Biofunctionality of Enzymatically Derived Peptides from Codfish (Gadus morhua) Frame: Bulk In Vitro Properties, Quantitative Proteomics, and Bioinformatic Prediction
Abstract
1. Introduction
2. Results and Discussion
2.1. Proteolysis Efficiency
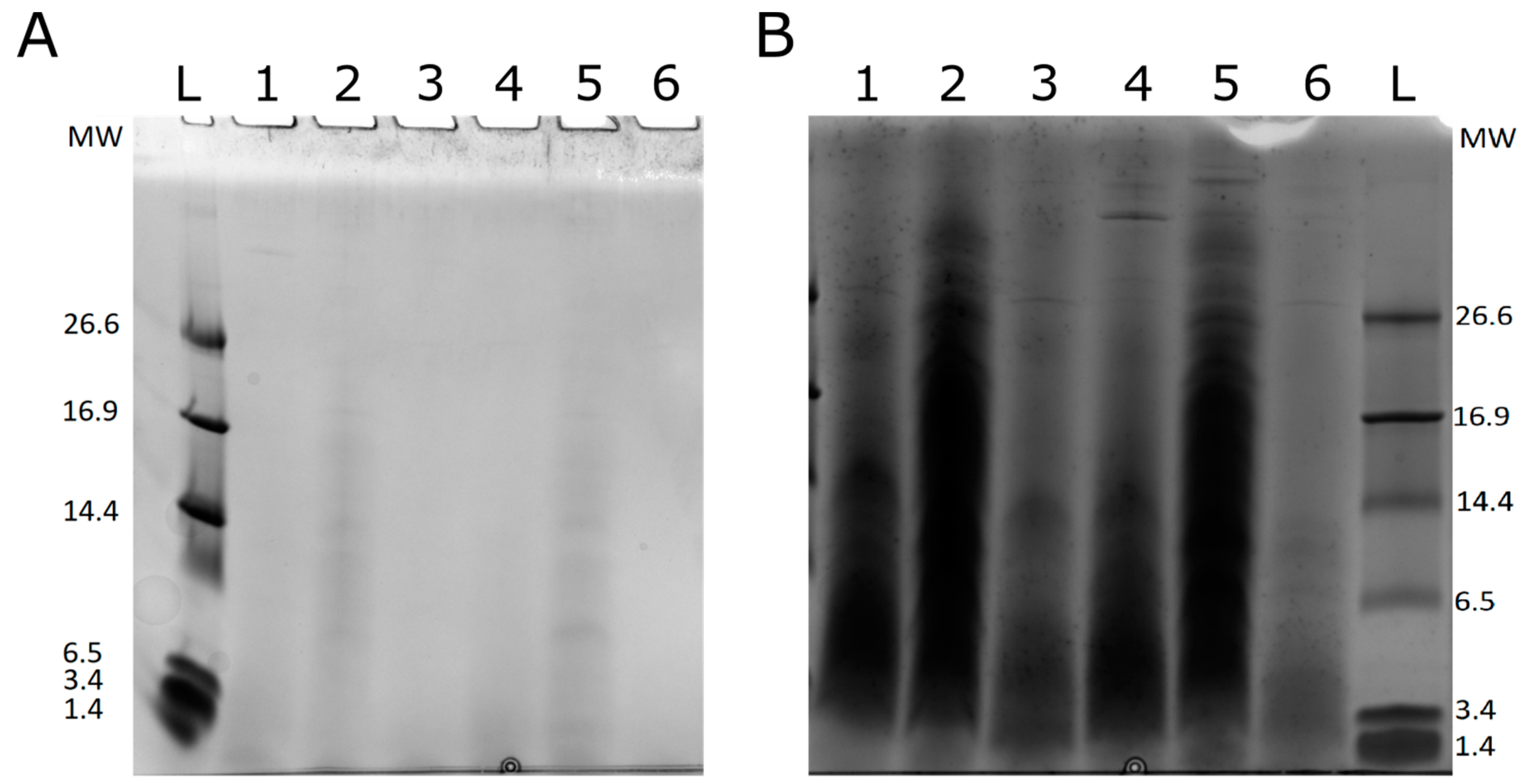
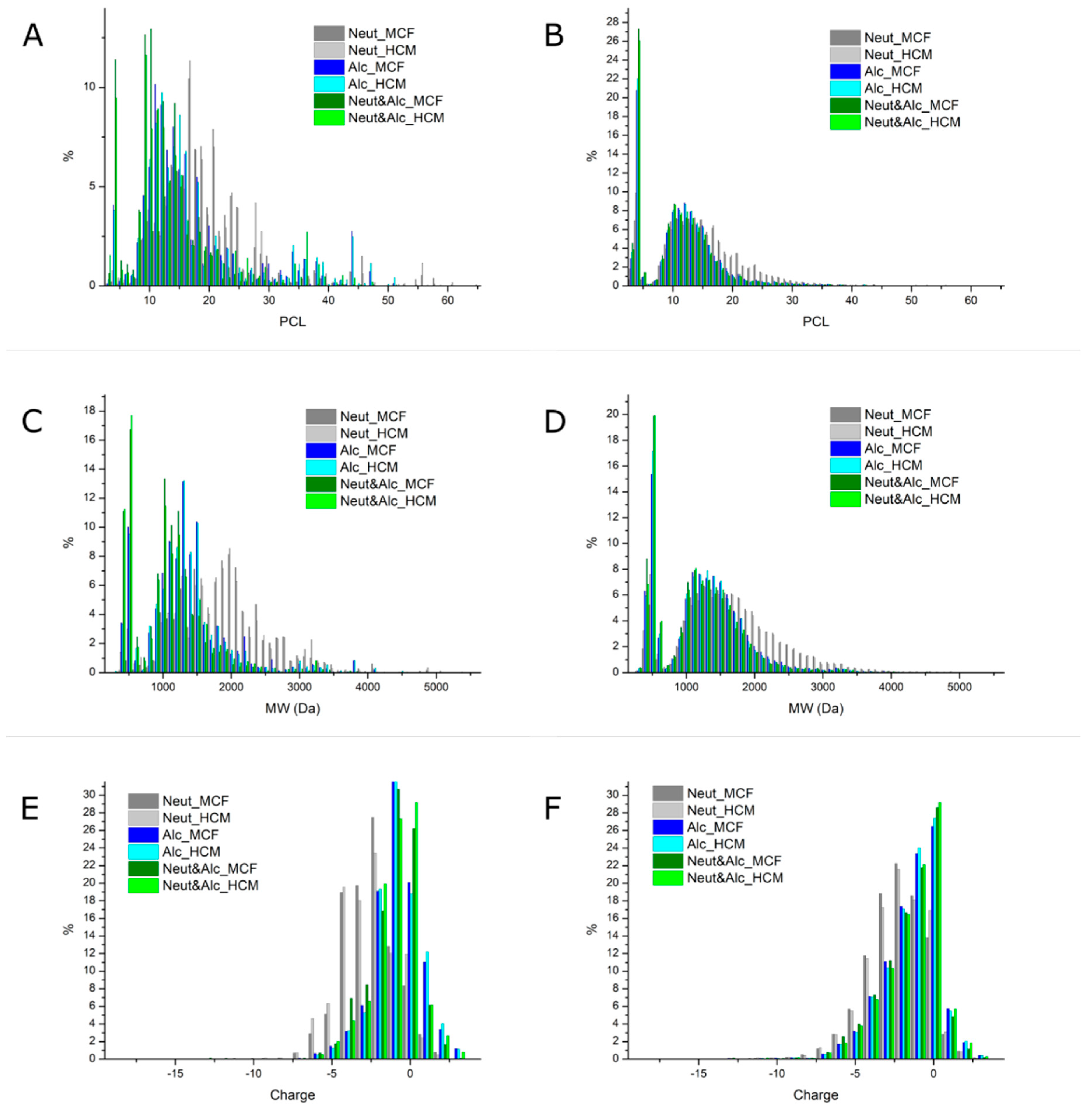
2.2. Proteomics Analysis and Methodological Limitations
2.3. Interfacial Properties
2.3.1. Bulk Emulsifying Properties
2.3.2. Bulk Foaming Properties
2.3.3. Interconnection of Bulk Interfacial Properties, Peptide Abundance, and Predicted Emulsifying Activity
2.4. Antioxidative Properties
2.4.1. Bulk Antioxdiative Properties
2.4.2. Interconnection of Bulk Antioxidative Properties, Peptide Abundance, and Predicted Antioxidative Activity
3. Materials and Methods
3.1. Enzymatic Hydrolysis Procedure
Degree of Hydrolysis (DH)
3.2. Emulsifying Properties
3.3. Foaming Properties
3.4. In Vitro Antioxidant Properties
3.4.1. Diphenyl-1-Picryhhydrazyl (DPPH) Radical Scavenging Activity (RSA)
3.4.2. Metal Chelating Activity (MCA)
3.5. 1D SDS-PAGE Analysis
3.6. Reference Proteome Construction
3.7. LC-MS/MS and Proteomics Data Analysis
3.7.1. Post-Processing of Proteomics Data
3.7.2. Prediction of Emulsification Activity
3.7.3. Prediction of Antioxidant Activity
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilson, N.; Shah, N.P. Microencapsulation of vitamins. Int. Food Res. J. 2007, 14, 1–14. [Google Scholar]
- Jafarpour, A.; Gomes, R.M.; Gregersen, S.; Sloth, J.J.; Jacobsen, C.; Moltke Sørensen, A.D. Characterization of cod (Gadus morhua) frame composition and its valorization by enzymatic hydrolysis. J. Food Compos. Anal. 2020, 89, 103469. [Google Scholar] [CrossRef]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef] [PubMed]
- Crynen, S. Bioactive Properties of Peptides Derived from Enzymatic Hydrolysis of Cod Muscle Myosin with Trypsin, Chymotrypsin and Elastase; University of Florida: Gainesville, FL, USA, 2011; Volume 546. [Google Scholar]
- Sabeena Farvin, K.H.; Andersen, L.L.; Nielsen, H.H.; Jacobsen, C.; Jakobsen, G.; Johansson, I.; Jessen, F. Antioxidant activity of Cod (Gadus morhua) protein hydrolysates: In vitro assays and evaluation in 5% fish oil-in-water emulsion. Food Chem. 2014, 149, 326–334. [Google Scholar] [CrossRef]
- Jeon, Y.-J.; Byun, H.-G.; Kim, S.-K. Improvement of functional properties of cod frame protein hydrolysates using ultrafiltration membranes. Process Biochem. 1999, 35, 471–478. [Google Scholar] [CrossRef]
- Aluko, R.E. Food protein-derived peptides: Production, isolation, and purification. In Proteins in Food Processing; Woodhead Publishing Series in Food Science, Technology and Nutrition: Cambridge, UK, 2018; pp. 389–412. [Google Scholar]
- Halim, N.R.A.; Yusof, H.M.; Sarbon, N.M. Functional and bioactive properties of fish protein hydolysates and peptides: A comprehensive review. Trends Food Sci. Technol. 2016, 51, 24–33. [Google Scholar] [CrossRef]
- Villamil, O.; Váquiro, H.; Solanilla, J.F. Fish viscera protein hydrolysates: Production, potential applications and functional and bioactive properties. Food Chem. 2017, 224, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Kristinsson, H.G.; Rasco, B.A. Fish protein hydrolysates: Production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 2000, 40, 43–81. [Google Scholar] [CrossRef]
- Awuor, O.L.; Edward Kirwa, M.; Betty, M.; Jackim, M.F. Optimization of Alcalase hydrolysis conditions for production of Dagaa (Rastrineobola argentea) Protein hydrolysate with antioxidative properties. Ind. Chem. 2017, 3. [Google Scholar] [CrossRef]
- Sila, A.; Bougatef, A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Zamora-Sillero, J.; Gharsallaoui, A.; Prentice, C. Peptides from Fish By-product Protein Hydrolysates and Its Functional Properties: An Overview. Mar. Biotechnol. 2018, 20, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Padial-Domínguez, M.; Espejo-Carpio, F.J.; García-Moreno, P.J.; Jacobsen, C.; Guadix, E.M. Protein derived emulsifiers with antioxidant activity for stabilization of omega-3 emulsions. Food Chem. 2020, 329, 127148. [Google Scholar] [CrossRef] [PubMed]
- Kunst, T.; Whitaker, J.R. Protein modification to optimize functionality: Protein hydrolysates. In Food Enzymology; Whitaker, J.R., Voragen, A.F.J., Wong, D.W.S., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2003; pp. 221–236. [Google Scholar]
