A Computer-Driven Approach to Discover Natural Product Leads for Methicillin-Resistant Staphylococcus aureus Infection Therapy †
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemical Space of the Anti-MRSA Models
2.1.1. Approach A
2.1.2. Approach B
2.2. Exploration of Empirical Molecular Descriptors and Fingerprints for QSAR Approach A
2.2.1. Exploration of Other State-of-the-art Machine Learning (ML) Techniques
2.2.2. Applicability Domain of the pMIC against MRSA Model
2.2.3. Application of the in silico Anti-MRSA Model in Virtual Screening
2.3. Exploration of NMR Descriptors for QSAR Approach B
Analysis of NMR Descriptors Identified as Relevant for Modeling Anti-MRSA Activity in the RF Model
3. Materials and Methods
3.1. Data Sets
3.1.1. Approach A
3.1.2. Approach B
3.2. Descriptors
3.2.1. Approach A
3.2.2. Approach B
3.3. Selection of Training and Test Sets
3.3.1. Approach A
3.3.2. Approach B
3.4. Machine Learning (ML) Techniques
3.4.1. Random Forests (RF)
3.4.2. Support Vector Machines (SVM)
3.4.3. Gaussian Processes (GPs)
3.4.4. Convolutional Neural Network (CNN)
3.5. Antibacterial Screening against MRSA Strain
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dzidic, S.; Suskovic, J.; Kos, B. Antibiotic resistance mechanisms in bacteria: Biochemical and genetic aspects. Food Technol. Biotechnol. 2008, 46, 11–21. [Google Scholar]
- Gallagher, R.; Motohashi, N.; Vanam, A.; Gollapudi, R. Global methicillin-resistant Staphylococcus aureus (MRSA) infections and current research trends. Arch. Gen. Intern. Med. 2017, 1, 3–11. [Google Scholar]
- Siddiqui, A.H.; Koirala, J. Methicillin Resistant Staphylococcus Aureus (MRSA); StatPearls Publishing LLC: Treasure Island, FL, USA, 2018. [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2013. Available online: http://www.cdc.gov/drugresistance/threat-report-2013/index.html (accessed on 10 October 2018).
- Khan, T.M.; Kok, Y.L.; Bukhsh, A.; Lee, L.-H.; Chan, K.-G.; Goh, B.-H. Incidence of methicillin resistant Staphylococcus aureus (MRSA) in burn intensive care unit: A systematic review. Germs 2018, 8, 113–125. [Google Scholar] [CrossRef] [PubMed]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Aires-de-Sousa, J. Computational methodologies in the exploration of marine natural product leads. Mar. Drugs 2018, 16, 236. [Google Scholar] [CrossRef] [PubMed]
- Koga, H.; Itoh, A.; Murayama, S.; Suzue, S.; Irikura, T. Structure-activity relationships of antibacterial 6,7- and 7,8-disubstituted 1-alkyl-1,4-dihydro-4-oxoquinoline-3-carboxylic acids. J. Med. Chem. 1980, 23, 1358–1363. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, M.; Siricilla, S.; Mitachi, K. Advances in MRSA drug discovery: Where are we and where do we need to be? Expert Opin. Drug Discov. 2013, 8, 1095–1116. [Google Scholar] [CrossRef]
