Prostaglandins Isolated from the Octocoral Plexaura homomalla: In Silico and In Vitro Studies Against Different Enzymes of Cancer
Abstract
1. Introduction
2. Results and Discussion
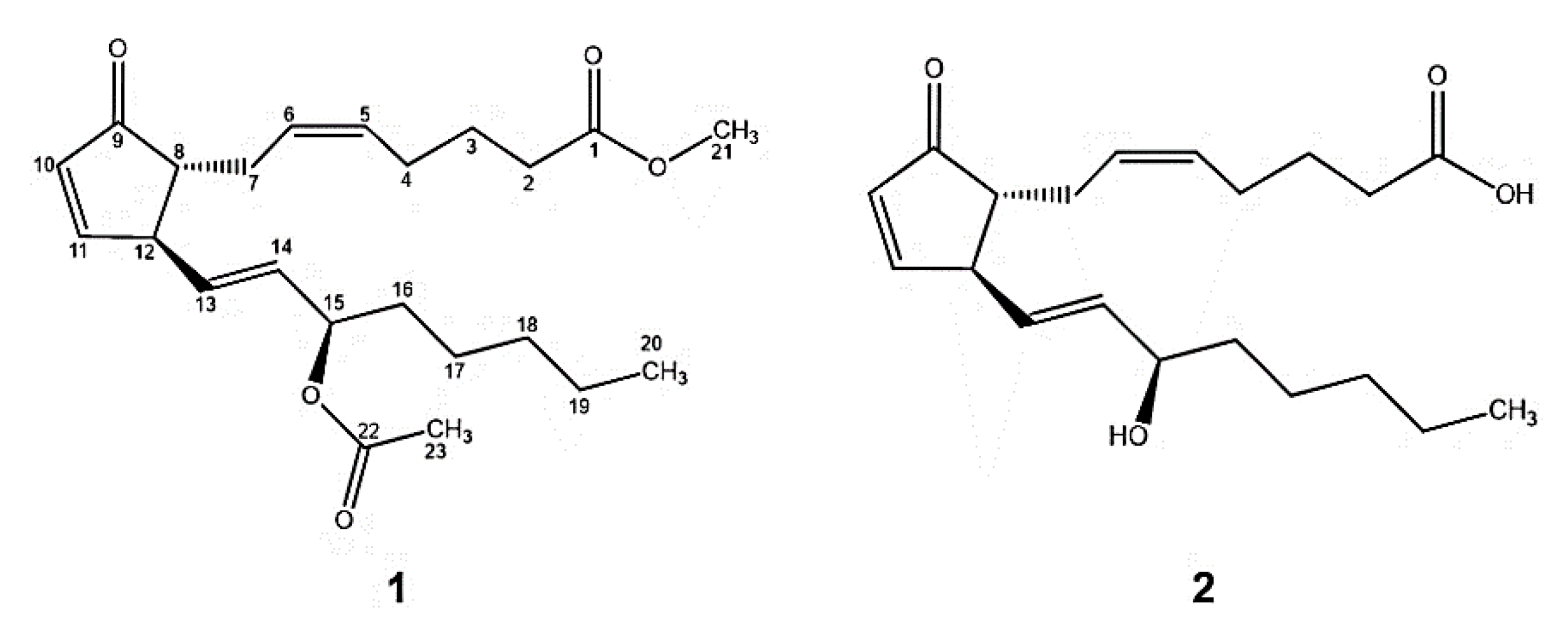
2.1. Extraction, Identification, and Structure Elucidation
