Purification and Evaluation of N-benzyl Cinnamamide from Red Seaweed Gracilaria fisheri as an Inhibitor of Vibrio harveyi AI-2 Quorum Sensing
Abstract
1. Introduction
2. Results
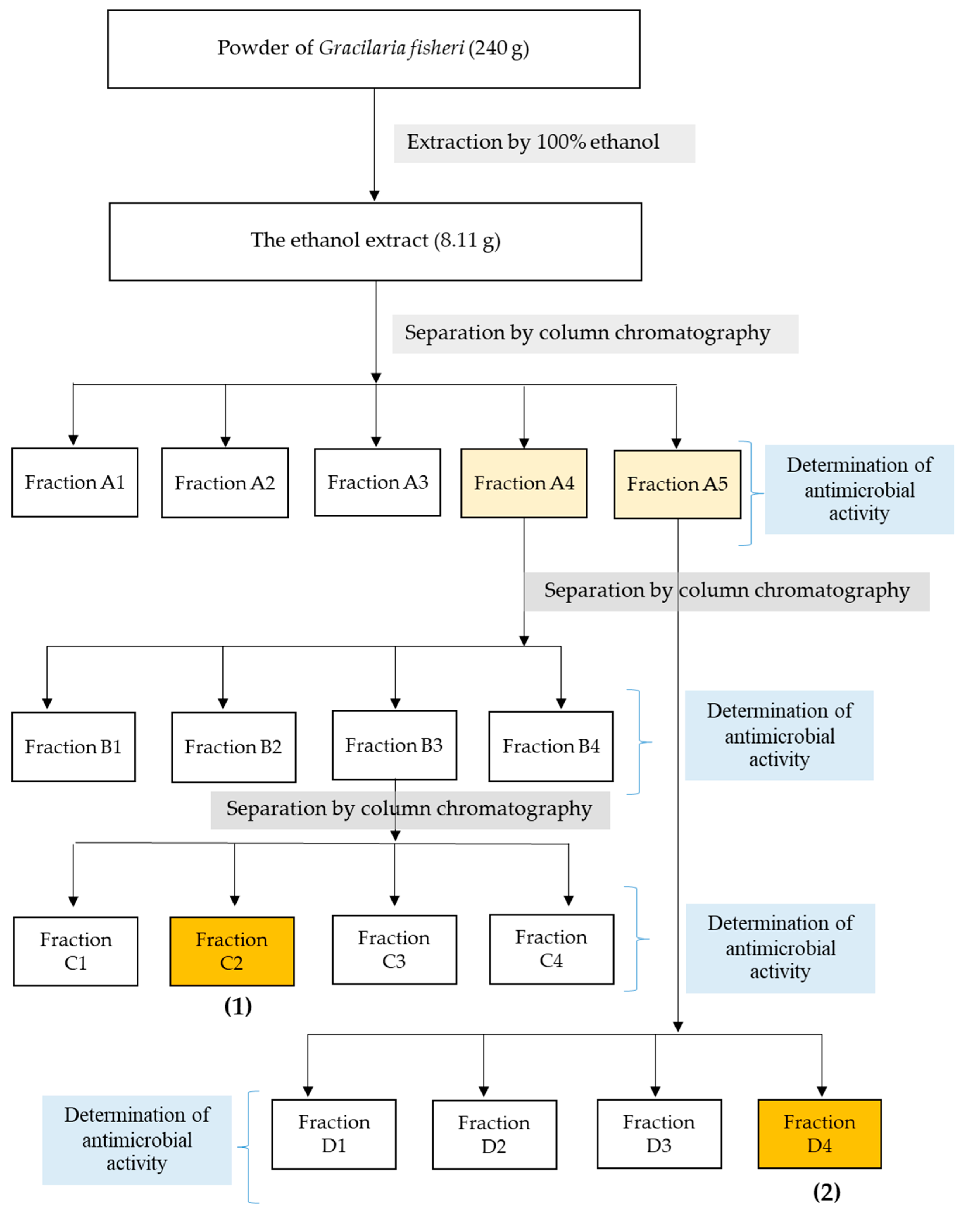
2.1. Fractionation of Antimicrobial Compounds from Ethanol Extract of G. fisheri
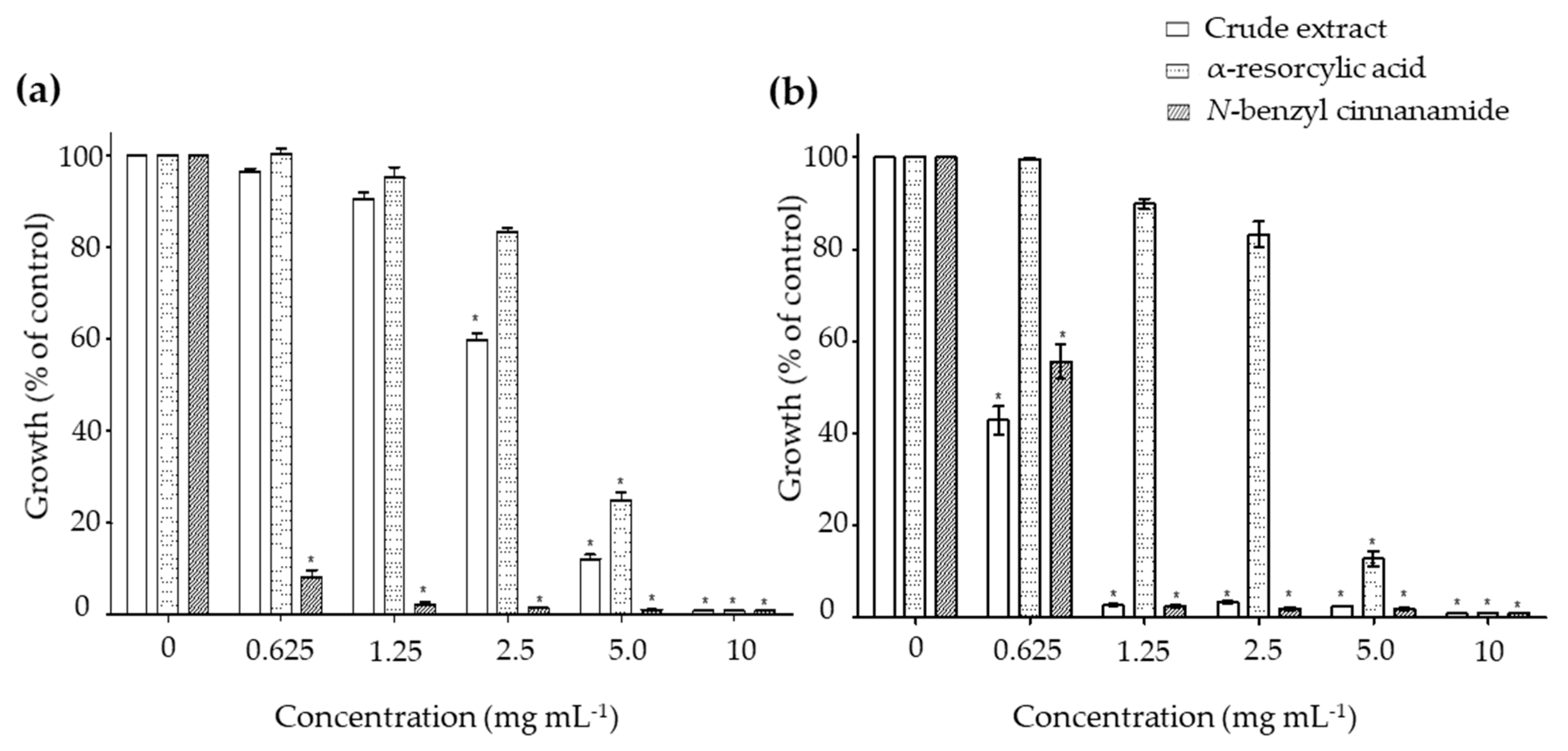
2.2. In Vitro Antibacterial Activity of the Pure Compounds
2.3. The Anti-Biofilm Biomass of the Pure Compounds
2.4. The Effect of the Pure Compounds on the Bioluminescence Production
2.5. The Effect of the Pure Compounds on AI-2 Communication
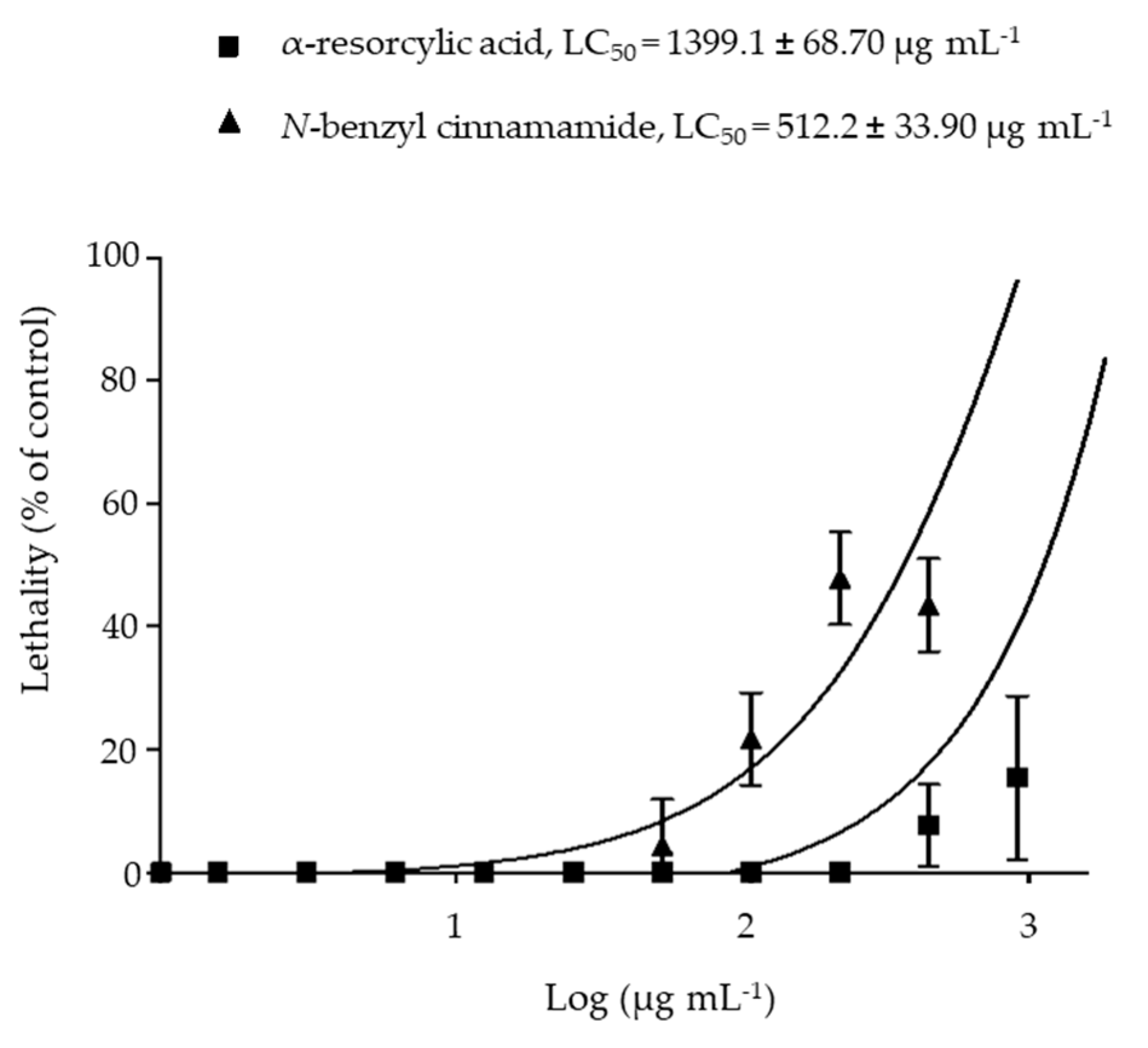
2.6. Toxicity of the Pure Compounds in the Brine Shrimp
3. Discussion
4. Materials and Methods
4.1. Extraction of the Ethanol Crude Extract
4.2. V. harveyi and Culture Conditions
4.3. Minimal Inhibitory Concentration (MIC) and Sub-MIC Concentration
4.4. Biofilm Biomass Inhibition
4.5. Activity Guided Purification of Anti-Microbial Compounds
4.6. Structural Elucidation
4.7. Quorum Sensing Inhibiting Activity
4.7.1. Bioluminescence Assay
4.7.2. Interference with AI-2 Communication
4.8. Artemia salina (A. salina) Lethality Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, P.C.; Lee, K.K. Cysteine protease is a major exotoxin of pathogenic luminous Vibrio harveyi in the tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 1999, 28, 428–430. [Google Scholar] [CrossRef]
- Austin, B.; Zhang, X.H. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrate. Lett. Appl. Microbiol. 2006, 43, 119–124. [Google Scholar] [CrossRef]
- Yang, Q.; Defoirdt, T. Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ. Microbial. 2015, 17, 960–968. [Google Scholar] [CrossRef]
- Winzer, K.; Williams, P. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol 2001, 291, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbial. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, J. Role and regulation of bacterial LuxR-like regulators. J. Cell. Biochem. 2011, 112, 2694–2702. [Google Scholar] [CrossRef] [PubMed]