- Foh, M.B.K.; Amadou, I.; Foh, B.M.; Kamara, M.T.; Xia, W. Functionality and antioxidant properties of tilapia (Oreochromis niloticus) as influenced by the degree of hydrolysis. Int. J. Mol. Sci. 2010, 11, 1851–1869. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, X.; Peng, Q.; Shan, Y.; Xue, F. Physicochemical properties of peanut protein isolate-glucomannan conjugates prepared by ultrasonic treatment. Ultrason. Sonochem. 2014, 21, 1722–1727. [Google Scholar] [CrossRef] [PubMed]
- Deb-Choudhury, S.; Haines, S.; Harland, D.; Clerens, S.; Van Koten, C.; Dyer, J. Effect of cooking on meat proteins: Mapping hydrothermal protein modification as a potential indicator of bioavailability. J. Agric. Food Chem. 2014, 62, 8187–8196. [Google Scholar] [CrossRef] [PubMed]
- Adler-Abramovich, L.; Vaks, L.; Carny, O.; Trudler, D.; Magno, A.; Caflisch, A.; Frenkel, D.; Gazit, E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat. Chem. Biol. 2012, 8, 701–706. [Google Scholar] [CrossRef]
- Anand, B.G.; Dubey, K.; Shekhawat, D.S.; Kar, K. Intrinsic property of phenylalanine to trigger protein aggregation and hemolysis has a direct relevance to phenylketonuria. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Liaset, B.; Nortvedt, R.; Lied, E.; Espe, M. Studies on the nitrogen recovery in enzymic hydrolysis of Atlantic salmon (Salmo salar, L.) frames by Protamex™ protease. Process Biochem. 2002, 37, 1263–1269. [Google Scholar] [CrossRef]
- Guérard, F. Enzymatic methods for marine by-products recovery. In Maximising the Value of Marine by-Products; Woodhead Publishing Limited: Cambridge, UK, 2007; pp. 107–143. ISBN 1845690133. [Google Scholar]
- Rutherfurd, S.M. Methodology for determining degree of hydrolysis of proteins in hydrolysates: A Review. J. AOAC Int. 2010, 93, 1515–1522. [Google Scholar] [CrossRef]
- Jacobsen, C.F.; Lenis, J.; Linderstrøm-Lann, K.; Ottesen, M. The pH-STAT and Its Use in Biochemistry. In Methods of Biochemica1 Analysis; Glick, D., Ed.; John Wiley & Sons (Interscience Publishers, Inc.): Hoboken, NJ, USA, 1957; pp. 171–209. [Google Scholar]
- Margot, A.; Flaschelb, E.; Renken, A.; Flaschel, E. Continuous Monitoring of Enzymatic Whey Protein Hydrolysis. Correlation of Base Consumption with Soluble Nitrogen Content. Process Biochem. 1994, 29, 257–262. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Enzymatic Hydrolysis of Food Proteins; Elsevier Applied Science Publishers Ltd.: London, UK, 1986. [Google Scholar]
- Sun, S.W.; Lin, Y.C.; Weng, Y.M.; Chen, M.J. Efficiency improvements on ninhydrin method for amino acid quantification. J. Food Compos. Anal. 2006, 19, 112–117. [Google Scholar] [CrossRef]
- Friedman, M. Applications of the Ninhydrin Reaction for Analysis of Amino Acids, Peptides, and Proteins to Agricultural and Biomedical Sciences. J. Agric. Food Chem. 2004, 52, 385–406. [Google Scholar] [CrossRef]
- Hurst, P.L.; Sinclair, B.K.; Jocelyn, R. Eason. Amino acids interfere with the ninhydrin assay for asparagine. Food Chem. 1995, 53, 467–469. [Google Scholar] [CrossRef]
- Chang, C.; Gao, Z.; Ying, W.; Fu, Y.; Zhao, Y.; Wu, S.; Li, M.; Wang, G.; Qian, X.; Zhu, Y.; et al. LFAQ: Toward Unbiased Label-Free Absolute Protein Quantification by Predicting Peptide Quantitative Factors. Anal. Chem. 2019, 91, 1335–1343. [Google Scholar] [CrossRef]
- Fusaro, V.A.; Mani, D.R.; Mesirov, J.P.; Carr, S.A. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 2009, 27, 190–198. [Google Scholar] [CrossRef]
- Gao, Z.; Chang, C.; Yang, J.; Zhu, Y.; Fu, Y. AP3: An Advanced Proteotypic Peptide Predictor for Targeted Proteomics by Incorporating Peptide Digestibility. Anal. Chem. 2019, 91, 8705–8711. [Google Scholar] [CrossRef]
- Jarnuczak, A.F.; Lee, D.C.H.; Lawless, C.; Holman, S.W.; Eyers, C.E.; Hubbard, S.J. Analysis of Intrinsic Peptide Detectability via Integrated Label-Free and SRM-Based Absolute Quantitative Proteomics. J. Proteome Res. 2016, 15, 2945–2959. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Tiwary, S.; Levy, R.; Gutenbrunner, P.; Salinas Soto, F.; Palaniappan, K.K.; Deming, L.; Berndl, M.; Brant, A.; Cimermancic, P.; Cox, J. High-quality MS/MS spectrum prediction for data-dependent and data-independent acquisition data analysis. Nat. Methods 2019, 16, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Bohrer, B.C.; Li, Y.F.; Reilly, J.P.; Clemmer, D.E.; DiMarchi, R.D.; Radivojac, P.; Tang, H.; Arnold, R.J. Combinatorial libraries of synthetic peptides as a model for shotgun proteomics. Anal. Chem. 2010, 82, 6559–6568. [Google Scholar] [CrossRef][Green Version]