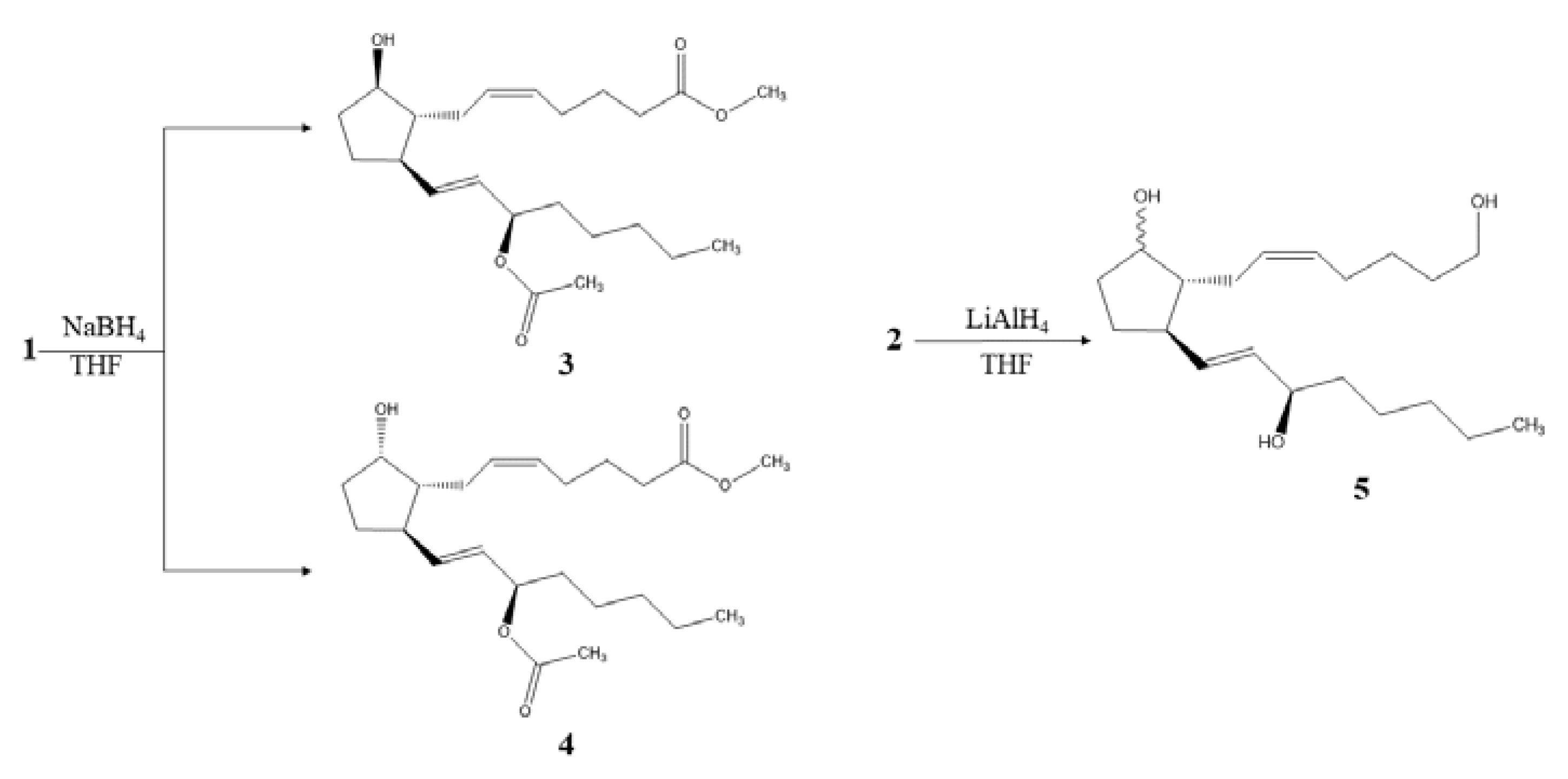
2.2. Semisynthetic Derivatives
2.3. Cytotoxic Activity
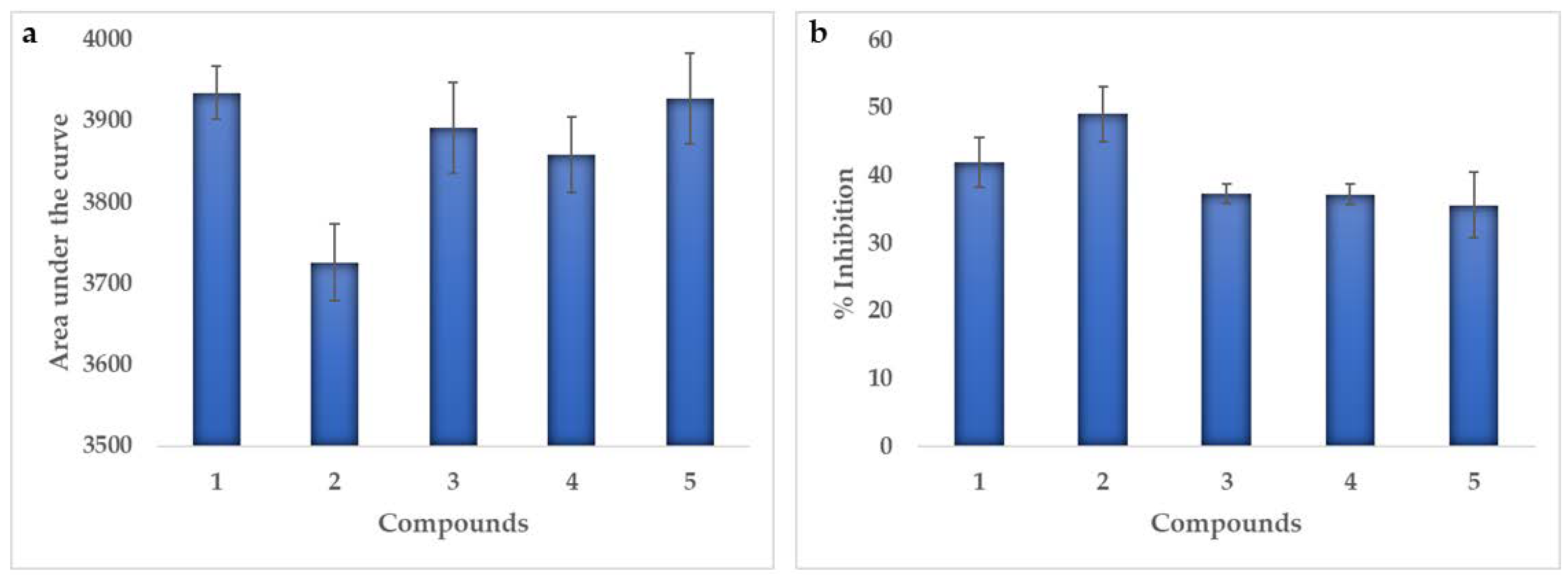
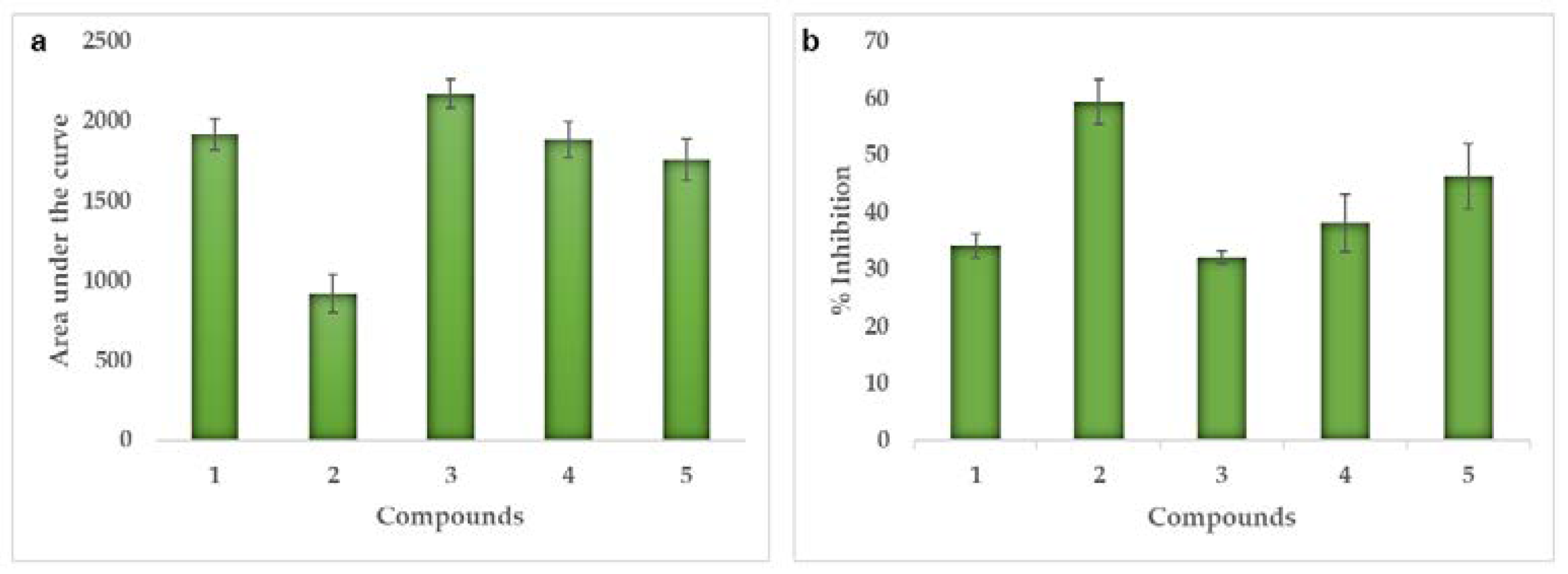
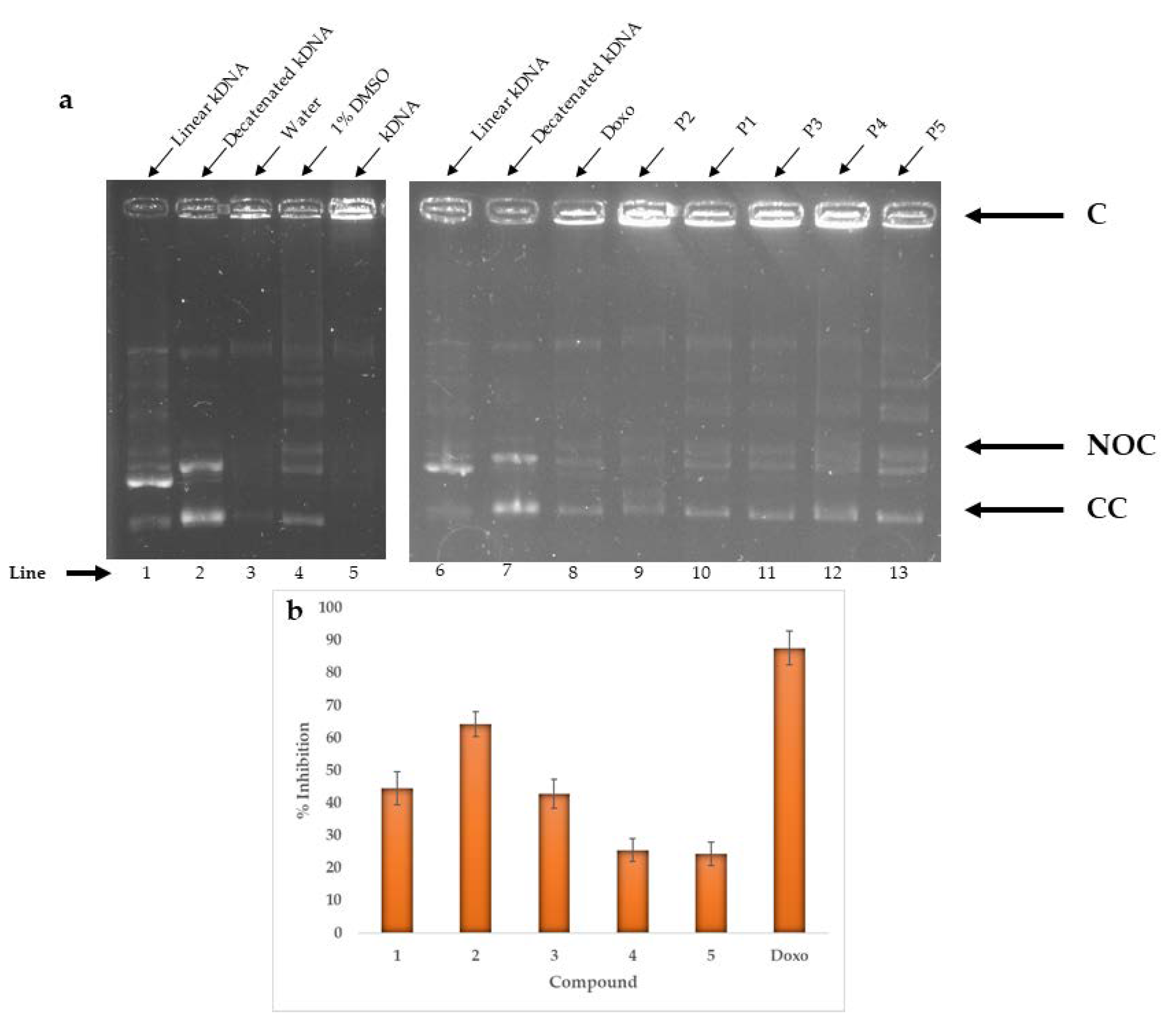