- Henke, J.M.; Bassler, B.L. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacterial. 2004, 186, 6902–6914. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Jesus, A.; Correia-da-Silva, M.; Afonso, C.; Pinto, M.; Cidade, H. Isolation and potential biological applications of haloaryl secondary metabolites from macroalgae. Mar. Drugs 2019, 17, 73. [Google Scholar] [CrossRef]
- Defoirdt, T.; Crab, R.; Wood, T.K.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Quorum sensing-disrupting brominated furanones protect the gnotobiotic brine shrimp Artemia franciscana from pathogenic Vibrio harveyi, Vibrio campbellii, and Vibrio parahaemolyticus isolates. Appl. Environ. Microbiol. 2006, 72, 6419–6423. [Google Scholar] [CrossRef]
- Brackman, G.; Celen, S.; Hillaert, U.; Van Calenbergh, S.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS ONE 2011, 6, e16084. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, D.J. Marine natural products. Nat. Prod. Rep. 2001, 18, 1R–49R. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.J.; Falqué, E.; Domínguez, H. Antimicrobial action of compounds from marine seaweed. Mar. Drugs 2016, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Kain, J.M.; Destombe, C. A review of the life history, reproduction and phenology of Gracilaria. J. Appl. Phycol. 1995, 7, 269–281. [Google Scholar]
- Reverter, M.; Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Use of plant extracts in fish aquaculture as an alternative to chemotherapy: Current status and future perspectives. Aquaculture 2014, 433, 50–61. [Google Scholar] [CrossRef]
- Kurniawati, I.; Adam, A. Antibacterial effect of Gracilaria verrucosa bioactive on fish pathogenic bacteria. Egypt. J. Aquat. Res. 2016, 42, 405–410. [Google Scholar]
- Kolanjinathan, K.; Saranraj, P. Pharmacological efficacy of marine seaweed Gracilaria edulis extracts against clinical pathogens. Glob. J. Pharmacol. 2014, 8, 268–274. [Google Scholar]
