 13 |
| Scaffold5 | 5.2 | T1PKS | 2,115,332–2,162,885 | polyketide | 50 |
| Scaffold6 | 6.1 | T1PKS | 252,626–300,167 | NRP+polyketide | 18 |
| Scaffold7 | 7.1 | fungal-RiPP | 280,898–318,613 | / | / |
| Scaffold7 | 7.2 | NRPS | 1,415,890–1,477,928 | NRP: Cyclic depsipeptide | 100 |
| Scaffold8 | 8.1 | T1PKS | 160,848–205,614 | polyketide | 100 |
| Scaffold8 | 8.2 | T1PKS | 918,158–961,487 | polyketide | 100 |
| Scaffold8 | 8.3 | NRPS | 1,667,861–1,723,346 | / | / |
| Scaffold9 | 9.1 | NRPS+indole | 74,519–134,677 | / | / |
| Scaffold9 | 9.2 | terpene | 1,083,778–1,105,346 | / | / |
| Scaffold9 | 9.3 | NRPS-like | 1,628,313–16,71,303 | / | / |
| Scaffold11 | 11.1 | NRPS | 383,909–428,735 | NRP+polyketide | 45 |
| Scaffold12 | 12.1 | NRPS-like | 821,562–865,410 | / | / |
| Scaffold17 | 17.1 | NRPS-like | 305,087–348,950 | / | / |
| Scaffold19 | 19.1 | NRPS | 260,997–319,962 | NRP | 100 |
| Scaffold21 | 21.1 | NRPS-like | 36,305–79,148 | / | / |
| Scaffold21 | 21.2 | T1PKS | 174,713–214,266 | polyketide | 62 |
| Gene | Gene Position | Hit | Similarity (%) |
|---|---|---|---|
| D1.t382 | 1,271,230–1,272,630 | polyketide synthase PksC [Alternaria alternata] OAG15814.1 | 100 |
| D5.t345 | 1105803–1,112,689 | polyketide synthase PksJ [A. alternata] OAG22978.1 | 84.5 |
| D5.t661 | 2135332–2,143,251 | polyketide synthase PksH [A. alternata] OAG13655.1 | 97.6 |
| D6.t72 | 272,318–280,167 | polyketide synthase PksG [A. alternata] | 95.7 |
| D6.t701 | 2,594,273–2,602,296 | putative polyketide synthase [Fusarium aywerte] | 56.8 |
| D8.t287 | 938,512–942,754 | ketoacyl-synt-domain-containing protein [A. alternata] OAG24819.1 | 85.3 |
| D11.t72 | 283,919–289,534 | polyketide synthase PksD [A. alternata] OAG18885.1 | 90.6 |
| D11.t100 | 403,909–416,371 | polyketide synthase PksB [A. alternata] | 98.3 |
| D21.t51 | 193,704–202,533 | polyketide synthase PksF [A. alternata] OAG16734.1 | 99.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Pan, R.; Bai, X.; Wei, B.; Chen, J.; Wang, H.; Zhang, H. Aromatic Polyketides from a Symbiotic Strain Aspergillus fumigatus D and Characterization of Their Biosynthetic Gene D8.t287. Mar. Drugs 2020, 18, 324. https://doi.org/10.3390/md18060324
Hua Y, Pan R, Bai X, Wei B, Chen J, Wang H, Zhang H. Aromatic Polyketides from a Symbiotic Strain Aspergillus fumigatus D and Characterization of Their Biosynthetic Gene D8.t287. Marine Drugs. 2020; 18(6):324. https://doi.org/10.3390/md18060324
Chicago/Turabian StyleHua, Yi, Rui Pan, Xuelian Bai, Bin Wei, Jianwei Chen, Hong Wang, and Huawei Zhang. 2020. "Aromatic Polyketides from a Symbiotic Strain Aspergillus fumigatus D and Characterization of Their Biosynthetic Gene D8.t287" Marine Drugs 18, no. 6: 324. https://doi.org/10.3390/md18060324
APA StyleHua, Y., Pan, R., Bai, X., Wei, B., Chen, J., Wang, H., & Zhang, H. (2020). Aromatic Polyketides from a Symbiotic Strain Aspergillus fumigatus D and Characterization of Their Biosynthetic Gene D8.t287. Marine Drugs, 18(6), 324. https://doi.org/10.3390/md18060324