- Rudnick, P.A.; Wang, X.; Yan, X.; Sedransk, N.; Stein, S.E. Improved normalization of systematic biases affecting ion current measurements in label-free proteomics data. Mol. Cell. Proteomics 2014, 13, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, R.; Ma, H.; Chen, S. Isolation and Identification of Dipeptidyl Peptidase IV-Inhibitory Peptides from Trypsin/Chymotrypsin-Treated Goat Milk Casein Hydrolysates by 2D-TLC and LC-MS/MS. J. Agric. Food Chem. 2015, 63, 8819–8828. [Google Scholar] [CrossRef] [PubMed]
- Schröder, A.; Berton-Carabin, C.; Venema, P.; Cornacchia, L. Interfacial properties of whey protein and whey protein hydrolysates and their influence on O/W emulsion stability. Food Hydrocoll. 2017, 73, 129–140. [Google Scholar] [CrossRef]
- García-Moreno, P.J.; Gregersen, S.; Nedamani, E.R.; Olsen, T.H.; Marcatili, P.; Overgaard, M.T.; Andersen, M.L.; Hansen, E.B.; Jacobsen, C. Identification of emulsifier potato peptides by bioinformatics: Application to omega-3 delivery emulsions and release from potato industry side streams. Sci. Rep. 2020, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Taheri, A.; Anvar, S.A.A.; Ahari, H.; Fogliano, V. Comparison the functional properties of protein Hydrolysates from poultry byproducts and rainbow trout (Onchorhynchus mykiss) viscera. Iran. J. Fish. Sci. 2013, 12, 154–169. [Google Scholar]
- McClements, D.J.; Jafari, S.M. Improving emulsion formation, stability and performance using mixed emulsifiers: A review. Adv. Colloid Interface Sci. 2018, 251, 55–79. [Google Scholar] [CrossRef]
- Padial-Domínguez, M.; Espejo-Carpio, F.J.; Pérez-Gálvez, R.; Guadix, A.; Guadix, E.M. Optimization of the emulsifying properties of food protein hydrolysates for the production of fish oil-in-water emulsions. Foods 2020, 9, 636. [Google Scholar] [CrossRef] [PubMed]
- Klompong, V.; Benjakul, S.; Kantachote, D.; Shahidi, F. Antioxidative activity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem. 2007, 102, 1317–1327. [Google Scholar] [CrossRef]
- Chi, C.-F.; Hu, F.-Y.; Wang, B.; Ren, X.-J.; Deng, S.-G.; Wu, C.-W. Purification and characterization of three antioxidant peptides from protein hydrolyzate of croceine croaker (Pseudosciaena crocea) muscle. Food Chem. 2015, 168, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Nalinanon, S.; Benjakul, S.; Kishimura, H.; Shahidi, F. Functionalities and antioxidant properties of protein hydrolysates from the muscle of ornate threadfin bream treated with pepsin from skipjack tuna. Food Chem. 2011, 124, 1354–1362. [Google Scholar] [CrossRef]
- Malcolm, A.S.; Dexter, A.F.; Middelberg, A.P.J. Foaming properties of a peptide designed to form stimuli-responsive interfacial films. Soft Matter 2006, 2, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Middelberg, A.P.J.; He, L.; Dexter, A.F.; Shen, H.H.; Holt, S.A.; Thomas, R.K. The interfacial structure and Young’s modulus of peptide films having switchable mechanical properties. J. R. Soc. Interface 2008, 5, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Dexter, A.F.; Middelberg, A.P.J. Peptides as functional surfactants. Ind. Eng. Chem. Res. 2008, 47, 6391–6398. [Google Scholar] [CrossRef]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 1982, 299, 371–374. [Google Scholar] [CrossRef]
- Feger, G.; Angelov, B.; Angelova, A. Prediction of Amphiphilic Cell-Penetrating Peptide Building Blocks from Protein-Derived Amino Acid Sequences for Engineering of Drug Delivery Nanoassemblies. J. Phys. Chem. B 2020, 124, 4069–4078. [Google Scholar] [CrossRef]
- Kilara, A.; Panyam, D. Peptides from Milk Proteins and Their Properties. Crit. Rev. Food Sci. Nutr. 2003, 43, 607–633. [Google Scholar] [CrossRef]
- Löwik, D.W.P.M.; Garcia-Hartjes, J.; Meijer, J.T.; Van Hest, J.C.M. Tuning secondary structure and self-assembly of amphiphilic peptides. Langmuir 2005, 21, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Martinek, T.A.; Hetényi, A.; Fülöp, L.; Mándity, I.M.; Tóth, G.K.; Dékány, I.; Fülöp, F. Secondary Structure Dependent Self-Assembly of β-Peptides into Nanosized Fibrils and Membranes. Angew. Chemie Int. Ed. 2006, 45, 2396–2400. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, H.; Hongo, K.; Hori, Y.; Yoshida, N.; Makabe, K. β-sheet elasticity of peptide self-assembly mimic, PSAM, with a grafted sequence characterized by comprehensive analyses of isomorphous crystals. J. Mol. Liq. 2019, 290, 111161. [Google Scholar] [CrossRef]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef]
