Purification and Characterization of a New CRISP-Related Protein from Scapharca broughtonii and Its Immunomodulatory Activity
Abstract
:1. Introduction
2. Results
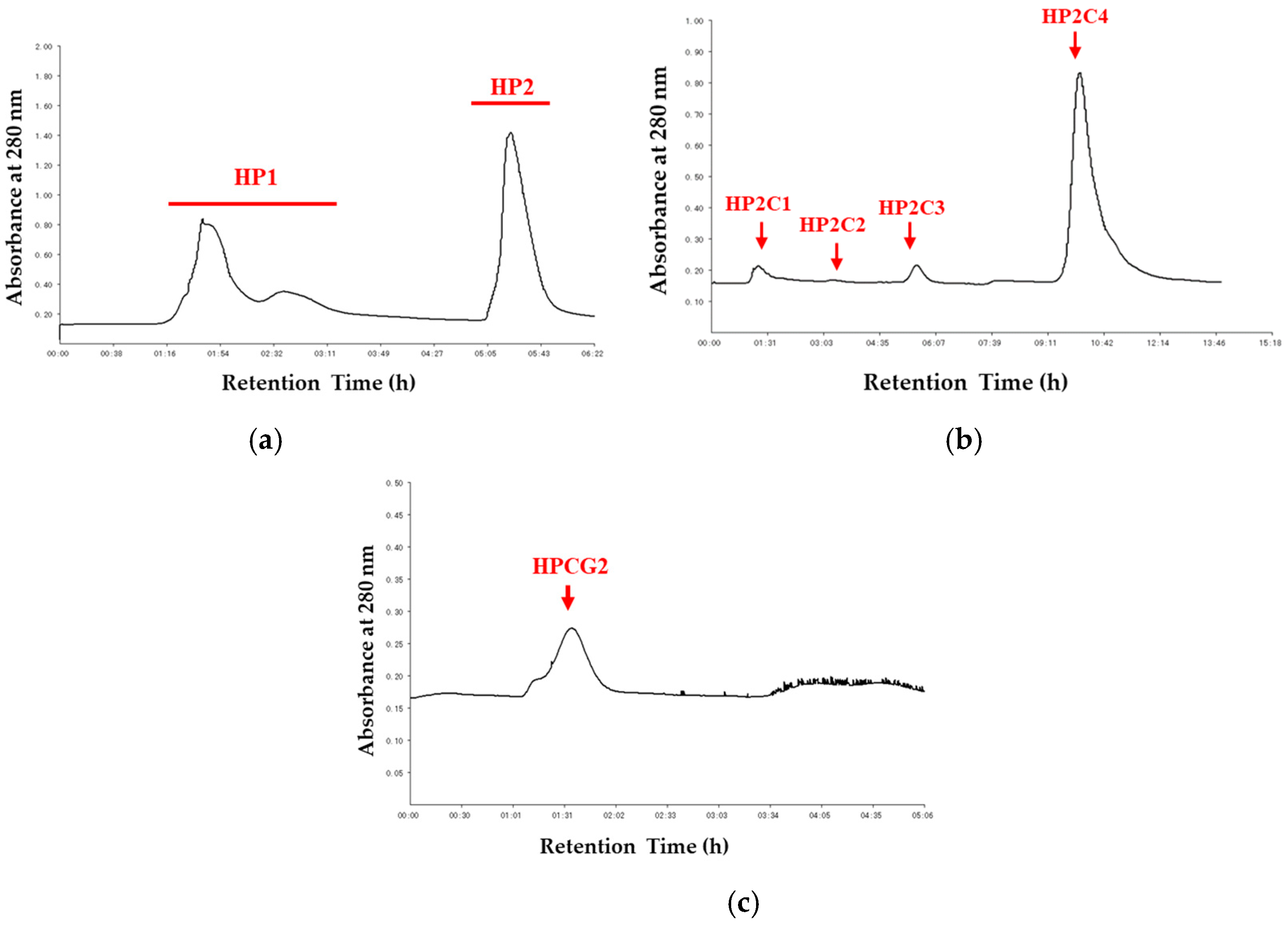
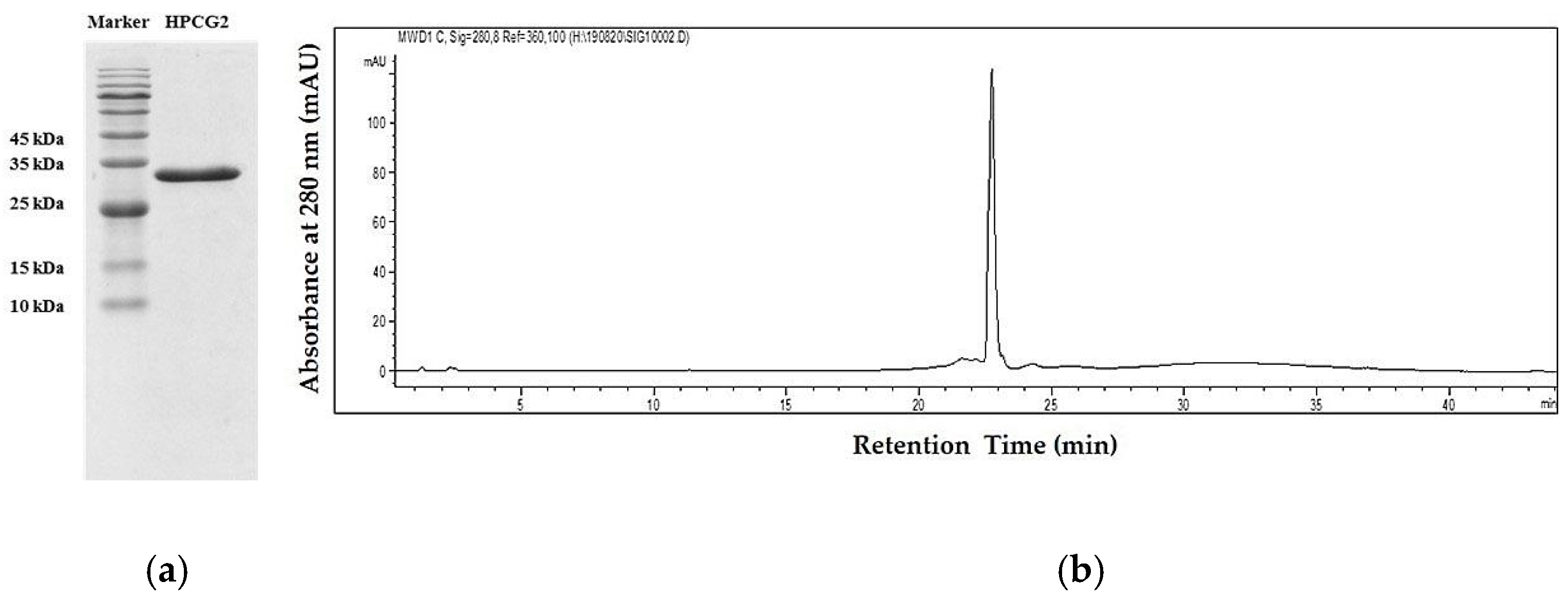
2.1. Purification of HPCG2
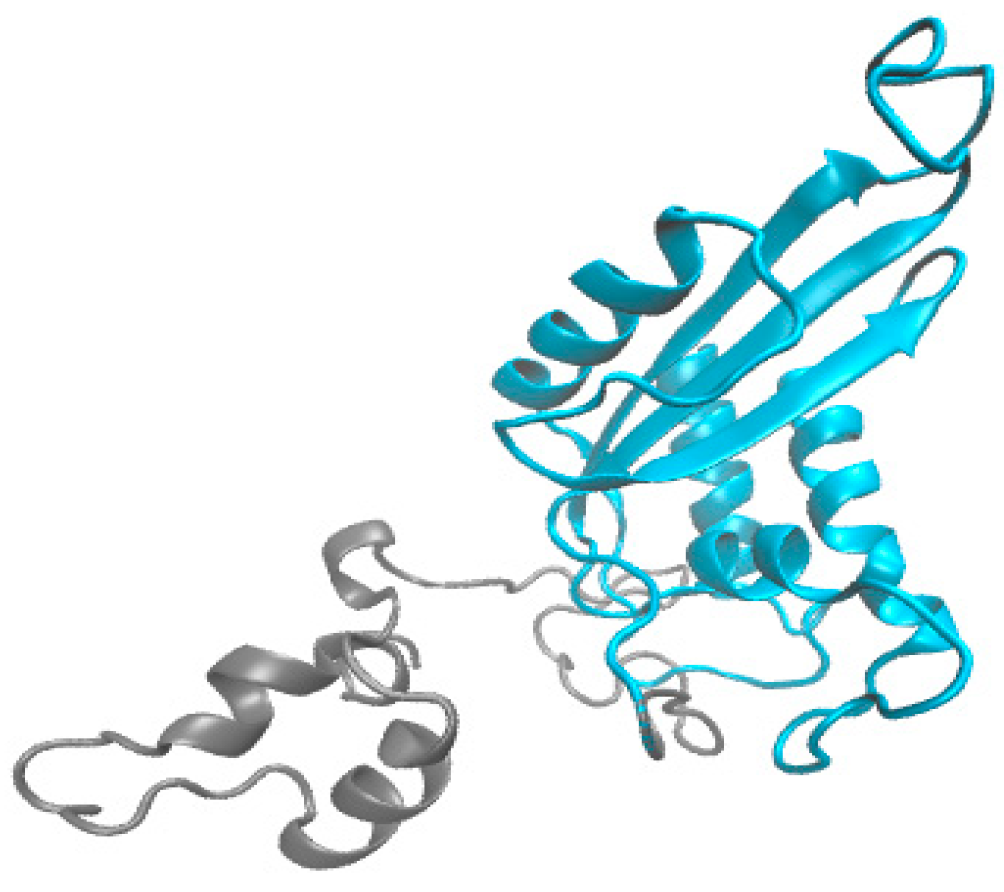
2.2. Structural Characterization of HPCG2
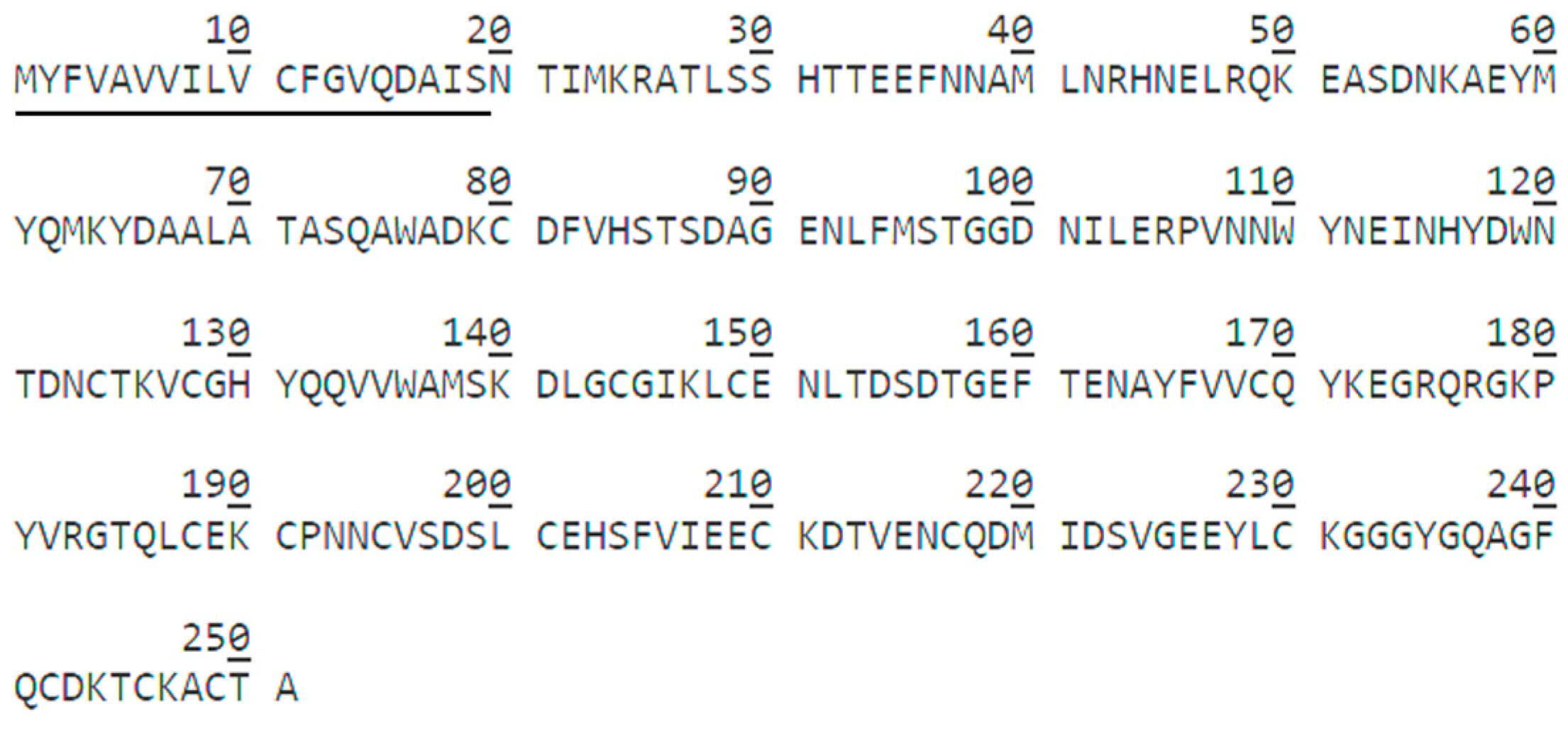
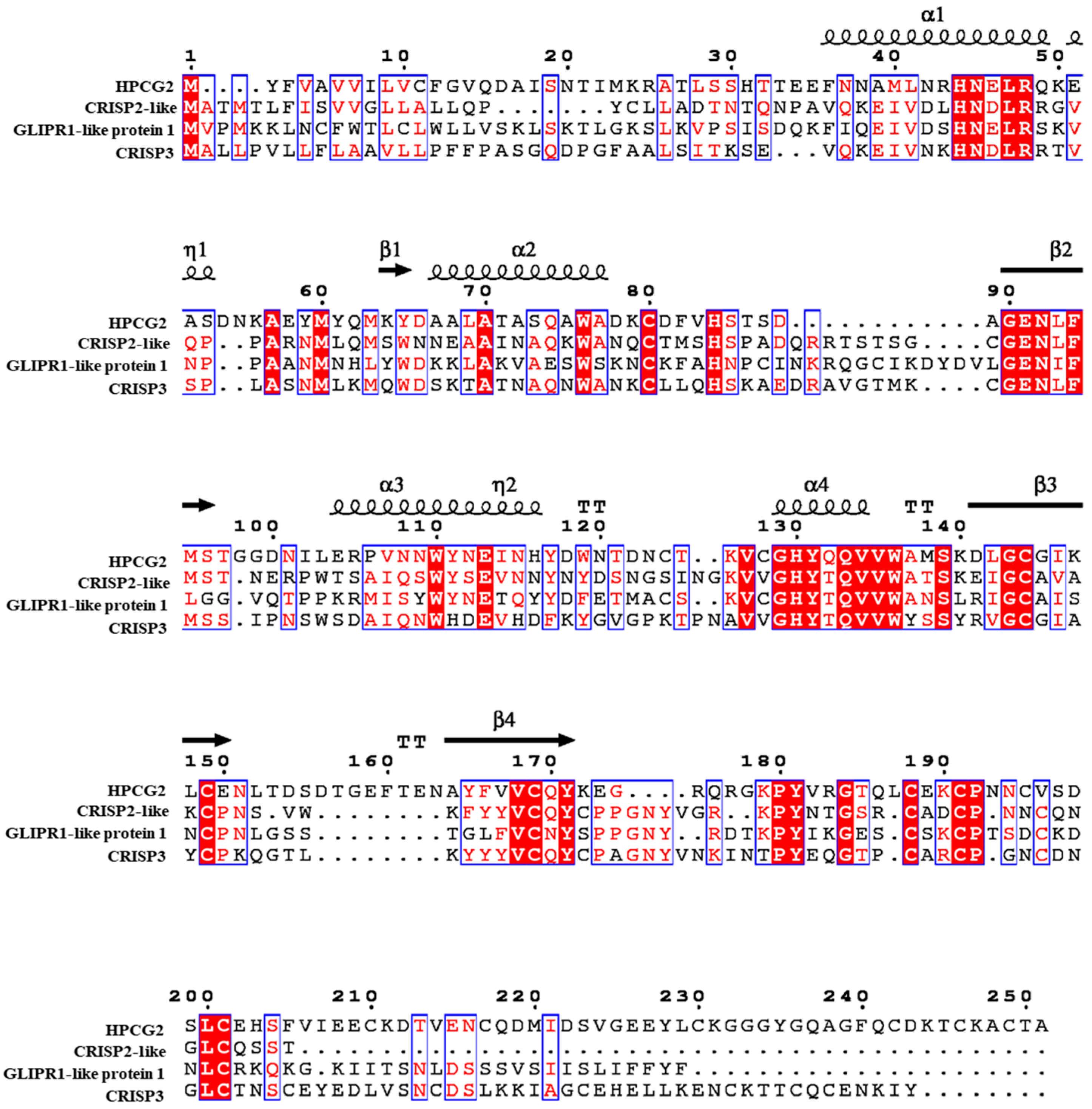
2.3. Sequence Analysis of HPCG2
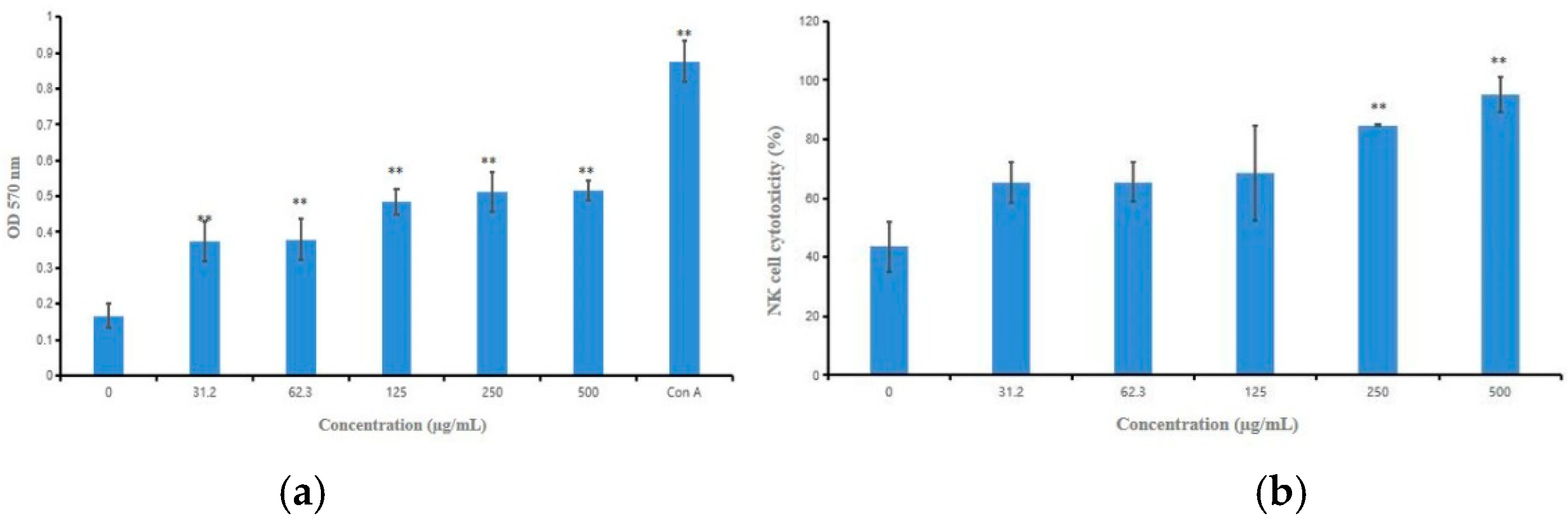
2.4. Effects of HPCG2 on Splenic Lymphocyte Proliferation and NK Cell Cytotoxicity
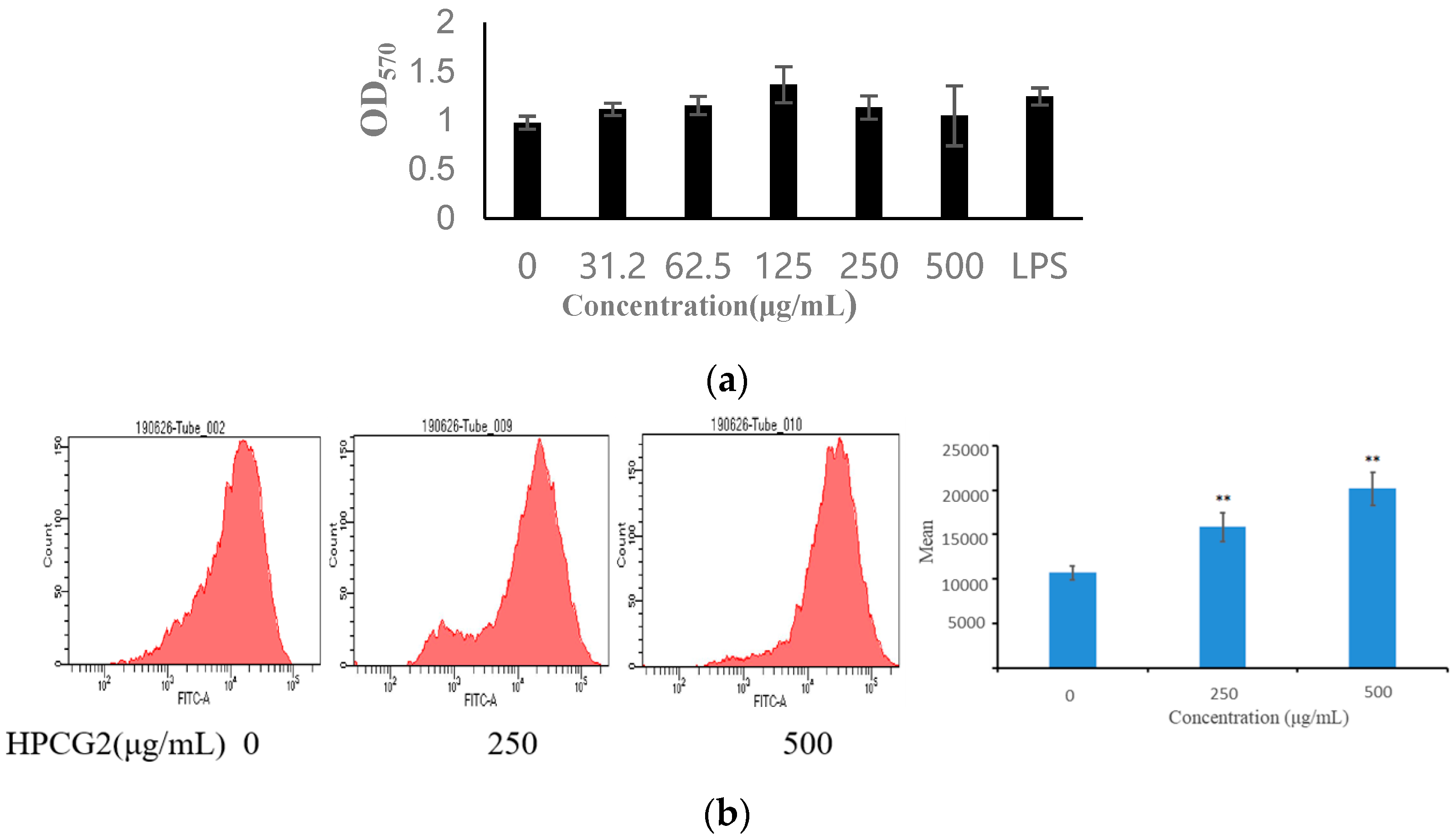
2.5. Effect of HPCG2 on the Viability of RAW264.7 Macrophages
2.6. Effect of HPCG2 on the Phagocytic Activity of Macrophages
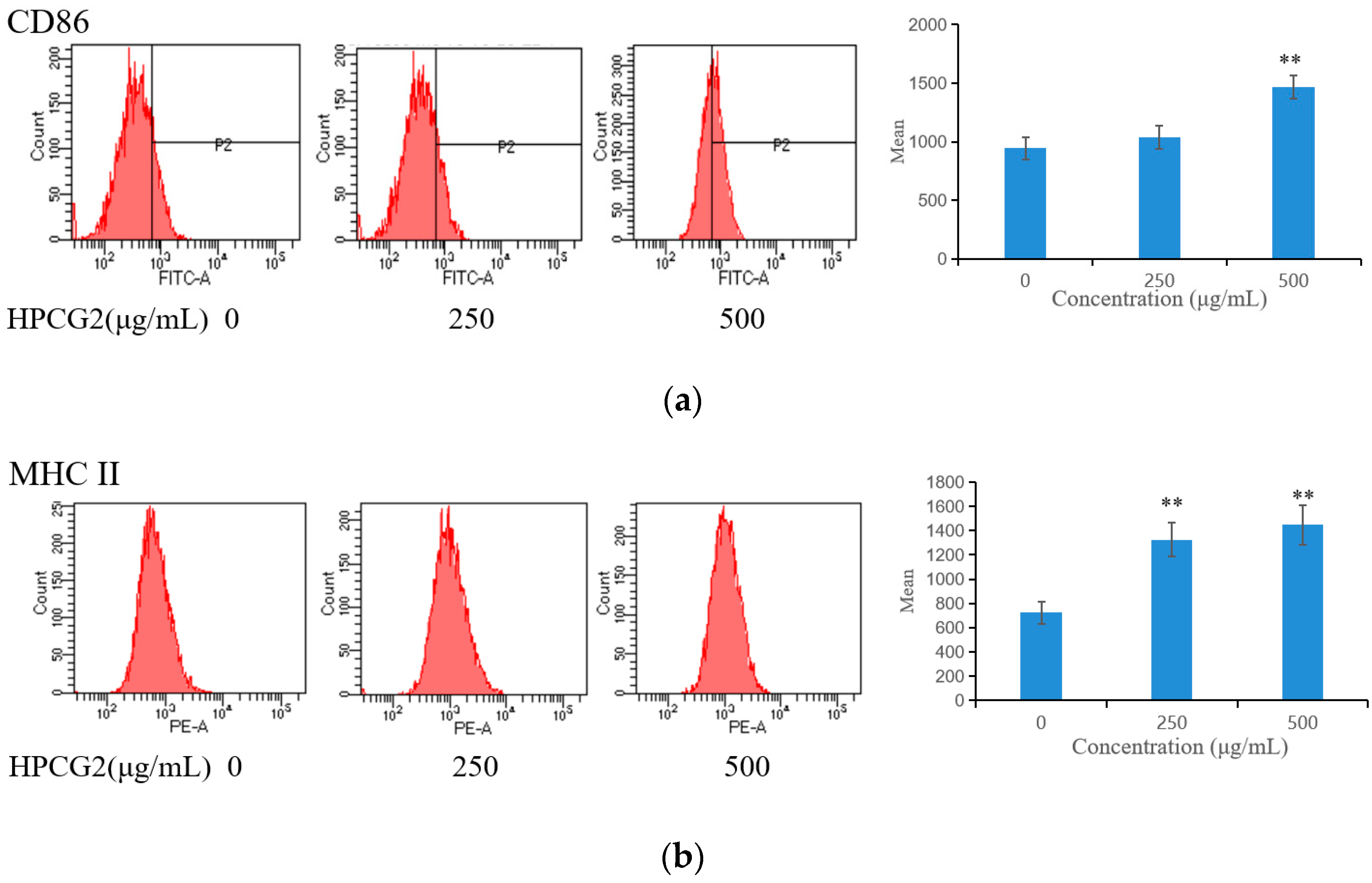
2.7. Effects of HPCG2 on the Expression of CD86 and MHC II on RAW264.7 Cells
2.8. Effect of HPCG2 on the Production of Nitric Oxide, IL-6, and TNF-α
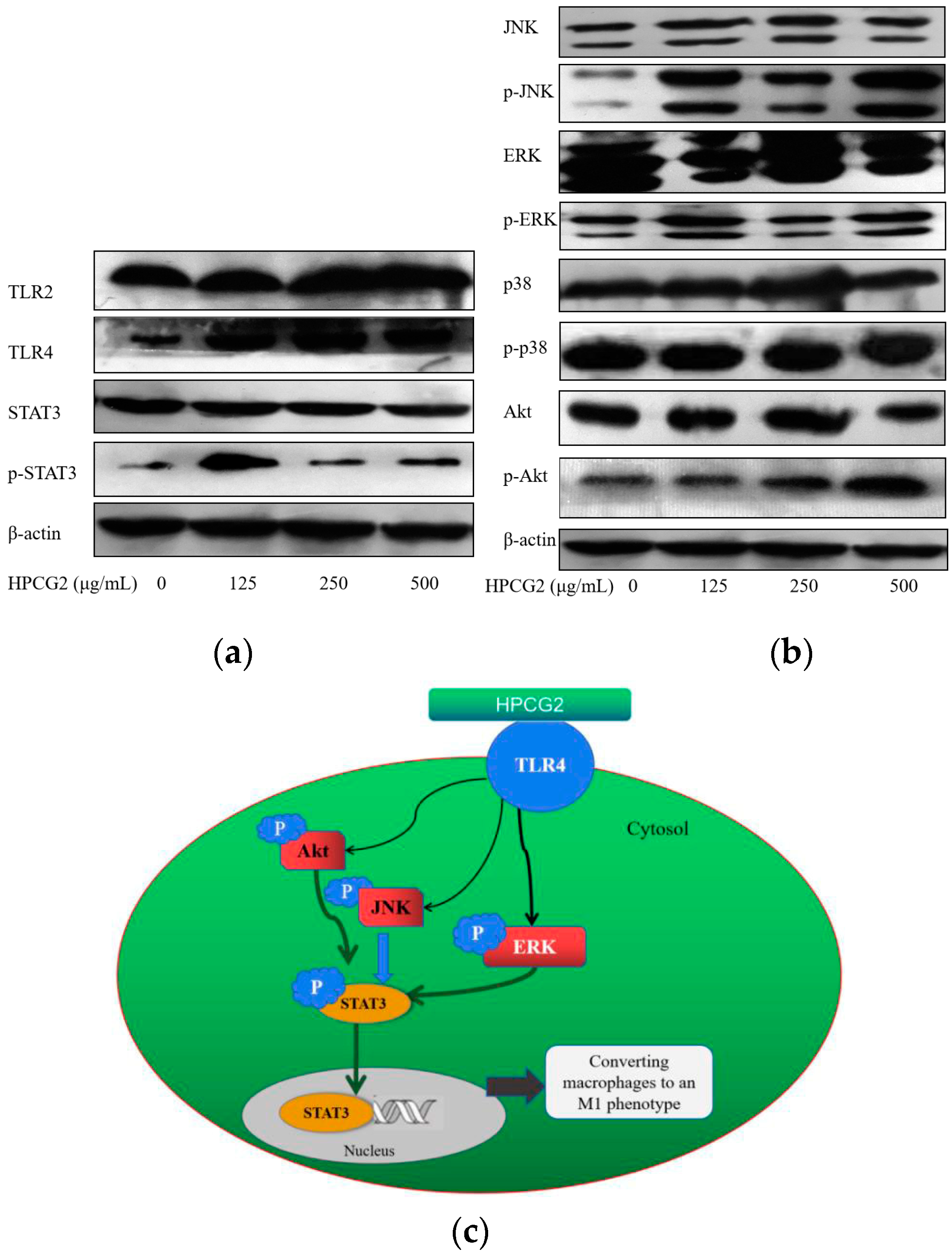
2.9. Effects of HPCG2 on the Expression of TLRs, Akt, MAPKs, and STAT3 in RAW264.7 Cells
3. Discussion
4. Materials and Methods
4.1. Biological Materials
4.2. Reagents and Cultures
4.3. Purification Process
4.4. Purity Determination
4.5. Measurement of Protein and Carbohydrate Concentration
4.6. Determination of Molecular Weight
4.7. Elucidation of Secondary Structure
4.8. Amino Acid Sequence of HPCG2
4.9. Cell Viability Assay
4.10. Secretion of Nitric Oxide and Cytokines
4.11. Expression of CD86 and MHC II
4.12. Phagocytosis Assay
4.13. Western Blot Assay