- Kumar, K.; Chopra, S. New drugs for methicillin-resistant Staphylococcus aureus: An update. J. Antimicrob. Chemother. 2013, 68, 1465–1470. [Google Scholar] [CrossRef]
- Indrasena, A.; Riyaz, S.; Malipeddi, P.L.; Padmaja, P.; Sridhar, B.; Dubey, P.K. Design, synthesis, and biological evaluation of indolylidinepyrazolones as potential anti-bacterial agents. Tetrahedron Lett. 2014, 55, 5014–5018. [Google Scholar] [CrossRef]
- Judge, V.; Narasimhan, B.; Ahuja, M.; Sriram, D.; Yogeeswari, P.; De Clercq, E.; Pannecouque, C.; Balzarini, J. Synthesis, antimycobacterial, antiviral, antimicrobial activity and QSAR studies of N-2-acyl isonicotinic acid hydrazide derivatives. Med. Chem. 2013, 9, 53–76. [Google Scholar] [CrossRef] [PubMed]
- Hadj-esfandiari, N.; Navidpour, L.; Shadnia, H.; Amini, M.; Samadi, N.; Faramarzi, M.A.; Shafiee, A. Synthesis, antibacterial activity, and quantitative structure-activity relationships of new (Z)-2-(nitroimidazolylmethylene)-3(2H)-benzofuranone derivatives. Bioorg. Med. Chem. Lett. 2007, 17, 6354–6363. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Jenssen, H.; Hilpert, K.; Cheung, W.A.; Pante, N.; Hancock, R.E.W.; Cherkasov, A. Identification of novel antibacterial peptides by chemoinformatics and machine learning. J. Med. Chem. 2009, 52, 2006–2015. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Mueller, A.T.; Gabernet, G.; Button, A.L.; Posselt, G.; Wessler, S.; Hiss, J.A.; Schneider, G. Hybrid network model for “deep learning” of chemical data: Application to antimicrobial peptides. Mol. Inform. 2017, 36, 1600011. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, R.; Luo, Z.; Wang, W.; Chen, J. Antimicrobial activity and molecular docking studies of a novel anthraquinone from a marine-derived fungus Aspergillus versicolor. Nat. Prod. Res. 2018, 32, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Altuntas, T.G.; Yilmaz, N.; Ece, A.; Altanlar, N.; Olgen, S. In vitro antibacterial and antifungal activity and computational evaluation of novel indole derivatives containing 4-substituted piperazine moieties. Lett. Drug Des. Discov. 2018, 15, 1079–1086. [Google Scholar] [CrossRef]