- Šližyte, R.; Mozuraityte, R.; Martínez-Alvarez, O.; Falch, E.; Fouchereau-Peron, M.; Rustad, T. Functional, bioactive and antioxidative properties of hydrolysates obtained from cod (Gadus morhua) backbones. Process Biochem. 2009, 44, 668–677. [Google Scholar] [CrossRef]
- Ktari, N.; Fakhfakh, N.; Balti, R.; Ben Khaled, H.; Nasri, M.; Bougatef, A. Effect of degree of hydrolysis and protease type on the antioxidant activity of protein hydrolysates from cuttlefish (Sepia officinalis) By-products. J. Aquat. Food Prod. Technol. 2013, 22, 436–448. [Google Scholar] [CrossRef]
- Centenaro, G.S.; Salas-Mellado, M.; Pires, C.; Batista, I.; Nunes, M.L.; Prentice, C. Fractionation of protein hydrolysates of fish and chicken using membrane ultrafiltration: Investigation of antioxidant activity. Appl. Biochem. Biotechnol. 2014, 172, 2877–2893. [Google Scholar] [CrossRef]
- Li, Z.; Wang, B.; Chi, C.; Gong, Y.; Luo, H.; Ding, G. Influence of average molecular weight on antioxidant and functional properties of cartilage collagen hydrolysates from Sphyrna lewini, Dasyatis akjei and Raja porosa. Food Res. Int. 2013, 51, 283–293. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Xing, R.; Chen, X.; Li, R.; Li, K.; Liu, S.; Li, P. Purification and identification of antioxidative peptides from mackerel (Pneumatophorus japonicus) protein. RSC Adv. 2018, 8, 20488–20498. [Google Scholar] [CrossRef]
- Atmaca, G. Antioxidant effects of sulfur-containing amino acids. Yonsei Med. J. 2004, 45, 776–788. [Google Scholar] [CrossRef]
- Rajapakse, N.; Mendis, E.; Byun, H.-G.; Kim, S.-K. Purification and in vitro antioxidative effects of giant squid muscle peptides on free radical-mediated oxidative systems. J. Nutr. Biochem. 2005, 16, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, E.; Jones, O.; Kim, Y.H.B.; San Martin-Gonzalez, F.; Liceaga, A.M. Impact of microwave-assisted enzymatic hydrolysis on functional and antioxidant properties of rainbow trout Oncorhynchus mykiss by-products. Fish. Sci. 2017, 83, 317–331. [Google Scholar] [CrossRef]
- Chalamaiah, M.; Dinesh kumar, B.; Hemalatha, R.; Jyothirmayi, T. Fish protein hydrolysates: Proximate composition, amino acid composition, antioxidant activities and applications: A review. Food Chem. 2012, 135, 3020–3038. [Google Scholar] [CrossRef]
- Suetsuna, K.; Ukeda, H.; Ochi, H. Isolation and characterization of free radical scavenging activities peptides derived from casein. J. Nutr. Biochem. 2000, 11, 128–131. [Google Scholar] [CrossRef]
- Ghelichi, S.; Sørensen, A.-D.M.; García-Moreno, P.J.; Hajfathalian, M.; Jacobsen, C. Physical and oxidative stability of fish oil-in-water emulsions fortified with enzymatic hydrolysates from common carp (Cyprinus carpio) roe. Food Chem. 2017, 237, 1048–1057. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morales-Medina, R.; Tamm, F.; Guadix, A.M.; Guadix, E.M.; Drusch, S. Functional and antioxidant properties of hydrolysates of sardine (S. pilchardus) and horse mackerel (T. mediterraneus) for the microencapsulation of fish oil by spray-drying. Food Chem. 2016, 194, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kong, B.; Xiong, Y.L.; Xia, X. Antioxidant activity and functional properties of porcine plasma protein hydrolysate as influenced by the degree of hydrolysis. Food Chem. 2010, 118, 403–410. [Google Scholar] [CrossRef]
- Rajabzadeh, M.; Pourashouri, P.; Shabanpour, B.; Alishahi, A. Amino acid composition, antioxidant and functional properties of protein hydrolysates from the roe of rainbow trout (Oncorhynchus mykiss). Int. J. Food Sci. Technol. 2018, 53, 313–319. [Google Scholar] [CrossRef]
- Hajfathalian, M.; Ghelichi, S.; García-Moreno, P.J.; Moltke Sørensen, A.D.; Jacobsen, C. Peptides: Production, bioactivity, functionality, and applications. Crit. Rev. Food Sci. Nutr. 2018, 58, 3097–3129. [Google Scholar] [CrossRef]
- Hernández-Ledesma, B.; García-Nebot, M.J.; Fernández-Tomé, S.; Amigo, L.; Recio, I. Dairy protein hydrolysates: Peptides for health benefits. Int. Dairy J. 2014, 38, 82–100. [Google Scholar] [CrossRef]
- Olsen, T.; Yesiltas, B.; Marin, F.I.; Pertseva, M.; García-Moreno, P.J.; Gregersen, S.; Overgaard, M.T.; Jacobsen, C.; Lund, O.; Hansen, E.B.; et al. AnOxPePred: Using deep learning for the prediction of antioxidative properties of peptides. Sci. Rep. (in press). Available online: https://services.healthtech.dtu.dk/service.php?AnOxPePred-1.0 (accessed on 31 March 2020).