4.14. Natural Killer Cell Cytotoxicity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, M.; Wang, G.; Lai, F.; Wu, H. Structural characterization and immunomodulatory activity of a novel polysaccharide from Lepidium meyenii. J. Agric. Food Chem. 2016, 64, 1921–1931. [Google Scholar] [CrossRef]
- Mebius, R.E.; Kraal, G. Structure and function of the spleen. Nat. Rev. Immunol. 2005, 5, 606–616. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Coffman, R.L. TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989, 7, 145–173. [Google Scholar] [CrossRef]
- Epelman, S.; Lavine, K.J.; Randolph, G.J. Origin and functions of tissue macrophages. Immunity 2014, 41, 21–35. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, A.; Allavena, P. The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 2015, 212, 435–445. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Kang, H.K.; Lee, H.H.; Seo, C.H.; Park, Y. Antimicrobial and immunomodulatory properties and applications of marine-derived proteins and peptides. Mar. Drugs 2019, 17, 350. [Google Scholar] [CrossRef] [Green Version]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef]
- Nishida, K.; Ishimura, T.; Suzuki, A.; Sasaki, T. Seasonal changes in the shell microstructure of the bloody clam, Scapharca broughtonii (Mollusca: Bivalvia: Arcidae). Palaeogeogr. Palaeocl. 2012, 363–364, 99–108. [Google Scholar] [CrossRef]
- Li, M.; Zhu, L.; Zhou, C.Y.; Sun, S.; Fan, Y.J.; Zhuang, Z.M. Molecular characterization and expression of a novel big defensin (Sb-BDef1) from ark shell Scapharca broughtonii. Fish Shellfish Immunol. 2012, 33, 1167–1173. [Google Scholar] [CrossRef]