- Zanni, R.; Galvez-Llompart, M.; Machuca, J.; Garcia-Domenech, R.; Recacha, E.; Pascual, A.; Rodriguez-Martinez, J.M.; Galvez, J. Molecular topology: A new strategy for antimicrobial resistance control. Eur. J. Med. Chem. 2017, 137, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Bueso-Bordils, J.I.; Perez-Gracia, M.T.; Suay-Garcia, B.; Duart, M.J.; Algarra, R.V.M.; Zamora, L.L.; Anton-Fos, G.M.; Lopez, P.A.A. Topological pattern for the search of new active drugs against methicillin resistant Staphylococcus aureus. Eur. J. Med. Chem. 2017, 138, 807–815. [Google Scholar] [CrossRef]
- Pereira, F.; Latino, D.A.R.S.; Gaudencio, S.P. A chemoinformatics approach to the discovery of lead-like molecules from marine and microbial sources en route to antitumor and antibiotic drugs. Mar. Drugs 2014, 12, 757–778. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Fernandes, J.; Gattass, C.R. Topological polar surface area defines substrate transport by multidrug resistance associated protein 1 (MRP1/ABCC1). J. Med. Chem. 2009, 52, 1214–1218. [Google Scholar] [CrossRef] [PubMed]
- Ebejer, J.-P.; Charlton, M.H.; Finn, P.W. Are the physicochemical properties of antibacterial compounds really different from other drugs? J. Cheminform. 2016, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Le, X.; Li, L.; Ju, Y.C.; Lin, Z.X.; Gu, Q.; Xu, J. Discovering new agents active against methicillin-resistant Staphylococcus aureus with ligand-based approaches. J. Chem. Inf. Model. 2014, 54, 3186–3197. [Google Scholar] [CrossRef] [PubMed]
- Berdy, J. Bioactive microbial metabolites—A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Drugs and drug candidates from marine sources: An assessment of the current “state of play”. Planta Med. 2016, 82, 775–789. [Google Scholar] [CrossRef]
- Choudhary, A.; Naughton, L.M.; Montanchez, I.; Dobson, A.D.W.; Rai, D.K. Current status and future prospects of marine natural products (MNPs) as antimicrobials. Mar. Drugs 2017, 15, 272. [Google Scholar] [CrossRef]
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; de Pascale, D. Antibiotics from deep-sea microorganisms: Current discoveries and perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef] [PubMed]
- Haste, N.M.; Hughes, C.C.; Tran, D.N.; Fenical, W.; Jensen, P.R.; Nizet, V.; Hensler, M.E. Pharmacological properties of the marine natural product marinopyrrole A against methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2011, 55, 3305–3312. [Google Scholar] [CrossRef] [PubMed]