- Studer, S.; Hansen, D.A.; Pianowski, Z.L.; Mittl, P.R.E.; Debon, A.; Guffy, S.L.; Der, B.S.; Kuhlman, B.; Hilvert, D. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science 2018, 362, 1285–1288. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, H.-B.; Ji, L.-N.; Mao, Z.-W. Insights into metalloenzyme microenvironments: Biomimetic metal complexes with a functional second coordination sphere. Chem. Soc. Rev. 2013, 42, 8360–8375. [Google Scholar] [CrossRef] [PubMed]
- Callaway, D.A.; Jiang, J.X. Reactive oxygen species and oxidative stress in osteoclastogenesis, skeletal aging and bone diseases. J. Bone Miner. Metab. 2015, 33, 359–370. [Google Scholar] [CrossRef]
- Halleen, J.M.; Räisänen, S.; Salo, J.J.; Reddy, S.V.; Roodman, G.D.; Hentunen, T.A.; Lehenkari, P.P.; Kaija, H.; Vihko, P.; Väänänen, H.K. Intracellular fragmentation of bone resorption products by reactive oxygen species generated by osteoclastic tartrate-resistant acid phosphatase. J. Biol. Chem. 1999, 274, 22907–22910. [Google Scholar] [CrossRef]
- Sui, X.; Bi, S.; Qi, B.; Wang, Z.; Zhang, M.; Li, Y.; Jiang, L. Impact of ultrasonic treatment on an emulsion system stabilized with soybean protein isolate and lecithin: Its emulsifying property and emulsion stability. Food Hydrocoll. 2017, 63, 727–734. [Google Scholar] [CrossRef]
- Bao, Z.J.; Zhao, Y.; Wang, X.Y.; Chi, Y.J. Effects of degree of hydrolysis (DH) on the functional properties of egg yolk hydrolysate with alcalase. J. Food Sci. Technol. 2017, 54, 669–678. [Google Scholar] [CrossRef]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H. Europe PMC Funders Group. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2013, 477, 207–210. [Google Scholar] [CrossRef]
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Jakobsen, K.S.; Jentoft, S. Data descriptor: Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci. Data 2017, 4, 1–13. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI) FASTA Tools Sequence Lengths. Available online: https://www.ncbi.nlm.nih.gov/CBBresearch/Spouge/html_ncbi/html/fasta/length.cgi (accessed on 6 August 2020).
- Lide, D.R. CRC Handbook of Chemistry and Physics, 87th ed.; Lide, D.R., Ed.; CRC Press/Taylor and Francis: Boca Raton, FL, USA, 2006; Volume 87, ISBN 0849304873. [Google Scholar]
- INKSCAPE. Available online: https://inkscape.org/ (accessed on 11 March 2018).
- García-Moreno, P.J.; Jacobsen, C.; Marcatili, P.; Gregersen, S.; Overgaard, M.T.; Andersen, M.L.; Sørensen, A.-D.M.; Hansen, E.B. Emulsifying peptides from potato protein predicted by bioinformatics: Stabilization of fish oil-in-water emulsions. Food Hydrocoll. 2020, 101, 105529. [Google Scholar] [CrossRef]
- EmulsiPred. Available online: https://github.com/MarcatiliLab/EmulsiPred (accessed on 31 March 2020).
- Gregersen, S. CodfishHydrolysatesMS. Mendeley Data V2 2020. [Google Scholar] [CrossRef]
| Substrate | Treatment | DH (%) * | EAI (m2/g) * | ESI (min) * | FC (%) | FS (%) | DPPH RSA (IC50 mg/mL) * | Fe2+ MCA (IC50 mg/mL) * |
|---|---|---|---|---|---|---|---|---|
| MCF | Neutrase | 27.0 ± 0.12 d | 80.75 ± 4.32 ab | 33.39 ± 0.56 cd | 40 | 88 | 4.36 ± 0.08 b | 0.53 ± 0.013 c |
| Alcalase | 36.1 ± 0.03 b | 88.18 ± 7.94 a | 29.38 ± 3.52 d | 30 | 87 | 4.97 ± 0.18 a | 0.77 ± 0.03 bc | |
| Neutrase & Alcalase | 39.2 ± 0.04 a | 87.54 ± 4.46 a | 36.10 ± 5.12 c | 4 | 100 | 4.31 ± 0.06 b | 0.77 ± 0.03 bc | |
| HCM | Neutrase | 18.7 ± 0.09 e | 66.41 ± 0.97 c | 46.82 ± 4.75 b | 54 | 37 | 3.36 ± 0.18 c | 0.69 ± 0.02 c |
| Alcalase | 30.2 ± 0.05 c | 68.35 ± 0.72 c | 65.15 ± 2.03 a | 23 | 78 | 3.58 ± 0.21 c | 1.01 ± 0.31 b | |
| Neutrase & Alcalase | 30.2 ± 0.04 c | 76.48 ± 3.99 b | 45.70 ± 1.94 b | 6 | 100 | 2.93 ± 0.05 d | 1.65 ± 0.07 a | |
| Control (SC) | - | N/A | 64.48 ± 3.33 c | 15.86 ± 1.15 e | N/A | N/A | N/A | N/A |
| Treatments | Weighted | Unweighted | DH-based | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrate | Treatment | Peptide IDs | PCLavg (AAs) 1 | PMWavg (Da) 1 | Pzavg 1 | PCLavg (AAs) 1 | PMWavg (Da) 1 | Pzavg 1 | PCLDH (AAs) * |
| MCF | Neutrase | 6268 | 17.4 ± 6.9 | 1921 ± 674 | −2.5 ± 1.7 | 15.3 ± 7.1 | 1686 ± 726 | −2.2 ± 1.9 | 3.7 ± 0.0 b |
| Alcalase | 4414 | 13.0 ± 7.3 | 1343 ± 635 | −0.95 ± 1.6 | 11.7 ± 6.5 | 1268 ± 636 | −1.5 ± 2.0 | 2.8 ± 0.0 d | |