- Mao, Y.; Zhou, C.; Zhu, L.; Huang, Y.; Yan, T.; Fang, J.; Zhu, W. Identification and expression analysis on bactericidal permeability-increasing protein (BPI)/lipopolysaccharide-binding protein (LBP) of ark shell, Scapharca broughtonii. Fish Shellfish Immunol. 2013, 35, 642–652. [Google Scholar] [CrossRef]
- Zheng, L.; Wu, B.; Liu, Z.; Tian, J.; Yu, T.; Zhou, L.; Sun, X.; Yang, A. A manganese superoxide dismutase (MnSOD) from ark shell, Scapharca broughtonii: Molecular characterization, expression and immune activity analysis. Fish Shellfish Immunol. 2015, 45, 656–665. [Google Scholar] [CrossRef]
- Li, C.; Zhu, J.; Wang, Y.; Chen, Y.; Song, L.; Zheng, W.; Li, J.; Yu, R. Antibacterial Activity of AI-Hemocidin 2, a Novel N-Terminal Peptide of Hemoglobin Purified from Arca inflata. Mar. Drugs 2017, 15, 205. [Google Scholar] [CrossRef]
- Wang, W.; Shi, H.; Zhu, J.; Li, C.; Song, L.; Yu, R. Purification and structural characterization of a novel antioxidant and antibacterial protein from Arca inflata. Int. J. Biol. Macromol. 2018, 116, 289–298. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Z.; Song, L.; Chen, L.; Zhu, J.; Lv, S.; Yu, R. A new in vitro anti-tumor polypeptide isolated from Arca inflata. Mar. Drugs 2013, 11, 4773–4787. [Google Scholar] [CrossRef]
- Rowley, A.F.; Powell, A. Invertebrate immune systems specific, quasi-specific, or nonspecific? J. Immunol. 2007, 179, 7209–7214. [Google Scholar] [CrossRef] [Green Version]