- Bister, B.; Bischoff, D.; Strobele, M.; Riedlinger, J.; Reicke, A.; Wolter, F.; Bull, A.T.; Zahner, H.; Fiedler, H.P.; Sussmuth, R.D. Abyssomicin C—A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed. 2004, 43, 2574–2576. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Liu, D.; Hu, X.; Proksch, P.; Shao, Z.; Lin, W. Spiromastixones A-O, antibacterial chlorodepsidones from a deep-sea-derived Spiromastix sp Fungus. J. Nat. Prod. 2014, 77, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.K.; Seo, C.H.; Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 2015, 13, 618–654. [Google Scholar] [CrossRef] [PubMed]
- Valliappan, K.; Sun, W.; Li, Z. Marine actinobacteria associated with marine organisms and their potentials in producing pharmaceutical natural products. Appl. Microbiol. Biotechnol. 2014, 98, 7365–7377. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Cheung, R.C.F.; Wong, J.H.; Bekhit, A.A.; Bekhit, A.E.-D. Antibacterial products of marine organisms. Appl. Microbiol. Biotechnol. 2015, 99, 4145–4173. [Google Scholar] [CrossRef] [PubMed]
- Cruz, S.; Gomes, S.E.; Borralho, P.M.; Rodrigues, C.M.P.; Gaudencio, S.P.; Pereira, F. In silico HCT116 human colon cancer cell-based models en route to the discovery of lead-like anticancer drugs. Biomolecules 2018, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Davo, A.; Dias, T.; Gomes, S.E.; Rodrigues, S.; Parera-Valadezl, Y.; Borralho, P.M.; Pereira, F.; Rodrigues, C.M.P.; Santos-Sanches, I.; Gaudencio, S.P. The Madeira Archipelago as a significant source of marine-derived actinomycete diversity with anticancer and antimicrobial potential. Front. Microbiol. 2016, 7, 1594. [Google Scholar] [CrossRef] [PubMed]
- Klementz, D.; Doering, K.; Lucas, X.; Telukunta, K.K.; Erxleben, A.; Deubel, D.; Erber, A.; Santillana, I.; Thomas, O.S.; Bechthold, A.; et al. StreptomeDB 2.0-an extended resource of natural products produced by streptomycetes. Nucleic Acids Res. 2016, 44, D509–D514. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.P. Generation of a set of simple, interpretable ADMET rules of thumb. J. Med. Chem. 2008, 51, 817–834. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.W. PaDEL-Descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Selzer, P.; Ertl, P. Identification and classification of GPCR ligands using self-organizing neural networks. QSAR Comb. Sci. 2005, 24, 270–276. [Google Scholar] [CrossRef]
- Aires-de-Sousa, J. JATOON: Java tools for neural networks. Chemom. Intell. Lab. Syst. 2002, 61, 167–173. [Google Scholar] [CrossRef]