2.4. Enzymatic Activity
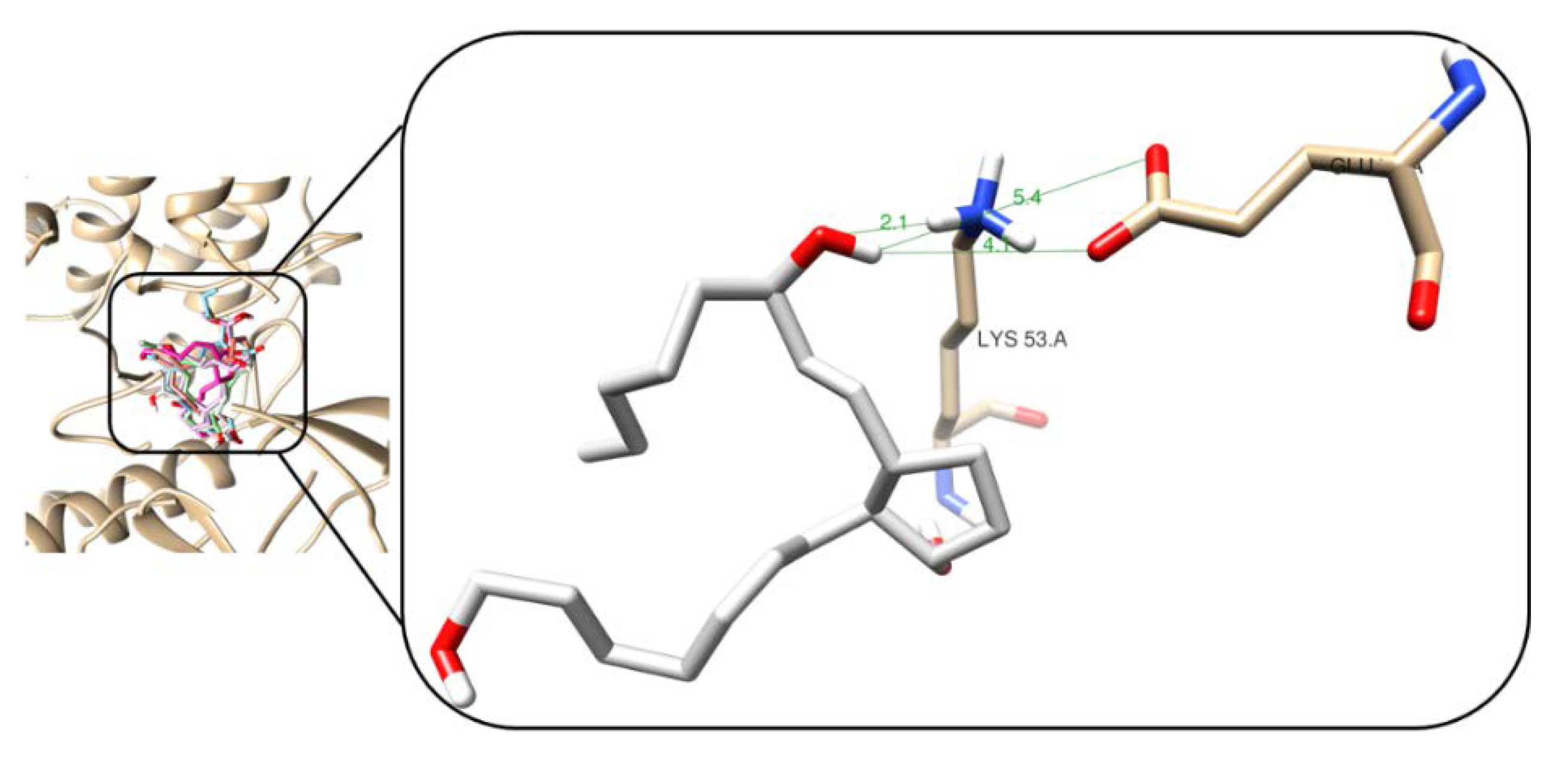
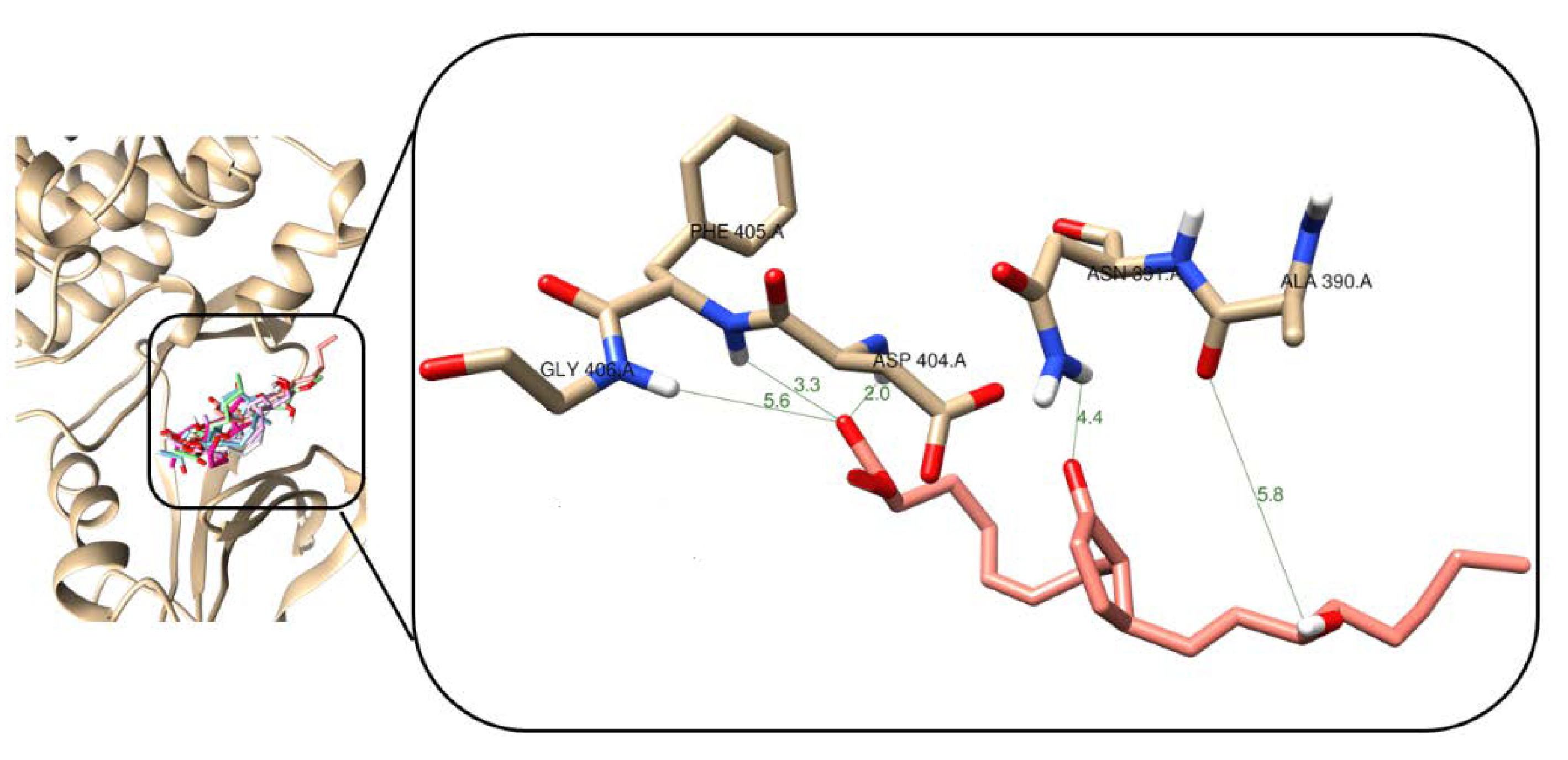
2.5. Binding Mode Analysis after Molecular Docking Studies
2.6. Comparison of In Vitro and In Silico Results
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Material, Extraction, and Separation of Compounds
3.3. Semisynthesis of Derivatives
3.3.1. Semi-synthesis of Derivates 3 and 4 from the Compound 1
3.3.2. Semi-synthesis of Derivate 5 from the Compound 2
3.4. Cytotoxic Activity
3.4.1. Cell Proliferation Assay
3.4.2. Anti-Proliferation Quantitative Analysis
3.5. Enzymatic Activity
3.5.1. Enzymatic Activity from p38α-Kinase and Src-Kinase
3.5.2. Enzymatic Activity from Topoisomerase IIα
3.6. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 19 May 2019).