- Bruno, D. Mass spectrometry and protein analysis. Science 2006, 5771, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. BBA-Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Pelton, J.T.; McLean, L.R. Spectroscopic methods for analysis of protein secondary structure. Anal. Biochem. 2000, 277, 167–176. [Google Scholar] [CrossRef]
- Xiaona, G. Purification and characterization of the antitumor protein from Chinese tartary buckwheat (Fagopyrum tataricum Gaertn.) water-soluble extracts. J. Agric. Food Chem. 2007, 17, 6958–6961. [Google Scholar] [CrossRef]
- Yang, J.; Yang, Z. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef]
- Cheng, A.; Wan, F.; Wang, J.; Jin, Z.; Xu, X. Macrophage immunomodulatory activity of polysaccharides isolated from Glycyrrhiza uralensis Fish. Int. Immunopharmacol. 2008, 8, 43–50. [Google Scholar] [CrossRef]
- Chen, D.; Xie, J.; Fiskesund, R.; Dong, W.; Liang, X.; Lv, J.; Jin, X.; Liu, J.; Mo, S.; Zhang, T.; et al. Publisher Correction: Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Bi, S.; Huang, W.; Chen, S.; Huang, C.; Li, C.; Guo, Z.; Yang, J.; Zhu, J.; Song, L.; Yu, R. Cordyceps militaris polysaccharide converts immunosuppressive macrophages into M1-like phenotype and activates T lymphocytes by inhibiting the PD-L1/PD-1 axis between TAMs and T lymphocytes. Int. J. Biol. Macromol. 2020, 150, 261–280. [Google Scholar] [CrossRef]