- Prasad, M.; Sushant, S.; Ganesh, R. Antibacterial activity of seaweed (Gracilaria species) extracts against infectious pathogens. Asian J. Biolog. Life Sci. 2012, 1, 219–222. [Google Scholar]
- Manefield, M.; de Nys, R.; Naresh, K.; Roger, R.; Givskov, M.; Peter, S.; Kjelleberg, S. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999, 145, 283–291. [Google Scholar] [CrossRef]
- Kanjana, K.; Radtanatip, T.; Asuvapongpatana, S.; Withyachumnarnkul, B.; Wongprasert, K. Solvent extracts of the red seaweed Gracilaria fisheri prevent Vibrio harveyi infections in the black tiger shrimp Penaeus monodon. Fish Shellfish Immunol. 2011, 30, 389–396. [Google Scholar] [CrossRef]
- Karchesy, Y.M.; Kelsey, R.G.; Constantine, G.; Karchesy, J.J. Biological screening of selected Pacific Northwest forest plants using the brine shrimp (Artemia salina) toxicity bioassay. Springerplus 2016, 5, 510. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chowdhury, M.; Huo, Y.; Gong, J. Phytogenic compounds as alternatives to in-feed antibiotics: Potentials and challenges in application. Pathogens 2015, 4, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Sijwali, P.S.; Pandey, K.C.; Rosenthal, P.J. Plasmodium falciparum: Biochemical characterization of the cysteine protease falcipain-2′. Exp. Parasitol. 2006, 112, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, Y.; Kinoshita, H.; Tanaka, T.; Nakamura, T.; Fujimoto, K.; Kashimoto, S.; Kojima, T.; Kato, S. Pleuromutilin derivatives having a purine ring. Part 2: Influence of the central spacer on the antibacterial activity against Gram-positive pathogens. Bioorg. Med. Chem. Lett. 2009, 19, 170–174. [Google Scholar] [CrossRef]
- Guzman, J.D. Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 2014, 19, 19292–19349. [Google Scholar] [CrossRef]
- Seelolla, G.; Cheera, P.; Ponneri, V. Synthesis, antimicrobial and antioxidant activities of novel series of cinnamamide derivatives having morpholine moiety. Med. Chem. 2014, 4, 778–783. [Google Scholar] [CrossRef]