- Granados García, M.; Arrieta Rodríguez, O.G.; Hinojosa Gómez, J. Tratamiento del Cancer: Oncología Médica, Quirúrgica y Radioterapia; Mexico, D.F., Ed.; Edició E. manual Moderno,1ra: Mexico City, Mexico, 2016. [Google Scholar]
- Ruiz-Torres, V.; Encinar, J.A.; Herranz-López, M.; Pérez-Sánchez, A.; Galiano, V.; Barrajón-Catalán, E.; Micol, V. An Updated Review on Marine Anticancer Compounds: The Use of Virtual Screening for the Discovery of Small-Molecule Cancer Drugs. Molecules 2017, 22, 1037. [Google Scholar] [CrossRef]
- Valmsen, K.; Jä, I.; Boeglin, W.E.; Lliki Varvas, K.; Koljak, R.; Nis Pehk, T.; Brash, A.R.; Samel, N. The origin of 15R-prostaglandins in the Caribbean coral Plexaura homomalla: Molecular cloning and expression of a novel cyclooxygenase. Proc. Natl. Acad. Sci. USA 2001, 98, 7700–7705. [Google Scholar] [CrossRef]
- Kavitha, T.; Velraj, G. Molecular structure, spectroscopic and docking analysis of 1,3-diphenylpyrazole-4-propionic acid: A good prostaglandin reductase inhibitor. J. Mol. Struct. 2018, 1155, 819–830. [Google Scholar] [CrossRef]
- Parker, J. Prostaglandin A, Protein Interactions and Inhibition of Cellular Proliferation. Prostaglandins 1995, 50, 359–375. [Google Scholar] [CrossRef]
- Agarwal, S.; Mehrotra, R. An overview of Molecular Docking. JSM Chem. 2016, 4, 1042–1045. [Google Scholar]
- Mukesh, B.; Rakesh, K. Molecular Docking: A Review. Int. J. Res. Ayurveda Pharm. 2011, 2, 1746–1751. [Google Scholar]
- Hegazy, M.-E.F.; Elshamy, A.I.; Mohamed, T.A.; Hamed, A.R.A.; Ibrahim, M.A.; Ohta, S.; Paré, P.W. Cembrene Diterpenoids with Ether Linkages from Sarcophyton ehrenbergi: An Anti-Proliferation and Molecular-Docking Assessment. Mar. Drugs 2017, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Reina Gamba, L.E. Estudio de Bioprospección de Octocorales de los Géneros Eunicea y Plexaura del Caribe Colombiano, Como Fuente de Metabolitos con Actividad Antiinflamatoria. Master’s Thesis, Universidad Nacional de Colombia, Bogota, Colombia, 2011. [Google Scholar]
- Gonzáles LAvaut, J.A.; Fernández Rijo, O.; Orret Cruz, E. Derivados de prostaglandinas partiendo de la prostaglandina A2: I. Estudio de la reacción de reducción del 15-acetato PGA2 metil ester. Rev. Cuba. Farm. 1987, 21, 305–312. [Google Scholar]
- Bundy Leonard, G.; Norman Allan, N. 11-Deoxyprostaglandine, 1974, CA 997338A,89. Available online: https://patents.google.com/patent/CA997338A/de (accessed on 20 June 2019).
- Garst, M.; Burk, R. Novel 7-(5-substituted cyclopentyl) and (5-subdtituted cyclopententyl) Heptyl Alcohols, Heptylamines and Heptanoic Acid Amines, and Method of Lowering Intraocular Pressure in the Eye of a Mammal by Administration of These Novel Compounds, 1994, WO 94/08587. Available online: https://patents.google.com/patent/CA2147502C/en (accessed on 20 June 2019).
- Elhassanny, A.E.M.; Ladin, D.A.; Soliman, E.; Albassam, H.; Morris, A.; Kobet, R.; Thayne, K.; Burns, C.; Danell, A.S.; Van Dross, R. Prostaglandin D 2 -ethanolamide induces skin cancer apoptosis by suppressing the activity of cellular antioxidants. Prostaglandins Other Lipid Mediat. 2019, 142, 9–23. [Google Scholar] [CrossRef]
- Ladin, D.A.; Soliman, E.; Escobedo, R.; Fitzgerald, T.L.; Yang, L.V.; Burns, C.; Van Dross, R. Synthesis and Evaluation of the Novel Prostamide, 15-Deoxy, Δ 12,14-Prostamide J 2, as a Selective Antitumor Therapeutic. Mol. Cancer Ther. 2017, 16, 838–849. [Google Scholar] [CrossRef]