- Feng, Y.; Mu, R.; Wang, Z.; Xing, P.; Zhang, J.; Dong, L.; Wang, C. A toll-like receptor agonist mimicking microbial signal to generate tumor-suppressive macrophages. Nat. Commun. 2019, 10, 2272. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.Z.; Hao, R.; Zha, X.Q.; Pan, L.H.; Liu, J.; Luo, J.P. Polysaccharide of Dendrobium huoshanense activates macrophages via toll-like receptor 4-mediated signaling pathways. Carbohydr. Polym. 2016, 146, 292–300. [Google Scholar] [CrossRef]
- Mao, Y.; Wang, B.; Xu, X.; Du, W.; Li, W.; Wang, Y. Glycyrrhizic acid promotes M1 macrophage polarization in murine bone marrow-derived macrophages associated with the activation of JNK and NF-kappaB. Mediat. Inflamm. 2015, 2015, 372931. [Google Scholar] [CrossRef] [Green Version]
- Greenhill, C.J.; Rose-John, S.; Lissilaa, R.; Ferlin, W.; Ernst, M.; Hertzog, P.J.; Mansell, A.; Jenkins, B.J. IL-6 trans-signaling modulates TLR4-dependent inflammatory responses via STAT3. J. Immunol. 2011, 186, 1199–1208. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Jiang, S.; Lian, L.-H.; Bai, T.; Cui, P.-H.; Sun, X.-T.; Jin, X.-J.; Wu, Y.-L.; Nan, J.-X. Betulinic acid and betulin ameliorate acute ethanol-induced fatty liver via TLR4 and STAT3 in vivo and in vitro. Int. Immunopharmacol. 2013, 17, 184–190. [Google Scholar] [CrossRef]
- Chikalovets, I.V.; Kondrashina, A.S.; Chernikov, O.V.; Molchanova, V.I.; Luk’Yanov, P.A. Isolation and general characteristics of lectin from the mussel Mytilus trossulus. Chem. Nat. Compd. 2013, 48, 1058–1061. [Google Scholar] [CrossRef]
- Kawabata, S.I.; Tsuda, R. Molecular basis of non-self recognition by the horseshoe crab tachylectins. BBA-Gen. Subj. 2002, 1572, 414–421. [Google Scholar] [CrossRef]