- Brackman, G.; Defoirdt, T.; Miyamoto, C.; Bossier, P.; Van Calenbergh, S.; Nelis, H.; Coenye, T. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008, 8, 149. [Google Scholar] [CrossRef]
- Joshi, J.R.; Burdman, S.; Lipsky, A.; Yariv, S.; Yedidia, I. Plant phenolic acids affect the virulence of P. ectobacterium aroidearum and P. carotovorum ssp. brasiliense via quorum sensing regulation. Mol. Plant Pathol. 2016, 17, 487–500. [Google Scholar]
- Solts, P.; Wright, C.W.; Anderson, M.M.; Gupta, M.P.; Phillipson, J.D. A microwell cytotoxicity assay using Artemia salina. Plant Med. 1993, 59, 250–252. [Google Scholar]
- Moshi, M.; Innocent, E.; Magadula, J.; Otieno, D.; Weisheit, A.; Mbabazi, P.; Nondo, R. Brine shrimp toxicity of some plants used as traditional medicines in Kagera region, north western Tanzania. Tanzan. J. Health Res. 2010, 12, 63–67. [Google Scholar] [CrossRef]
- Lee, M.-N.; Kim, S.-K.; Li, X.-H.; Lee, J.-H. Bacterial virulence analysis using brine shrimp as an infection model in relation to the importance of quorum sensing and proteases. J. Gen. Appl. Microbiol. 2014, 60, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Qin, S.; Li, F.; Chi, X.; Laatsch, H. Isolation, antimicrobial activity, and metabolites of fungus Cladosporium sp. associated with red alga Porphyra yezoensis. Curr. Microbial. 2008, 56, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.L.; Yhebra, R.S.; Sardiñas, I.G.; Buela, L.I. Comparative study of the assay of Artemia salina L. and the estimate of the medium lethal dose (LD50 value) in mice, to determine oral acute toxicity of plant extracts. Phytomedicine 2001, 8, 395–400. [Google Scholar]
- Karnjana, K.; Soowannayan, C.; Wongprasert, K. Ethanolic extract of red seaweed Gracilaria fisheri and furanone eradicate Vibrio harveyi and Vibrio parahaemolyticus biofilms and ameliorate the bacterial infection in shrimp. Fish Shellfish Immunol. 2019, 88, 91–101. [Google Scholar] [CrossRef]