- Amin, K.M.; Syam, Y.M.; Anwar, M.M.; Ali, H.I.; Abdel-Ghani, T.M.; Serry, A.M. Synthesis and molecular docking study of new benzofuran and furo[3,2-g]chromone-based cytotoxic agents against breast cancer and p38α MAP kinase inhibitors. Bioorg. Chem. 2018, 76, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Magid, A.F. Allosteric Modulators: An Emerging Concept in Drug Discovery. ACS Med. Chem. Lett. 2015, 6, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Karim, S.S.; Anwar, M.M.; Mohamed, N.A.; Nasr, T.; Elseginy, S.A. Design, synthesis, biological evaluation and molecular docking studies of novel benzofuran-pyrazole derivatives as anticancer agents. Bioorg. Chem. 2015, 63, 1–12. [Google Scholar] [CrossRef]
- Manda, S.; Sharma, S.; Wani, A.; Joshi, P.; Kumar, V.; Guru, S.K.; Bharate, S.S.; Bhushan, S.; Vishwakarma, R.A.; Kumar, A.; et al. Discovery of a marine-derived bis-indole alkaloid fascaplysin, as a new class of potent P-glycoprotein inducer and establishment of its structure-activity relationship. Eur. J. Med. Chem. 2016, 107, 1–11. [Google Scholar] [CrossRef]
- Abdelhafez, O.M.; Ahmed, E.Y.; Abdel Latif, N.A.; Arafa, R.K.; Abd Elmageed, Z.Y.; Ali, H.I. Design and molecular modeling of novel P38α MAPK inhibitors targeting breast cancer, synthesized from oxygen heterocyclic natural compounds. Bioorg. Med. Chem. 2019, 27, 1308–1319. [Google Scholar] [CrossRef]
- Jemimah Naine, S.; Subathra Devi, C.; Mohanasrinivasan, V.; George Priya Doss, C.; Thirumal Kumar, D. Binding and molecular dynamic studies of sesquiterpenes (2R-acetoxymethyl-1,3,3-trimethyl-4t-(3-methyl-2-buten-1-yl)-1t- cyclohexanol) derived from marine Streptomyces sp. VITJS8 as potential anticancer agent. Appl. Microbiol. Biotechnol. 2015, 100, 2869–2882. [Google Scholar] [CrossRef]
- Jackson, P.A.; Widen, J.C.; Harki, D.A.; Brummond, K.M. Covalent Modifiers: A Chemical Perspective on the Reactivity of α,β-Unsaturated Carbonyls with Thiols via Hetero-Michael Addition Reactions. J. Med. Chem. 2017, 60, 839–885. [Google Scholar] [CrossRef]
- Tello, E.; Castellanos, L.; Arevalo-Ferro, C.; Duque, C. Cembranoid diterpenes from the Caribbean sea whip Eunicea knighti. J. Nat. Prod. 2009, 72, 1595–1602. [Google Scholar] [CrossRef]
- Yin Di, S.; Tzu-Rong, S.; Zhi Hong, W.; Tsong-Long, H.; Lee-Shing, F.; Jih-Jung, C.; Yang-Chang, W.; Jyh Horng, S.; Ping Jyun, S. Briarenolides K and L, New Anti-Inflammatory Briarane Diterpenoids from an Octocoral Briareum sp. (Briareidae). Mar. Drugs 2015, 13, 1037–1050. [Google Scholar]
- Nel, M.; Joubert, A.M.; Dohle, W.; Potter, B.V.L.; Theron, A.E. Modes of cell death induced by tetrahydroisoquinoline-based analogs in MDA-MB-231 breast and A549 lung cancer cell lines. Drug Des. Devel. Ther. 2018, 12, 1881–1904. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Sebaugh, J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011, 10, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Worzella, T.; Butzler, M.; Hennek, J.; Hanson, S.; Simdon, L.; Goueli, S.; Cowan, C.; Zegzouti, H. A Flexible Workflow for Automated Bioluminescent Kinase Selectivity Profiling. SLAS Technol. Transl. Life Sci. Innov. 2017, 22, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Zegzouti, H.; Alves, J.; Worzella, T.; Vidugiris, G.; Cameron, G.; Vidugiriene, J.; Goueli, S. Screening and Profiling Kinase Inhibitors with a Luminescent ADP Detection Platform. Available online: https://worldwide.promega.com/resources/pubhub/screening-and-profiling-kinase-inhibitors-with-a-luminescent-adp-detection-platform/ (accessed on 22 May 2019).
- TopoGEN Inc. Topoisomerase II Assay Kit User Manual; TopoGEN Inc.: Port Orange, FL, USA, 2011. [Google Scholar]

| No. | 3 | 4 | 5 | |||
|---|---|---|---|---|---|---|
| δC | δH, Multi, (J in Hz) | δC | δH, Multi, (J in Hz) | δC | δH, Multi, (J in Hz) | |
| 1 | 175.6 | ----- | 174.1 | ----- | 62.9 | 3.92 d (4.8) |
| 2 | 34.5 | 2.33 t (7.4) | 34.4 | 2.32 t (8.0) | 32.3 | 1.44 s; 1.58 s |
| 3 | 24.8 | 1.70 dd (14.6, 7.4) | 26.6 | 1.69 m | 25.9 | 1.55 s |
| 4 | 26.6 | 2.09 d (7.9) | 29.1 | 2.09 d, (7.3) | 29.9 | 2.09 q (6.8, 6.3) |