- Miles, D.; Roché, H.; Martin, M.; Perren, T.J.; Cameron, D.A.; Glaspy, J.; Dodwell, D.; Parker, J.; Mayordomo, J.; Tres, A.; et al. Phase III multicenter clinical trial of the Sialyl-TN (STn)-keyhole limpet hemocyanin (KLH) vaccine for metastatic breast cancer. Oncologist 2011, 16, 1092–1100. [Google Scholar] [CrossRef] [Green Version]
- Arancibia, S.; Espinoza, C.; Salazar, F.; Del Campo, M.; Tampe, R.; Zhong, T.Y.; De Ioannes, P.; Moltedo, B.; Ferreira, J.; Lavelle, E.C.; et al. A novel immunomodulatory hemocyanin from the limpet Fissurella latimarginata promotes potent anti-tumor activity in melanoma. PLoS ONE 2014, 9, e87240. [Google Scholar] [CrossRef]
- Silva, O.N.; de la Fuente-Núñez, C.; Haney, E.F.; Fensterseifer, I.C.M.; Ribeiro, S.M.; Porto, W.F.; Brown, P.; Faria-Junior, C.; Rezende, T.M.B.; Moreno, S.E. An anti-infective synthetic peptide with dual antimicrobial and immunomodulatory activities. Sci. Rep.-UK 2016, 6, 35465. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Liu, X.; Bu, L.; Li, H.; Zhang, S. Antimicrobial–immunomodulatory activities of zebrafish phosvitin-derived peptide Pt5. Peptides 2012, 37, 309–313. [Google Scholar] [CrossRef]
- Seo, J.K.; Crawford, J.M.; Stone, K.L.; Noga, E.J. Purification of a novel arthropod defensin from the American oyster, Crassostrea virginica. Biochem. Biophys. Res. Commun. 2005, 338, 1998–2004. [Google Scholar] [CrossRef]