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacterial. 1997, 179, 4043–4045. [Google Scholar] [CrossRef]
- Surette, M.G.; Bassler, B.L. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbial. 1999, 31, 585–595. [Google Scholar] [CrossRef]
- Pasharawipas, T.; Thaikua, S.; Sriurairatana, S.; Ruangpan, L.; Direkbusarakum, S.; Manopvisetcharean, J.; Flegel, T.W. Partial characterization of a novel bacteriophage of Vibrio harveyi isolated from shrimp culture ponds in Thailand. Virus Res. 2005, 114, 63–69. [Google Scholar] [CrossRef]
- Valgas, C.; de Souza, S.M.; Smânia, E.F.; Smânia, A., Jr. Screening methods to determine antibacterial activity of natural products. Braz. J. Microbiol. 2007, 38, 369–380. [Google Scholar] [CrossRef]
- Van den Driessche, F.; Rigole, P.; Brackman, G.; Coenye, T. Optimization of resazurin-based viability staining for quantification of microbial biofilms. J. Microbiol. Methods 2014, 98, 31–34. [Google Scholar] [CrossRef]
- Pettit, R.K.; Weber, C.A.; Kean, M.J.; Hoffmann, H.; Pettit, G.R.; Tan, R.; Franks, K.S.; Horton, M.L. Microplate Alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob. Agents Chemother. 2005, 49, 2612–2617. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef] [PubMed]
- 3,5-Dihydroxybenzoic Acid. Available online: http://www.chemspider.com/Chemical-Structure.7146.html?rid=766eba99-201a-4ed0-bf90-66f72be804bc (accessed on 24 January 2020).
- Harard, G.; Gunther, J. 1H nuclear magnetic resonance spectra of cyclic monoenes: Hydrocarbons, ketones, heterocycles, and benzo derivatives. Chem. Rev. 1977, 77, 599–637. [Google Scholar]
- Scott, K.N. Carbon-13 nuclear magnetic resonance of biologically important aromatic acids. I. Chemical shifts of benzoic acid and derivatives. J. Am. Chem. Soc. 1972, 94, 8564–8568. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.N. NMR parameters of biologically important aromatic acids I. Benzoic acid and derivatives. J. Magn. Resonance 1970, 2, 361–376. [Google Scholar]