| Neutrase & Alcalase | 3702 | 9.36 ± 4.8 | 999 ± 463 | −1.3 ± 1.6 | 10.3 ± 6.1 | 1128 ± 614 | −1.6 ± 2.0 | 2.6 ± 0.0 e | |
| HCM | Neutrase | 5871 | 16.7 ± 7.3 | 1853 ± 722 | −2.5 ± 1.8 | 14.8 ± 7.4 | 1635 ± 755 | −2.2 ± 2.0 | 5.4 ± 0.0 a |
| Alcalase | 4020 | 13.1 ± 7.7 | 1334 ± 647 | −0.88 ± 1.6 | 11.3 ± 6.5 | 1228 ± 622 | −1.4 ± 1.9 | 3.3 ± 0.0 c | |
| Neutrase & Alcalase | 4361 | 10.2 ± 6.6 | 1082 ± 591 | −1.1 ± 1.6 | 10.6 ± 6.3 | 1157 ± 611 | −1.5 ± 2.0 | 3.3 ± 0.0 c | |
| MCF | ||||||||||||||
| Neut. | RI (%) | Emulsifying Scores | Alc | RI (%) | Emulsifying Scores | Neut&Alc | RI (%) | Emulsifying Scores | ||||||
| α | β | γ | α | β | γ | α | β | γ | ||||||
| LQGEVEDLMVDVERANG | 1.94 | 1.38 | −0.45 | 0.55 | GFAGDDAPRAVFPS | 2.30 | 0.29 | −0.86 | 0.71 | IIAPPERKYS | 3.49 | −0.59 | −0.62 | 3.43 |
| LEQQVDDLEGSLEQEKK | 1.92 | 2.18 | −0.87 | 1.26 | RVAPEEHPTLL | 2.13 | 0.36 | −0.09 | 1.79 | GVDNPGHPF | 2.00 | 1.13 | −0.49 | 0.08 |
| IITNWDDMEK | 1.78 | 0.75 | −1.28 | 2.95 | AGDDAPRAVFPS | 2.09 | 0.97 | −1.33 | 1.23 | VAPEEHPTL | 1.59 | −0.18 | −0.07 | 1.04 |
| VQHELEEAEERADIAETQVNK | 1.39 | 2.21 | −0.41 | 0.64 | AGPAGPSGPRGPAGIA | 1.56 | 0.16 | −0.17 | 0.47 | VLYPPVVEE | 1.53 | −0.43 | −1.16 | 0.55 |
| LTKLEEAEKAADESERGMK | 1.17 | 1.53 | −0.55 | 1.15 | KSYELPDGQVITIG | 1.33 | −0.78 | −0.79 | 2.64 | GADPEDVIVA | 1.43 | −1.29 | −1.00 | 3.10 |
| LEDQLSEIKAKSDENARQ | 1.00 | 1.54 | 0.10 | 1.33 | GAAGPAGPSGPRGPAGIA | 1.30 | 0.16 | −0.51 | 0.23 | VIDQDKSGFIE | 1.35 | 0.37 | −0.43 | 0.88 |
| LEKSYELPDGQVIT | 0.92 | −0.55 | −0.43 | 0.82 | DIDIRKDLYAN | 1.16 | −1.24 | 3.65 | 0.83 | AGDDAPRAVFPS | 1.20 | 0.97 | −1.33 | 1.23 |
| VAPEEHPTL | 0.90 | −0.18 | −0.07 | 1.04 | VAPEEHPTLL | 1.12 | −0.53 | −0.81 | 1.14 | KSYELPDGQ | 1.19 | −1.09 | −0.94 | −0.55 |
| IIDQNRDGIISKDDLRD | 0.86 | −0.51 | −0.56 | 0.70 | AGPSGPRGPAGIA | 1.08 | −0.23 | −0.12 | 1.26 | GERGEQGPGGPGGF | 1.08 | −0.79 | −0.73 | 1.36 |
| LDDLQAEEDKVNT | 0.81 | 0.55 | −1.02 | 1.17 | NWDDMEKIWHH | 1.06 | 0.75 | −0.27 | 1.02 | IIDQNRDGIIS | 0.83 | 0.68 | −0.86 | 1.67 |
| LEKTIDDLEDELYAQK | 0.78 | 1.51 | −0.63 | 0.12 | GQKDSYVGDEAQSKRGILTL | 0.92 | −0.71 | −0.30 | 1.27 | AGPAGPSGPRGPAG | 0.81 | −0.56 | −0.48 | 0.21 |
| LKGTEDELDKYSEALKDAQEKLE | 0.77 | 2.96 | 0.22 | 0.47 | LRVAPEEHPTL | 0.88 | 0.64 | 1.15 | 1.24 | GLPGPSGPSGPRGR | 0.78 | −0.81 | −1.11 | 0.80 |
| LTEEMASQDESVAK | 0.76 | 1.01 | −0.17 | 0.11 | RGDSGPAGPPGEQGML | 0.84 | −1.16 | −0.62 | 0.13 | RGEQGPGGPGGF | 0.76 | −1.42 | −0.18 | 1.04 |
| LKGADPEDVIVAA | 0.76 | −1.53 | −0.19 | 2.78 | ELPDGQVITIG | 0.82 | −0.99 | 0.01 | 2.07 | GFAGDDAPRA | 0.69 | 0.45 | −0.67 | 1.06 |
| LADWKQKYEEGQAELEGSLKEARS | 0.71 | 0.70 | −0.08 | 0.43 | SGSAGKDGMSGLPGPSGPSGPRGR | 0.67 | −0.23 | −1.12 | 0.32 | AINDPFIDL | 0.67 | 0.83 | −0.47 | 0.47 |
| SKYETDAIQRTEELEESKKK | 0.69 | 0.71 | −0.69 | 1.57 | LLPVLYPPVVEE | 0.67 | 0.24 | −1.23 | 1.33 | KAGDSDGDGAIGVD | 0.59 | −0.30 | 0.65 | 1.81 |
| LKAGDSDGDGAIGVDEWAV | 0.69 | 0.26 | −0.72 | 1.09 | SGPGGPTGPSGM | 0.64 | −1.24 | −0.74 | −0.49 | GFAGDDAPRAVFPS | 0.58 | 0.29 | −0.86 | 0.71 |
| LTDAETKAF | 0.65 | 0.43 | −0.14 | −0.44 | KSYELPDGQVITIGNE | 0.62 | −0.17 | −0.82 | 1.35 | LEDQLSELK | 0.54 | 1.36 | −1.20 | −0.22 |
| LKAGDSDGDGAIGVDEWAVLVKA | 0.61 | −0.98 | −1.16 | 1.53 | GPAGPSGPRGPAGIA | 0.59 | −0.10 | −0.37 | 0.91 | KILDPEAT | 0.52 | 0.59 | −0.71 | 0.96 |
| VDDIIQTGVDNPGHPFIMT | 0.59 | 0.89 | 0.34 | 0.07 | FAGDDAPRAVFPS | 0.58 | 0.35 | −0.89 | 0.70 | GKDGMSGLPGPSGPSGPRGR | 0.52 | −0.08 | −0.72 | 0.47 |
| HCM | ||||||||||||||
| Neut. | RI (%) | α | β | γ | Alc | RI (%) | α | β | γ | Neut&Alc | RI (%) | α | β | γ |
| LEQQVDDLEGSLEQEKK | 2.38 | 2.18 | −0.87 | 1.26 | AGDDAPRAVFPS | 3.13 | 0.97 | −1.33 | 1.23 | VAPEEHPTL | 1.91 | −0.18 | −0.07 | 1.04 |
| LQGEVEDLMVDVERANG | 2.28 | 1.38 | −0.45 | 0.55 | GPAGPSGPRGPAGIA | 2.64 | −0.10 | −0.37 | 0.91 | VLYPPVVEE | 1.75 | −0.43 | −1.16 | 0.55 |
| IITNWDDMEK | 2.23 | 0.75 | −1.28 | 2.95 | AGPAGPSGPRGPAGIA | 1.90 | 0.16 | −0.17 | 0.47 | IIDQNRDGIIS | 1.45 | 0.68 | −0.86 | 1.67 |
| VQHELEEAEERADIAETQVNK | 1.46 | 2.21 | −0.41 | 0.64 | VAPEEHPTLL | 1.87 | −0.53 | −0.81 | 1.14 | ELPDGQVIT | 1.14 | −1.76 | −0.72 | 1.69 |
| LEKSYELPDGQVIT | 1.27 | −0.55 | −0.43 | 0.82 | GFAGDDAPRAVFPS | 1.75 | 0.29 | −0.86 | 0.71 | VIDQDKSGFIE | 1.13 | 0.37 | −0.43 | 0.88 |