- Li, S.Q.; Fedorowicz, A.; Singh, H.; Soderholm, S.C. Application of the random forest method in studies of local lymph node assay based skin sensitization data. J. Chem. Inf. Model. 2005, 45, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrian-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Sterling, T.; Irwin, J.J. ZINC 15-Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zheng, F.; Fartaria, R.; Latino, D.A.R.S.; Qu, X.; Campos, T.; Zhao, T.; Aires-de-Sousa, J. A QSPR approach for the fast estimation of DFT/NBO partial atomic charges. Chemom. Intell. Lab. Syst. 2014, 134, 158–163. [Google Scholar] [CrossRef]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sc. 2003, 43, 1947–1958. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org (accessed on 10 October 2018).
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; DeBoy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef] [PubMed]


| Clusters 1 | Training Set 2 | Test Set 2 | Average/Maximum pMIC 3 |
|---|---|---|---|
A—Indole derivative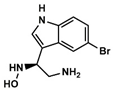 | 1123 | 324 | 4.72/7.80 |
B—1H-2-Benzopyran derivative | 1657 | 517 | 4.59/7.81 |
C—2-Oxazolidone derivative | 879 | 253 | 5.09/11.82 |
D—Triazole derivative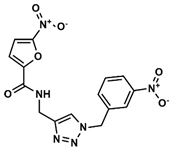 | 913 | 270 | 5.12/9.00 |
E—Cephalosporin derivative | 540 | 169 | 5.18/8.68 |
| Actinobacteria Genera | Set (Number/Sample Types) | Activity Class/Average MIC 1 |
|---|---|---|
| Actinomadura | Tr set 2 (2, cr 4) | cr: inactive/>250 |
| Te set 3 (1, cr 4) | cr: inactive/>250 | |
| Brevibacterium | - | - |
| Te set 3 (1, cr 4) | cr: inactive/>250 | |
| Micromonospora | Tr set 2 (23, 3 cr 4, 13 fr 5, 7 pu 6) | cr: inactive/>250 fr: 5 active/31; 8 inactive/≥250 pu: inactive/>250 |
| Te set 3 (9, 3 cr 4, 3 fr 5, 3 pu 6) | cr: 2 active/35; 1 inactive/>250 fr: 1 active/8; 2 inactive/>250 pu: inactive/>250 | |
| Salinispora | Tr set 2 (29, 11 cr 4, 10 fr 5, 8 pu 6) | cr: 1 active/63; 10 inactive/>250 fr: 2 active/23; 8 inactive/≥250 pu: inactive/>250 |
| Te set 3 (10, 4 cr 4, 4 fr 5, 2 pu 6) | cr: inactive/>250 fr: 1 active/8; 3 inactive/>250 pu: inactive/>250 | |
| Streptomyces | Tr set 2 (62, 21 cr 4, 18 fr 5, 23 pu 6) | cr: 2 active/5; 19 inactive/243 fr: 12 active/11; 6 inactive/229 pu: 14 active/12; 9 inactive/222 |
| Te set 3 (18, 4 cr 4, 7 fr 5, 7 pu 6) | cr: inactive/219 fr: 5 active /23; 2 inactive/188 pu: 4 active/11; 3 inactive/188 |
| Descriptors (#) | R2 | RMSE 1 | MAE 2 | % Error ≥ 0.5/% Error < 0.5 3 |
|---|---|---|---|---|
| MACCS (166) 4 | 0.555 | 0.649 | 0.477 | 37/63 |
| Sub (307) 4 | 0.509 | 0.681 | 0.505 | 39/61 |
| SubC (307) 4 | 0.563 | 0.642 | 0.468 | 36/64 |
| PubChem (881) 4 | 0.561 | 0.643 | 0.467 | 36/64 |
| CDK (1024) 4 | 0.551 | 0.652 | 0.471 | 36/64 |
| CDK Ext (1024) 4 | 0.572 | 0.636 | 0.456 | 34/66 |
| 1D2D (218) 5 | 0.364 | 0.775 | 0.600 | 49/51 |
| Descriptors (#) | R2 | RMSE | MAE | % Error ≥ 0.5/% Error < 0.5 1 |