- Ren, W.; Yamane, M. Mo(CO)6-Mediated Carbamoylation of Aryl Halides. J. Org. Chem. 2010, 75, 8410–8415. [Google Scholar] [CrossRef]
- Vikram, A.; Jesudhasan, P.R.; Jayaprakasha, G.K.; Pillai, S.D.; Patil, B.S. Citrus limonoids interfere with Vibrio harveyi cell-cell signalling and biofilm formation by modulating the response regulator LuxO. Microbiology 2011, 157, 99–110. [Google Scholar] [CrossRef]
- Nunes, B.S.; Carvalho, F.D.; Guilhermino, L.M.; Van Stappen, G. Use of the genus Artemia in ecotoxicity testing. Environ. Pollut. 2006, 144, 453–462. [Google Scholar] [CrossRef]
- Meyer, B.; Ferrigni, N.; Putnam, J.; Jacobsen, L.; Nichols, D.E.; McLaughlin, J.L. Brine shrimp: A convenient general bioassay for active plant constituents. Planta Med. 1982, 45, 31–34. [Google Scholar] [CrossRef]




| Bacteria | Compound | Minimum Inhibitory Concentration (MIC, mg mL−1) |
|---|---|---|
| V. harveyi (1114) | Crude extract | 8.08 ± 0.06 |
| α-Resorcylic acid | 11.27 ± 0.18 | |
| N-Benzyl cinnamamide | 1.66 ± 0.08 | |
| V. harveyi (BAA 1116) | Crude extract | 2.40 ± 0.07 |
| α-Resorcylic acid | 11.25 ± 0.21 | |
| N-Benzyl cinnamamide | 2.46 ± 0.17 |
| Strain | Relevant Feature | Reference |
|---|---|---|
| BAA 1116 (BB120) | Wild type | [35] |
| BAA 1119 (BB152) | luxM::Tn5, (Sensors 1+, 2+ and Auto inducers 1−, 2+) | [35] |
| BAA 1121 (MM32) | luxN::CmR luxS::Tn5, (Sensors 1−, 2+ and Auto inducers 1+, 2−) | [36] |
| 1114 | Isolated from Thai shrimp ponds | [37] |
| α-Resorcylic Acid | |||
|---|---|---|---|
| Position | δC, Type | δH, (J in Hz) | HMBC |
| 1 | 170.19 C=O | ||
| 2 | 133.79 (C) | ||
| 3 | 109.19 (CH) | 6.46 t (2.3) | 1, 2, 4, |
| 4 | 159.82 (C) | ||
| 5 | 108.24 (CH) | 6.94 d (2.3 | 4, 6 |
| 6 | 159.82 (C) | ||
| 7 | 109.19 (CH) | 6.46 t (2.3) | 1, 2, 6 |
| N-benzyl Cinnamamide | |||
|---|---|---|---|
| Position | δC, Type | δH, (J in Hz) | HMBC |
| 1 | 166.08 (C=O) | - | 2, 3, 2/ |