- King, T.P.; Sobotka, A.K.; Alagon, A.; Kochoumian, L.; Lichtenstein, L.M. Protein allergens of white-faced hornet, yellow hornet, and yellow jacket venoms. Biochemistry 1978, 17, 5165–5174. [Google Scholar] [CrossRef]
- Hoffman, D.R. Allergens in hymenoptera venom XV: The immunologic basis of vespid venom cross-reactivity. J. Allergy Clin. Immunol. 1985, 75, 611–613. [Google Scholar] [CrossRef]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Hunt, M.D. Systemic acquired resistance. Plant Physiol. 1996, 8, 1809–1819. [Google Scholar] [CrossRef] [Green Version]
- Dixon, D.C.; Cutt, J.R.; Klessig, D.F. Differential targeting of the tobacco PR-1 pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J. 1991, 10, 1317–1324. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immun. 2008, 9, 503–510. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 866. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, X.; Li, Y. A modified Coomassie Brilliant Blue staining method at nanogram sensitivity compatible with proteomic analysis. Biotechnol. Lett. 2007, 29, 1599–1603. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.; Lee, Y.C. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell Proteomics 2012, 11. [Google Scholar] [CrossRef] [Green Version]
- Lv, S.; Gao, J.; Liu, T.; Zhu, J.; Xu, J.; Song, L.; Liang, J.; Yu, R. Purification and partial characterization of a new antitumor protein from Tegillarca granosa. Mar. Drugs 2015, 13, 1466–1480. [Google Scholar] [CrossRef] [Green Version]