| 5 | 129.6 | 5.38 d (7.2) | 130.1 | 5.40 d (7.4) | 131,4 | 5.45 m |
| 6 | 128.6 | 5.34 d (7.1) | 128.2 | 5.36 d (7.2) | 128.1 | 5.45 m |
| 7 | 29.7 | 2.09 d (7.9); 2.17 s | 29.5 | 2.13 d (8.0); 2.17 s | 27.1 | 2.25 dd (13.9, 4.9); 2.04 s |
| 8 | 51.7 | 1.70 dd (14.6, 7.4) | 54.1 | 1,69 m | 55.9 | 1.91 dt (7.5, 3.7) |
| 9 | 74.8 | 3.89 dd (11.6, 6.9) | 78.1 | 4.21 t (4.8) | 78.5 | 4.05 dd (6.4, 3.9) |
| 10 | 33.4 | 1.53 d (6.6); 1.59 d (7.5) | 33.3 | 1.64 d (9.6); 1.69 m | 33.6 | 1.76 s |
| 11 | 29.9 | 1.33 s; 1.59 d (7.5) | 29.7 | 1.33 s | 29.3 | 1.46 s; 1.60 s |
| 12 | 45.5 | 2.33 t (7.4) | 47.0 | 2.32 t (8.0) | 52.7 | 2,32 s |
| 13 | 137.4 | 5.62 dd (15.3, 8.3) | 137.1 | 5.54 dd (15.3, 8.7) | 135.1 | 5.77 d (20.6) |
| 14 | 129.3 | 5.46 dd (13.3, 7.0) | 128.7 | 5.44 dd (11.2, 5.4) | 133.2 | 5.58 dd (15.5, 8.0) |
| 15 | 73.9 | 5.19 dd (13.4, 6.7) | 74.7 | 5.19 dd (13.5, 6.8) | 72.9 | 4.05 dd (6.4, 3.9) |
| 16 | 33.5 | 1.59 d (7.5); 1.53 d (6.6) | 33.4 | 1.64 d (9.6); 1.69 m | 37.5 | 1.44 s; 1.58 s |
| 17 | 25.1 | 1.25 m | 24.8 | 1.25 s | 25.3 | 1.28 s |
| 18 | 31.5 | 1.25 m | 31.5 | 1.25 s | 31.9 | 1.28 s |
| 19 | 22.5 | 1.25 m | 22.5 | 1.25 s | 22.8 | 1.28 s |
| 20 | 14.0 | 0.88 s | 14.0 | 0.88 s | 14.2 | 0.88 s |
| 21 | 51.5 | 3.67 s | 51.5 | 3.67 s | ----- | ----- |
| 22 | 170.4 | ----- | 170.4 | ----- | ----- | ----- |
| 23 | 21.3 | 2.04 s | 21.4 | 2.04 s | ----- | ----- |
| Compound | 1 IC50 (µg/mL) | |||
|---|---|---|---|---|
| A549 | MDA-MB-231 | HDFa | L929 | |
| 1 | >100 | 27.53 | 28.28 | 71.74 |
| 2 | 25.20 | 16.46 | 17.87 | 40.39 |
| 3 | >100 | >100 | >100 | >100 |
| 4 | >100 | >100 | 50.68 | 54.33 |
| 5 | >100 | >100 | >100 | >100 |
| Compound | 1 | 2 | 3 | 4 | 5 | ||
|---|---|---|---|---|---|---|---|
| Cytotoxicity | MDA-MB-231 (breast cancer) | 27.53 | 16.46 | >100 | >100 | >100 | |
| A549 (lung cancer) | >100 | 25.20 | >100 | >100 | >100 | ||
| p38α-Kinase | In vivo (% inhibition) | 42 | 49 | 37 | 37 | 36 | |
| In silico | Scores (kcal/mol) | −8.3 | −8.0 | −8.1 | −8.1 | −8.5 | |
| Ki (µM) | 0.82 | 1.37 | 1.16 | 1.16 | 0.59 | ||
| Topoisomerase Iiα | In vivo (% inhibition) | 45 | 64 | 43 | 26 | 24 | |
| In silico | Scores (kcal/mol) | −8.2 | −8.7 | −8.5 | −8.6 | −8.0 | |
| Ki (µM) | 0.98 | 0.42 | 0.59 | 0.50 | 1.37 | ||
| Src-Kinase | In vivo (% inhibition) | 34 | 59 | 32 | 38 | 46 | |
| In silico | Scores (kcal/mol) | −8.3 | −8.9 | −7.9 | −8.1 | −8.1 | |
| Ki (µM) | 0.82 | 0.30 | 1.62 | 1.16 | 1.16 | ||
| Kinase | Substrate | Concentrations of ATP (µM) | SB10 (ng) 1 |
|---|---|---|---|
| p38α | p38 substrate | 150 | 4 |
| Src | Src substrate | 50 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hurtado, D.X.; Castellanos, F.A.; Coy-Barrera, E.; Tello, E. Prostaglandins Isolated from the Octocoral Plexaura homomalla: In Silico and In Vitro Studies Against Different Enzymes of Cancer. Mar. Drugs 2020, 18, 141. https://doi.org/10.3390/md18030141
Hurtado DX, Castellanos FA, Coy-Barrera E, Tello E. Prostaglandins Isolated from the Octocoral Plexaura homomalla: In Silico and In Vitro Studies Against Different Enzymes of Cancer. Marine Drugs. 2020; 18(3):141. https://doi.org/10.3390/md18030141
Chicago/Turabian StyleHurtado, Diana Ximena, Fabio A. Castellanos, Ericsson Coy-Barrera, and Edisson Tello. 2020. "Prostaglandins Isolated from the Octocoral Plexaura homomalla: In Silico and In Vitro Studies Against Different Enzymes of Cancer" Marine Drugs 18, no. 3: 141. https://doi.org/10.3390/md18030141
APA StyleHurtado, D. X., Castellanos, F. A., Coy-Barrera, E., & Tello, E. (2020). Prostaglandins Isolated from the Octocoral Plexaura homomalla: In Silico and In Vitro Studies Against Different Enzymes of Cancer. Marine Drugs, 18(3), 141. https://doi.org/10.3390/md18030141