| 2 | 120.73 (CH) | 6.45, d, (15.6) | 3, 2/ |
| 3 | 141.51 (CH) | 7.65, d, (15.6) | 2, 5, 9 |
| 4 | 134.99 (C) | - | 2, 3, 5, 6, 8, 9 |
| 5 | 129.80 (CH) | 7.49, m | 2, 3, 6, 8 |
| 6 | 128.97 (CH) | 7.28, m | 5, 9, 6, 8 |
| 7 | 129.86(CH) | 7.28, m | 5, 6, 8 |
| 8 | 128.97 (CH) | 7.28, m | 5, 6, 8, 9 |
| 9 | 129.80. (CH) | 7.49, m | 2, 3, 6, 8 |
| NH | 6.37 (NH) | 2, 3, 6 | |
| 1′ | 44.02 (CH2) | 4.57, d, (5.7) | 5, 6, 7 |
| 2′ | 138.40 (C) | - | 1′, 4′ |
| 3′ | 127.98 (CH) | 7.35, m | 1′, 4′, 5′, 6′, 7′ |
| 4′ | 128.06 (CH) | 7.35, m | 1′, 4′, 5′, 6′, 7′ |
| 5′ | 127.71 (CH) | 7.35, m | 1′, 4′, 5′, 6′, 7′ |
| 6′ | 128.06 (CH) | 7.35, m | 1′, 4′, 5′, 6′, 7′ |
| 7′ | 127.98 (CH) | 7.35, m | 1′, 4′, 5′, 6′, 7′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karnjana, K.; Nobsathian, S.; Soowannayan, C.; Zhao, W.; Tang, Y.-J.; Wongprasert, K. Purification and Evaluation of N-benzyl Cinnamamide from Red Seaweed Gracilaria fisheri as an Inhibitor of Vibrio harveyi AI-2 Quorum Sensing. Mar. Drugs 2020, 18, 80. https://doi.org/10.3390/md18020080
Karnjana K, Nobsathian S, Soowannayan C, Zhao W, Tang Y-J, Wongprasert K. Purification and Evaluation of N-benzyl Cinnamamide from Red Seaweed Gracilaria fisheri as an Inhibitor of Vibrio harveyi AI-2 Quorum Sensing. Marine Drugs. 2020; 18(2):80. https://doi.org/10.3390/md18020080
Chicago/Turabian StyleKarnjana, Kulwadee, Saksit Nobsathian, Chumporn Soowannayan, Wei Zhao, Ya-Jie Tang, and Kanokpan Wongprasert. 2020. "Purification and Evaluation of N-benzyl Cinnamamide from Red Seaweed Gracilaria fisheri as an Inhibitor of Vibrio harveyi AI-2 Quorum Sensing" Marine Drugs 18, no. 2: 80. https://doi.org/10.3390/md18020080
APA StyleKarnjana, K., Nobsathian, S., Soowannayan, C., Zhao, W., Tang, Y.-J., & Wongprasert, K. (2020). Purification and Evaluation of N-benzyl Cinnamamide from Red Seaweed Gracilaria fisheri as an Inhibitor of Vibrio harveyi AI-2 Quorum Sensing. Marine Drugs, 18(2), 80. https://doi.org/10.3390/md18020080