- Bi, S.; Jing, Y.; Zhou, Q.; Hu, X.; Zhu, J.; Guo, Z.; Song, L.; Yu, R. Structural elucidation and immunostimulatory activity of a new polysaccharide from Cordyceps militaris. Food Funct. 2018, 9, 279–293. [Google Scholar] [CrossRef]
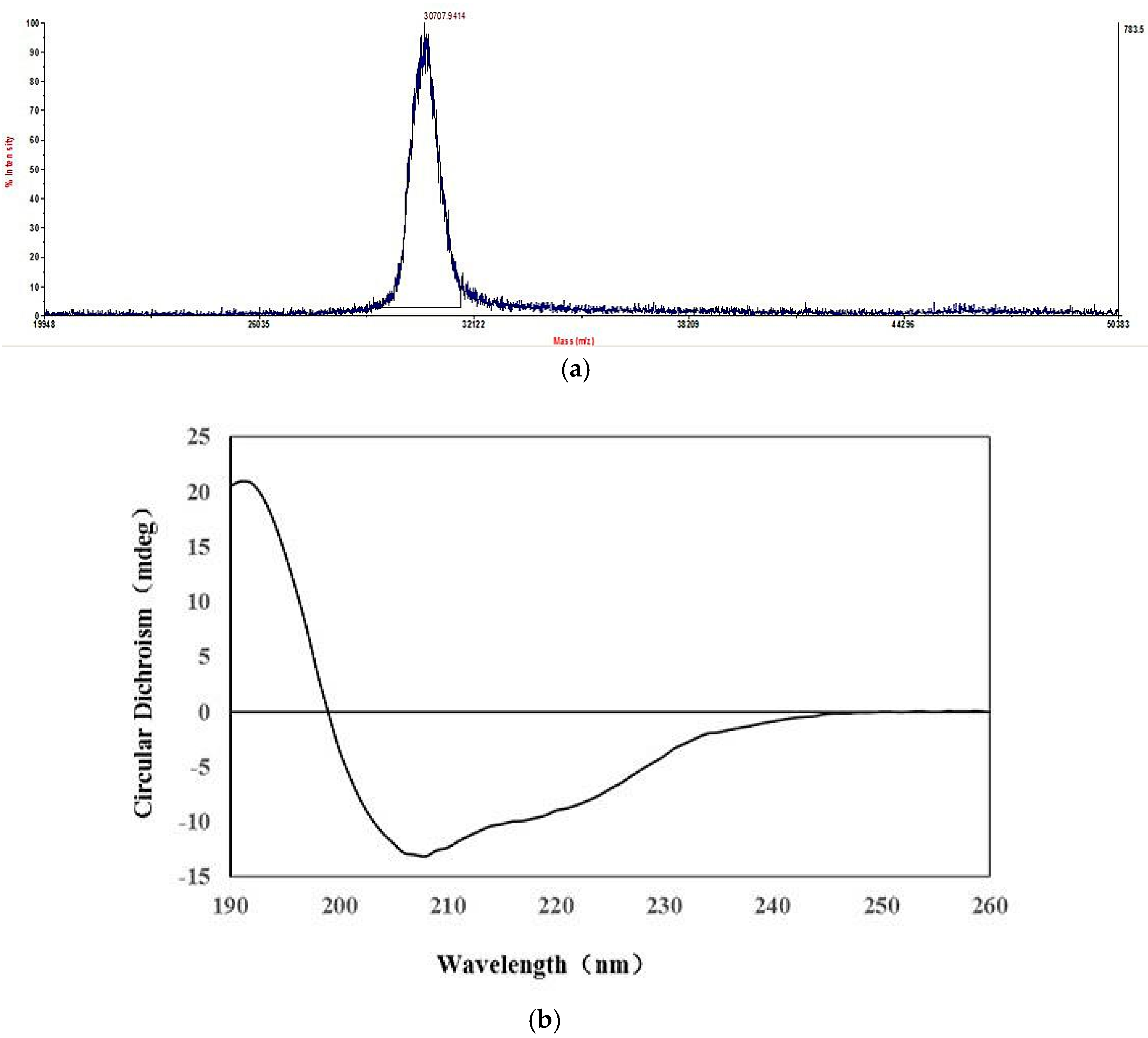

| Secondary Structure | α-Helix | β-Sheet | β-Turn | Random Coil |
|---|---|---|---|---|
| Ratio (%) | 28.4 | 26.0 | 18.6 | 29.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Bi, S.; Li, C.; Zheng, H.; Guo, Z.; Luo, Y.; Ou, X.; Song, L.; Zhu, J.; Yu, R. Purification and Characterization of a New CRISP-Related Protein from Scapharca broughtonii and Its Immunomodulatory Activity. Mar. Drugs 2020, 18, 299. https://doi.org/10.3390/md18060299
Liu W, Bi S, Li C, Zheng H, Guo Z, Luo Y, Ou X, Song L, Zhu J, Yu R. Purification and Characterization of a New CRISP-Related Protein from Scapharca broughtonii and Its Immunomodulatory Activity. Marine Drugs. 2020; 18(6):299. https://doi.org/10.3390/md18060299
Chicago/Turabian StyleLiu, Wanying, Sixue Bi, Chunlei Li, Hang Zheng, Zhongyi Guo, Yuanyuan Luo, Xiaozheng Ou, Liyan Song, Jianhua Zhu, and Rongmin Yu. 2020. "Purification and Characterization of a New CRISP-Related Protein from Scapharca broughtonii and Its Immunomodulatory Activity" Marine Drugs 18, no. 6: 299. https://doi.org/10.3390/md18060299
APA StyleLiu, W., Bi, S., Li, C., Zheng, H., Guo, Z., Luo, Y., Ou, X., Song, L., Zhu, J., & Yu, R. (2020). Purification and Characterization of a New CRISP-Related Protein from Scapharca broughtonii and Its Immunomodulatory Activity. Marine Drugs, 18(6), 299. https://doi.org/10.3390/md18060299