| VETEKTEIQSALEEAEGTLEHEESKILR | 1.00 | 2.29 | −0.80 | 0.25 | GAAGPAGPSGPRGPAGIA | 1.62 | 0.16 | −0.51 | 0.23 | IIAPPERKYS | 1.11 | −0.59 | −0.62 | 3.43 |
| LDDLQAEEDKVNT | 0.94 | 0.55 | −1.02 | 1.17 | RVAPEEHPTLL | 1.53 | 0.36 | −0.09 | 1.79 | LEDQLSELK | 1.06 | 1.36 | −1.20 | −0.22 |
| LADWKQKYEEGQAELEGSLKEARS | 0.74 | 0.70 | −0.08 | 0.43 | ELPDGQVITIG | 1.34 | −0.99 | 0.01 | 2.07 | GVDNPGHPF | 1.00 | 1.13 | −0.49 | 0.08 |
| VAPEEHPTL | 0.72 | -0.18 | −0.07 | 1.04 | KSYELPDGQVITIG | 1.21 | −0.78 | −0.79 | 2.64 | GADPEDVIVA | 0.95 | −1.29 | −1.00 | 3.10 |
| LDDVIQTGVDNPGHPFIMT | 0.71 | 0.83 | 0.33 | −0.02 | LLPVLYPPVVEE | 1.09 | 0.24 | −1.23 | 1.33 | AAGPAGPSGPRGPAG | 0.75 | −0.56 | −0.75 | 0.55 |
| LEDECSELKKDIDDLELT | 0.71 | 1.84 | −0.42 | 0.82 | LGEQIDNL | 1.03 | 2.57 | −0.40 | −0.40 | AGDDAPRAVFPS | 0.71 | 0.97 | −1.33 | 1.23 |
| ARIEELEEELEAERA | 0.68 | −0.65 | −0.93 | 0.54 | RGDSGPAGPPGEQGML | 0.94 | −1.16 | −0.62 | 0.13 | LDKNKDPLNDSVVQ | 0.68 | 1.41 | −0.79 | 1.18 |
| LTEEMASQDESVAK | 0.67 | 1.01 | −0.17 | 0.11 | GSAGPRGPSGNIGMPGMTGPQ | 0.85 | −1.75 | −0.73 | 0.14 | FAGDDAPRA | 0.67 | 0.49 | −0.69 | 1.15 |
| IEELEEELEAERA | 0.65 | 0.22 | 0.05 | 1.05 | TIIDQNRDGIIS | 0.76 | 0.70 | −0.99 | 1.49 | ELPDGQVITI | 0.67 | −0.93 | 0.00 | 2.37 |
| LDFENEMAT | 0.65 | 0.10 | 1.04 | 0.59 | GEKLKGADPEDVIVA | 0.74 | −1.01 | −0.41 | 2.36 | KSYELPDGQVITIG | 0.63 | −0.78 | −0.79 | 2.64 |
| VDDIIQTGVDNPGHPFIMT | 0.63 | 0.89 | 0.34 | 0.07 | SGSAGKDGMSGLPGPSGPSGPRGR | 0.67 | −0.23 | −1.12 | 0.32 | GFNPPDLDIM | 0.60 | −0.75 | −0.19 | 1.20 |
| LKAGDSDGDGAIGVDEWA | 0.61 | 0.26 | −0.18 | 0.56 | SGPGGPTGPSGM | 0.67 | −1.24 | −0.74 | −0.49 | ELPDGQVI | 0.60 | −1.92 | −0.85 | 0.68 |
| LVQVQGEVDDSVQEARNAEEKAKKA | 0.60 | 2.64 | 1.54 | 2.42 | GEQIDNL | 0.67 | 1.96 | −1.14 | 0.44 | NPPKYDKIEDM | 0.55 | −0.15 | −0.70 | 1.79 |
| ALEEAEGTLEHEESKLLR | 0.58 | 1.83 | −0.16 | 0.57 | ELPDGQVI | 0.66 | −1.92 | −0.85 | 0.68 | KSYELPDGQVITI | 0.53 | −0.70 | −0.82 | 2.76 |
| IEELEEELEAERAAR | 0.56 | −0.70 | 0.82 | 0.31 | KSYELPDGQVITI | 0.65 | −0.70 | −0.82 | 2.76 | KSYELPDGQVIT | 0.51 | −1.28 | −1.17 | 1.13 |
| MCF | |||||||||||
| Neut. | Rel. Int. (%) | Score | Alc | Rel. Int. (%) | Score | Neut&Alc | Rel. Int. (%) | Score | |||
| FRS | MCA | FRS | MCA | FRS | MCA | ||||||
| LQGEVEDLMVDVERANG | 1.94 | 0.33 | 0.17 | GFAGDDAPRAVFPS | 2.30 | 0.51 | 0.25 | IIAPPERKYS | 3.49 | 0.40 | 0.23 |
| LEQQVDDLEGSLEQEKK | 1.92 | 0.38 | 0.19 | RVAPEEHPTLL | 2.13 | 0.51 | 0.26 | GVDNPGHPF | 2.00 | 0.56 | 0.28 |
| IITNWDDMEK | 1.78 | 0.38 | 0.19 | AGDDAPRAVFPS | 2.09 | 0.41 | 0.24 | LDLL | 1.75 | 0.35 | 0.28 |
| LTKLEEAEKAADESERGMK | 1.17 | 0.29 | 0.17 | AGPAGPSGPRGPAGIA | 1.56 | 0.51 | 0.21 | VGPF | 1.65 | 0.47 | 0.27 |
| LEDQLSEIKAKSDENARQ | 1.00 | 0.23 | 0.17 | FLGM | 1.34 | 0.43 | 0.28 | VAPEEHPTL | 1.59 | 0.51 | 0.25 |
| LEKSYELPDGQVIT | 0.92 | 0.48 | 0.23 | KSYELPDGQVITIG | 1.33 | 0.35 | 0.21 | VLYPPVVEE | 1.53 | 0.52 | 0.21 |
| VAPEEHPTL | 0.90 | 0.51 | 0.25 | GAAGPAGPSGPRGPAGIA | 1.30 | 0.49 | 0.20 | GADPEDVIVA | 1.43 | 0.32 | 0.20 |
| IIDQNRDGIISKDDLRD | 0.86 | 0.30 | 0.22 | DIDIRKDLYAN | 1.16 | 0.27 | 0.17 | VIDQDKSGFIE | 1.35 | 0.30 | 0.19 |
| LDDLQAEEDKVNT | 0.81 | 0.34 | 0.23 | VAPEEHPTLL | 1.12 | 0.56 | 0.26 | AGDDAPRAVFPS | 1.20 | 0.41 | 0.24 |
| LEKTIDDLEDELYAQK | 0.78 | 0.35 | 0.22 | AGPSGPRGPAGIA | 1.08 | 0.47 | 0.20 | KSYELPDGQ | 1.19 | 0.44 | 0.23 |
| LKGTEDELDKYSEALKDAQEKLE | 0.77 | 0.34 | 0.16 | NWDDMEKIWHH | 1.06 | 0.34 | 0.20 | VAVL | 1.17 | 0.33 | 0.23 |
| LTEEMASQDESVAK | 0.76 | 0.31 | 0.21 | GQKDSYVGDEAQSKRGILTL | 0.92 | 0.30 | 0.18 | LFPE | 1.17 | 0.44 | 0.30 |
| LKGADPEDVIVAA | 0.76 | 0.38 | 0.19 | NWDDME | 0.88 | 0.40 | 0.21 | GLVL | 1.16 | 0.37 | 0.24 |
| LADWKQKYEEGQAELEGSLKEARS | 0.71 | 0.30 | 0.15 | LRVAPEEHPTL | 0.85 | 0.45 | 0.25 | LLEM | 1.12 | 0.39 | 0.27 |
| SKYETDAIQRTEELEESKKK | 0.69 | 0.40 | 0.14 | RGDSGPAGPPGEQGML | 0.84 | 0.51 | 0.29 | GERGEQGPGGPGGF | 1.08 | 0.52 | 0.23 |
| LKAGDSDGDGAIGVDEWAV | 0.69 | 0.35 | 0.18 | ELPDGQVITIG | 0.74 | 0.31 | 0.20 | LGVL | 0.95 | 0.38 | 0.25 |
| LTDAETKAF | 0.65 | 0.33 | 0.24 | SGSAGKDGMSGLPGPSGPSGPRGR | 0.67 | 0.55 | 0.25 | LDFENE | 0.87 | 0.37 | 0.27 |