|---|---|---|---|---|
| MACCS_3D_CDK (232) 2 | 0.441 | 0.731 | 0.569 | 47/53 |
| MACCS_3D_RDF (550) 3 | 0.426 | 0.745 | 0.587 | 45/55 |
| SubC_3D_CDK (373) 2 | 0.466 | 0.715 | 0.556 | 47/53 |
| SubC_3D_RDF (691) 3 | 0.448 | 0.730 | 0.574 | 48/52 |
| PubChem_3D_CDK (947) 2 | 0.479 | 0.705 | 0.546 | 45/55 |
| PubChem_3D_RDF (1265) 3 | 0.465 | 0.719 | 0.564 | 48/52 |
| CDK_Ext_3D_CDK (1090) 2 | 0.537 | 0.667 | 0.507 | 41/59 |
| CDK_Ext_3D_RDF (1408) 3 | 0.507 | 0.690 | 0.533 | 44/56 |
| Model/n.° Descriptors | R2 | RMSE | MAE | % Error ≥ 0.5/% Error < 0.5 1 |
|---|---|---|---|---|
| Training set 2 | ||||
| SubC/75 3 | 0.556 | 0.647 | 0.471 | 36/64 |
| SubC/100 3 | 0.563 | 0.642 | 0.467 | 36/64 |
| SubC/150 3 | 0.562 | 0.643 | 0.467 | 36/64 |
| Pubchem/75 3 | 0.528 | 0.667 | 0.496 | 39/61 |
| PubChem/100 3 | 0.544 | 0.656 | 0.484 | 38/62 |
| PubChem/150 3 | 0.558 | 0.645 | 0.471 | 37/63 |
| CDK Ext/75 3 | 0.545 | 0.655 | 0.481 | 37/63 |
| CDK Ext/100 3 | 0.566 | 0.641 | 0.467 | 36/64 |
| CDK Ext/150 3 | 0.574 | 0.635 | 0.460 | 35/65 |
| Test set | ||||
| SubC/75 3 | 0.637 | 0.626 | 0.464 | 36/64 |
| SubC/100 3 | 0.645 | 0.620 | 0.459 | 36/64 |
| SubC/150 3 | 0.644 | 0.621 | 0.460 | 36/64 |
| Pubchem/75 3 | 0.603 | 0.654 | 0.494 | 39/61 |
| Pubchem/100 3 | 0.620 | 0.639 | 0.475 | 37/63 |
| Pubchem/150 3 | 0.632 | 0.629 | 0.471 | 37/63 |
| CDK Ext/75 3 | 0.609 | 0.650 | 0.483 | 38/62 |
| CDK Ext/100 3 | 0.632 | 0.631 | 0.467 | 37/63 |
| CDK Ext/150 3 | 0.641 | 0.623 | 0.458 | 34/66 |
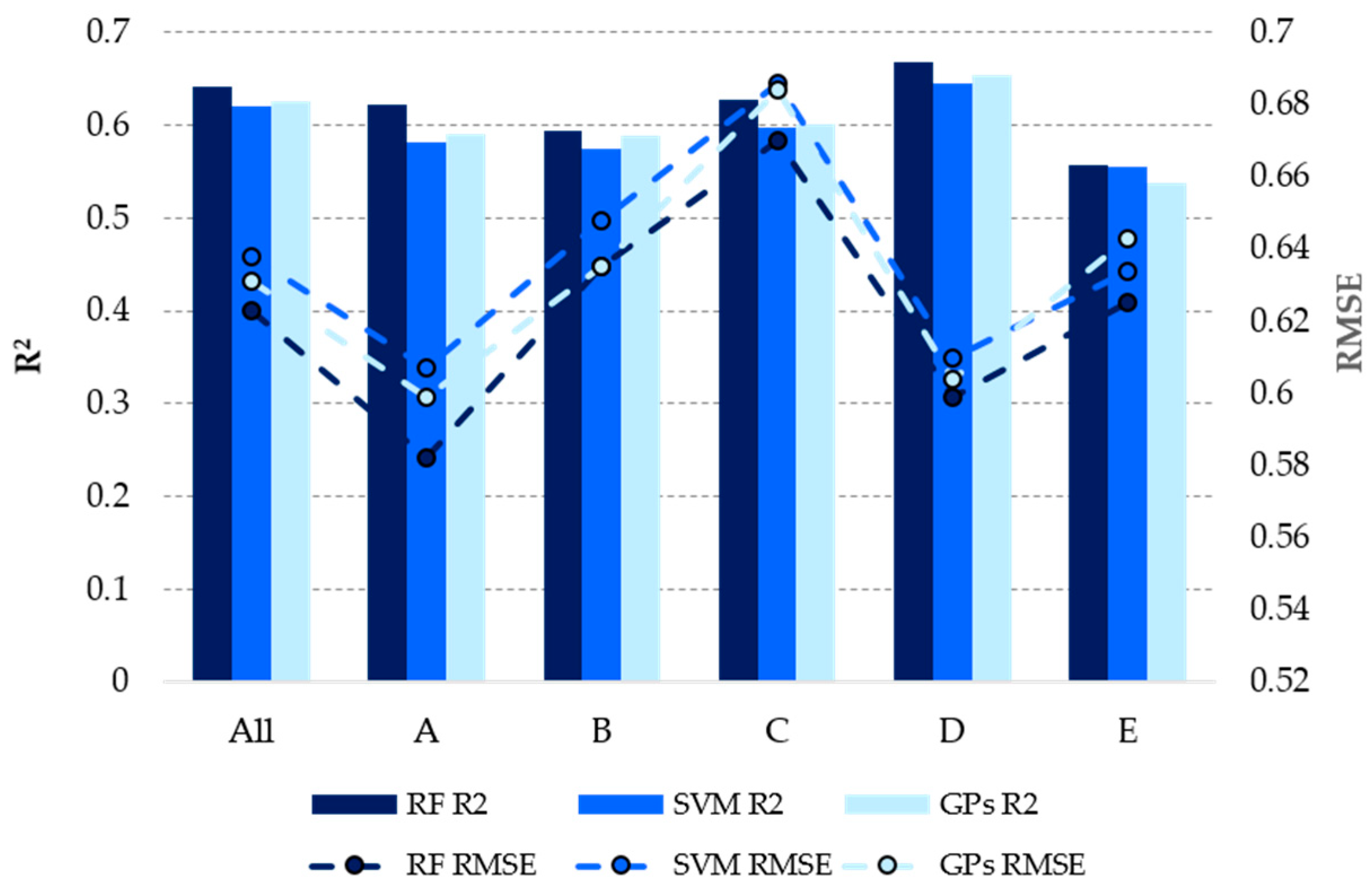
| ML | R2 | RMSE | MAE | |
|---|---|---|---|---|
| RF 1 | All | 0.574 | 0.635 | 0.460 |
| A | 0.583 | 0.555 | 0.416 | |
| B | 0.549 | 0.622 | 0.458 | |
| C | 0.445 | 0.757 | 0.520 | |
| D | 0.594 | 0.641 | 0.467 | |
| E | 0.575 | 0.600 | 0.449 | |
| SVM 2 | All | 0.564 | 0.645 | 0.457 |
| A | 0.584 | 0.556 | 0.407 | |
| B | 0.518 | 0.649 | 0.463 | |
| C | 0.466 | 0.743 | 0.508 | |
| D | 0.569 | 0.659 | 0.465 | |
| E | 0.570 | 0.606 | 0.443 | |
| GPs 2 | All | 0.568 | 0.638 | 0.465 |
| A | 0.590 | 0.548 | 0.415 | |
| B | 0.528 | 0.636 | 0.466 | |
| C | 0.450 | 0.752 | 0.530 | |
| D | 0.583 | 0.647 | 0.474 | |
| E | 0.577 | 0.599 | 0.442 |
| Model | R2 | RMSE | MAE | % Error ≥ 0.5/% Error < 0.5 1 |
|---|---|---|---|---|
| Training set | ||||
| CM1 2 | 0.587 | 0.624 | 0.450 | 33/67 |
| CM2 3 | 0.601 | 0.617 | 0.453 | 34/66 |
| Test set | ||||
| CM1 | 0.644 | 0.617 | 0.453 | 33/67 |
| CM2 | 0.683 | 0.593 | 0.444 | 35/65 |
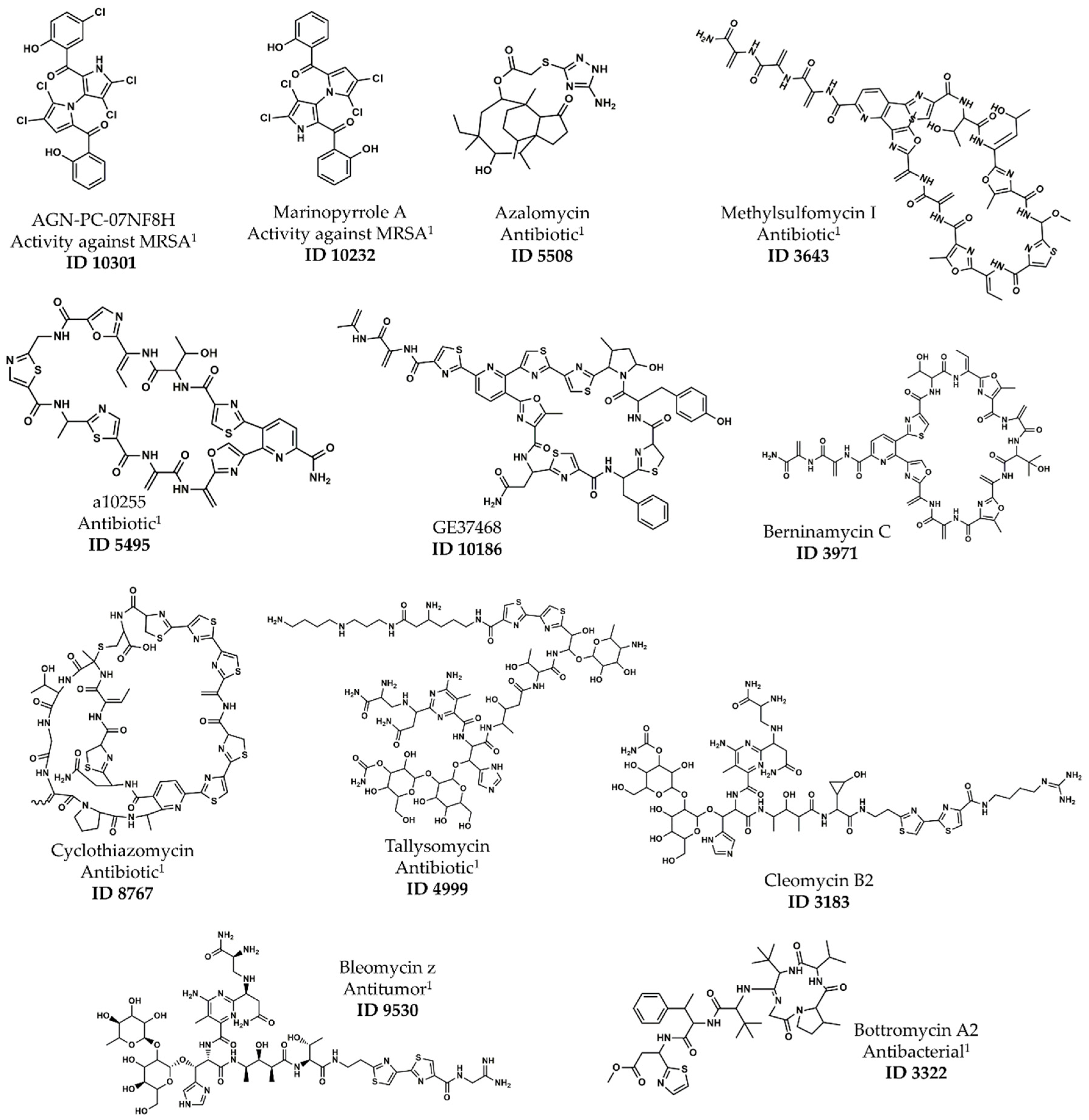
| ID 1 | Name | Type | pMIC 2 | Cluster 3 | ASD |
|---|---|---|---|---|---|
| 10301 | AGN-PC-07NF8H | Bis-pyrrole | 6.06 | A | 0.23 |
| 10232 | Marinopyrrole A | Bis-pyrrole | 5.92 | A | 0.23 |
| 5508 | Azalomycin | Spiro-tricyclic | 5.51 | A | 0.39 |
| 3643 | Methylsulfomycin I | Pyridine-containing 4 | 7.08 | E | 0.33 |
| 5495 | a10255 | Pyridine-containing 4 | 6.51 | E | 0.31 |
| 10186 | GE37468 | Pyridine-containing 4 | 6.41 | E | 0.35 |
| 3971 | Berninamycin C | Pyridine-containing 4 | 6.23 | E | 0.38 |
| 8767 | Cyclothiazomycin | Polythiazole-containing 5 | 6.02 | E | 0.34 |
| 4999 | Tallysomycin | Glycopeptide | 5.42 | E | 0.39 |
| 3183 | Cleomycin B2 | Glycopeptide | 5.36 | E | 0.37 |
| 9530 | Bleomycin z | Glycopeptide | 5.32 | E | 0.39 |
| 3322 | Bottromycin A2 | Macrocyclic peptide | 5.30 | E | 0.31 |
| Model | # 1 | TP 2 | TN 3 | FP 4 | FN 5 | SE 6 | SP 7 | Q 8 | MCC 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Training set 10 | ||||||||||
| 13C | 0.5 | 400 | 12 | 72 | 8 | 24 | 0.33 | 0.90 | 0.72 | 0.29 |
| 1 | 200 | 9 | 71 | 9 | 27 | 0.25 | 0.89 | 0.69 | 0.18 | |
| 1.5 | 133 | 12 | 72 | 8 | 24 | 0.33 | 0.90 | 0.72 | 0.29 | |
| 1H | 0.05 | 240 | 10 | 73 | 7 | 26 | 0.28 | 0.91 | 0.72 | 0.25 |
| 0.1 | 120 | 12 | 72 | 8 | 24 | 0.33 | 0.90 | 0.72 | 0.29 | |
| 0.2 | 61 | 14 | 71 | 9 | 22 | 0.39 | 0.89 | 0.73 | 0.32 | |
| 0.5 | 23 | 8 | 69 | 11 | 28 | 0.22 | 0.86 | 0.66 | 0.11 | |
| 13C 1H | 0.5 0.2 | 461 | 13 | 75 | 5 | 23 | 0.36 | 0.94 | 0.76 | 0.38 |
| Test set | ||||||||||
| 13C | 0.5 | 400 | 4 | 24 | 2 | 9 | 0.31 | 0.92 | 0.72 | 0.30 |
| 1 | 200 | 2 | 25 | 1 | 11 | 0.15 | 0.96 | 0.69 | 0.20 | |
| 1.5 | 133 | 1 | 22 | 4 | 12 | 0.08 | 0.85 | 0.59 | 0.11 | |
| 1H | 0.05 | 240 | 7 | 22 | 4 | 6 | 0.54 | 0.85 | 0.74 | 0.40 |
| 0.1 | 120 | 7 | 21 | 5 | 6 | 0.54 | 0.81 | 0.72 | 0.36 | |
| 0.2 | 61 | 8 | 21 | 5 | 5 | 0.62 | 0.81 | 0.74 | 0.42 | |
| 0.5 | 23 | 7 | 22 | 4 | 6 | 0.54 | 0.85 | 0.74 | 0.40 | |
| 13C 1H | 0.5 0.2 | 461 | 7 | 24 | 2 | 6 | 0.54 | 0.92 | 0.79 | 0.52 |
| ML | SE 1 | SP 2 | Q 3 | MCC 4 |
|---|---|---|---|---|
| Training set | ||||
| RF 5 | 0.56 | 0.91 | 0.80 | 0.51 |
| SVM 6 | 0.72 | 0.81 | 0.78 | 0.52 |
| CNN 6 | 0.61 | 0.89 | 0.80 | 0.52 |
| Test set | ||||
| RF | 0.46 | 0.92 | 0.77 | 0.45 |
| SVM | 0.69 | 0.73 | 0.72 | 0.41 |
| CNN | 0.62 | 0.81 | 0.74 | 0.42 |
| Code | Actinobacteria Genera | Structural Family | Activity Class 1 | MIC (μg/mL) 2 | Activity Class 2 |
|---|---|---|---|---|---|
| PTM-290 F7,F26 | Salinispora | Diketopiperazine | InAct 3 | >250 | InAct 3 |
| PTM-290 F7,F27 | Salinispora | Diketopiperazine | InAct 3 | >250 | InAct 3 |