| LKAGDSDGDGAIGVDEWAVLVKA | 0.61 | 0.36 | 0.13 | LDLL | 0.67 | 0.35 | 0.28 | IIDQNRDGIIS | 0.83 | 0.29 | 0.19 |
| VDDIIQTGVDNPGHPFIMT | 0.59 | 0.42 | 0.20 | LLPVLYPPVVEE | 0.65 | 0.64 | 0.21 | AGPAGPSGPRGPAG | 0.81 | 0.49 | 0.22 |
| VIDQDKSGFIEEDELKLF | 0.57 | 0.36 | 0.19 | SGPGGPTGPSGM | 0.64 | 0.50 | 0.23 | LLLS | 0.79 | 0.34 | 0.29 |
| HCM | |||||||||||
| Neut. | Rel. Int. (%) | FRS | MCA | Alc. | Rel. Int. (%) | FRS | MCA | Netu&Alc | Rel. Int. (%) | FRS | MCA |
| LEQQVDDLEGSLEQEKK | 2.38 | 0.38 | 0.19 | AGDDAPRAVFPS | 3.13 | 0.41 | 0.24 | VAPEEHPTL | 1.91 | 0.51 | 0.25 |
| LQGEVEDLMVDVERANG | 2.28 | 0.33 | 0.17 | GPAGPSGPRGPAGIA | 2.64 | 0.49 | 0.21 | VLYPPVVEE | 1.75 | 0.52 | 0.21 |
| IITNWDDMEK | 2.23 | 0.38 | 0.19 | AGPAGPSGPRGPAGIA | 1.90 | 0.51 | 0.21 | IIDQNRDGIIS | 1.45 | 0.29 | 0.19 |
| VQHELEEAEERADIAETQVNK | 1.46 | 0.33 | 0.15 | VAPEEHPTLL | 1.87 | 0.56 | 0.26 | ELPDGQVIT | 1.14 | 0.36 | 0.23 |
| LEKSYELPDGQVIT | 1.27 | 0.48 | 0.23 | GFAGDDAPRAVFPS | 1.75 | 0.51 | 0.25 | VIDQDKSGFIE | 1.13 | 0.30 | 0.19 |
| VETEKTEIQSALEEAEGTLEHEESKILR | 1.00 | 0.37 | 0.13 | GAAGPAGPSGPRGPAGIA | 1.62 | 0.49 | 0.20 | IIAPPERKYS | 1.11 | 0.40 | 0.23 |
| LDDLQAEEDKVNT | 0.94 | 0.34 | 0.23 | FLGM | 1.53 | 0.43 | 0.28 | FLPM | 1.07 | 0.46 | 0.31 |
| LADWKQKYEEGQAELEGSLKEARS | 0.74 | 0.30 | 0.15 | RVAPEEHPTLL | 1.42 | 0.51 | 0.26 | LEDQLSELK | 1.06 | 0.34 | 0.25 |
| VAPEEHPTL | 0.72 | 0.51 | 0.25 | ELPDGQVITIG | 1.34 | 0.31 | 0.20 | GVDNPGHPF | 1.00 | 0.56 | 0.28 |
| LFQPSF | 0.72 | 0.42 | 0.28 | KSYELPDGQVITIG | 1.21 | 0.35 | 0.21 | GADPEDVIVA | 0.95 | 0.32 | 0.20 |
| LDDVIQTGVDNPGHPFIMT | 0.71 | 0.41 | 0.21 | LLPVLYPPVVEE | 1.09 | 0.64 | 0.21 | AAGPAGPSGPRGPAG | 0.75 | 0.49 | 0.22 |
| LEDECSELKKDIDDLELT | 0.71 | 0.26 | 0.18 | LGEQIDNL | 1.03 | 0.35 | 0.22 | AGDDAPRAVFPS | 0.71 | 0.41 | 0.24 |
| ARIEELEEELEAERA | 0.68 | 0.36 | 0.20 | NWDDME | 0.94 | 0.40 | 0.21 | LDKNKDPLNDSVVQ | 0.68 | 0.29 | 0.27 |
| LTEEMASQDESVAK | 0.67 | 0.31 | 0.21 | RGDSGPAGPPGEQGML | 0.90 | 0.51 | 0.29 | FAGDDAPRA | 0.67 | 0.37 | 0.22 |
| FLPM | 0.66 | 0.46 | 0.31 | FETF | 0.85 | 0.40 | 0.25 | ELPDGQVITI | 0.67 | 0.33 | 0.20 |
| IEELEEELEAERA | 0.65 | 0.36 | 0.20 | GSAGPRGPSGNIGMPGMTGPQ | 0.84 | 0.48 | 0.23 | KSYELPDGQVITIG | 0.63 | 0.35 | 0.21 |
| LDFENEMAT | 0.65 | 0.37 | 0.25 | TIIDQNRDGIIS | 0.76 | 0.29 | 0.20 | LDFENE | 0.60 | 0.37 | 0.27 |
| VDDIIQTGVDNPGHPFIMT | 0.63 | 0.42 | 0.20 | GEKLKGADPEDVIVA | 0.70 | 0.38 | 0.17 | GFNPPDLDIM | 0.60 | 0.39 | 0.25 |
| LKAGDSDGDGAIGVDEWA | 0.61 | 0.35 | 0.19 | SGSAGKDGMSGLPGPSGPSGPRGR | 0.67 | 0.55 | 0.25 | ELPDGQVI | 0.60 | 0.36 | 0.25 |
| LVQVQGEVDDSVQEARNAEEKAKKA | 0.60 | 0.34 | 0.11 | SGPGGPTGPSGM | 0.67 | 0.50 | 0.23 | LDLL | 0.58 | 0.35 | 0.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jafarpour, A.; Gregersen, S.; Marciel Gomes, R.; Marcatili, P.; Hegelund Olsen, T.; Jacobsen, C.; Overgaard, M.T.; Sørensen, A.-D.M. Biofunctionality of Enzymatically Derived Peptides from Codfish (Gadus morhua) Frame: Bulk In Vitro Properties, Quantitative Proteomics, and Bioinformatic Prediction. Mar. Drugs 2020, 18, 599. https://doi.org/10.3390/md18120599
Jafarpour A, Gregersen S, Marciel Gomes R, Marcatili P, Hegelund Olsen T, Jacobsen C, Overgaard MT, Sørensen A-DM. Biofunctionality of Enzymatically Derived Peptides from Codfish (Gadus morhua) Frame: Bulk In Vitro Properties, Quantitative Proteomics, and Bioinformatic Prediction. Marine Drugs. 2020; 18(12):599. https://doi.org/10.3390/md18120599
Chicago/Turabian StyleJafarpour, Ali, Simon Gregersen, Rocio Marciel Gomes, Paolo Marcatili, Tobias Hegelund Olsen, Charlotte Jacobsen, Michael Toft Overgaard, and Ann-Dorit Moltke Sørensen. 2020. "Biofunctionality of Enzymatically Derived Peptides from Codfish (Gadus morhua) Frame: Bulk In Vitro Properties, Quantitative Proteomics, and Bioinformatic Prediction" Marine Drugs 18, no. 12: 599. https://doi.org/10.3390/md18120599
APA StyleJafarpour, A., Gregersen, S., Marciel Gomes, R., Marcatili, P., Hegelund Olsen, T., Jacobsen, C., Overgaard, M. T., & Sørensen, A.-D. M. (2020). Biofunctionality of Enzymatically Derived Peptides from Codfish (Gadus morhua) Frame: Bulk In Vitro Properties, Quantitative Proteomics, and Bioinformatic Prediction. Marine Drugs, 18(12), 599. https://doi.org/10.3390/md18120599