| PTM-420 F4,F15 | Streptomyces | Unknown | InAct 3 | >250 | InAct 3 |
| PTM-420 F5,F45 | Streptomyces | Napyradiomycin | InAct 3 | 62.5 | MAct 4 |
| H or C (# 1) | NMR Range (ppm) | Ranking 2 | Importance for Classes | Pattern Identification | |
|---|---|---|---|---|---|
| InAct 3 | MAct 4 | ||||
| H (19) | 11.2393–11.5676 | 1st | 9.23 | 6.59 | Hydrogen bond CO and –NH and –OH; heteroaromatic NH; COOH |
| H (8) | 13.8656–14.1939 | 2nd | 6.22 | 4.86 | Hydrogen bond CO and –NH and –OH; heteroaromatic NH |
| H (22) | 10.5828–10.9111 | 3rd | 3.92 | 4.86 | Hydrogen bond CO and –NH and –OH; heteroaromatic NH; COOH; C=N–OH |
| H (28) | 9.5979–9.9262 | 8th | 2.30 | 2.46 | Aldehyde CHO |
| C (318) | 127.4927–127.9927 | 9th | 3.08 | −0.02 | Aromatic; olefinic; nitrile |
| H (58) | 1.3909–1.7191 | 10th | 2.91 | 1.77 | Saturated alkane |
| H (48) | 7.3000–7.6282 | 11th | 2.17 | 1.05 | Aromatic; conjugated olefinic |
| C (410) | 175.9927–176.4927 | 14th | 1.85 | 0.08 | COX; X: O, N, Cl a,b unsat. COX; X: O, N, Cl |
| C (321) | 33.9927–34.4927 | 15th | 2.61 | 0.58 | –CH2COX; –NHCH3; CH3CH2CH2– |
| C (350) | 168.9927–169.4927 | 16th | 1.97 | 0.43 | COX; X: O, N, Cl,b unsat. COX; X: O, N, Cl |
| H (36) | 5.6585–5.9868 | 18th | 2.66 | 0.48 | Vinylic |
| C (141) | 52.4927–52.9927 | 19th | 2.24 | 1.00 | –CHCl; –CH2Cl |
| C (329) | 123.9927–124.4927 | 20th | 1.86 | 0.93 | Vinylic |
| C (415) | 171.9927–172.4927 | 21th | 2.14 | 3.40 | COX; X: O, N, Cl,b unsat. COX; X: O, N, Cl |
| C (401) | 178.4927–178.9927 | 23th | 2.57 | 0.18 | COX; X: O, N, Cl,b unsat. COX; X: O, N, Cl |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, T.; Gaudêncio, S.P.; Pereira, F. A Computer-Driven Approach to Discover Natural Product Leads for Methicillin-Resistant Staphylococcus aureus Infection Therapy. Mar. Drugs 2019, 17, 16. https://doi.org/10.3390/md17010016
Dias T, Gaudêncio SP, Pereira F. A Computer-Driven Approach to Discover Natural Product Leads for Methicillin-Resistant Staphylococcus aureus Infection Therapy. Marine Drugs. 2019; 17(1):16. https://doi.org/10.3390/md17010016
Chicago/Turabian StyleDias, Tiago, Susana P. Gaudêncio, and Florbela Pereira. 2019. "A Computer-Driven Approach to Discover Natural Product Leads for Methicillin-Resistant Staphylococcus aureus Infection Therapy" Marine Drugs 17, no. 1: 16. https://doi.org/10.3390/md17010016
APA StyleDias, T., Gaudêncio, S. P., & Pereira, F. (2019). A Computer-Driven Approach to Discover Natural Product Leads for Methicillin-Resistant Staphylococcus aureus Infection Therapy. Marine Drugs, 17(1), 16. https://doi.org/10.3390/md17010016








