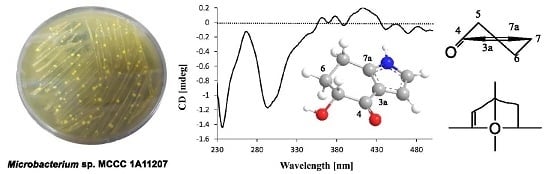
Microindolinone A, a Novel 4,5,6,7-Tetrahydroindole, from the Deep-Sea-Derived Actinomycete Microbacterium sp. MCCC 1A11207
Abstract
:1. Introduction
2. Results and Discussion
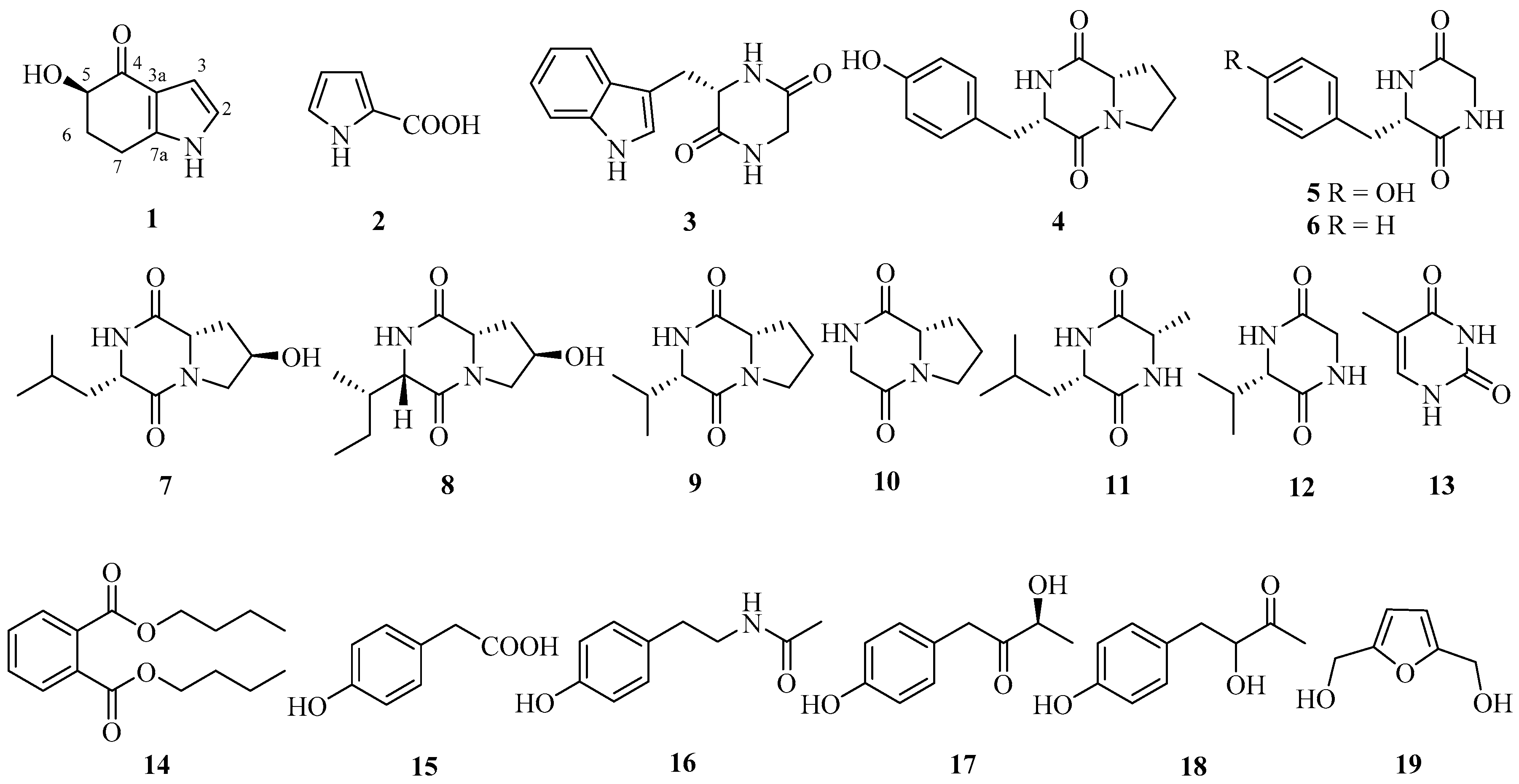
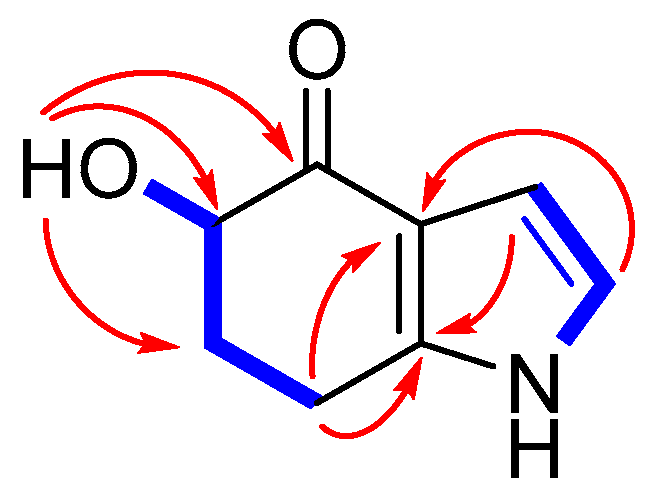
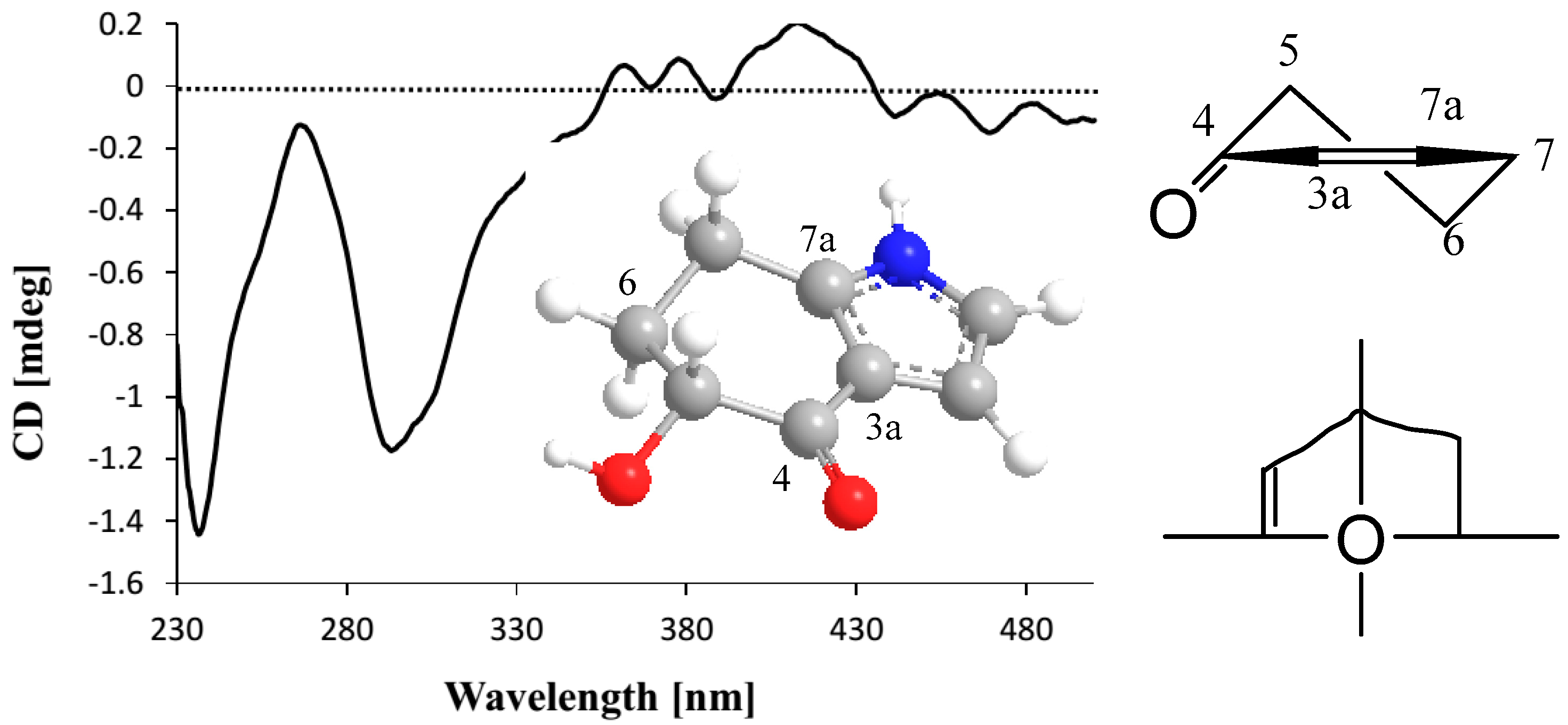
2.1. Structure Elucidation
2.2. Anti-Proliferative Activity of 1 against RBL-2H3 Cells
2.3. Anti-Allergic Activity of 1
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Bacterial Material
3.3. Cultivation and Extraction
3.4. Isolation and Purification
3.5. Anti-Proliferative Assay
3.6. Anti-Allergic Test
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Diminic, J.; Starcevic, A.; Lisfi, M.; Baranasic, D.; Gacesa, R.; Hranueli, D.; Long, P.F.; Cullum, J.; Zucko, J. Evolutionary concepts in natural products discovery: What actinomycetes have taught us. J. Ind. Microbiol. Biotechnol. 2014, 41, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus salinospora. Angew. Chem. Int. Ed. Engl. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, C.; Huang, C.; Zhang, L.; Zhang, H.; Zhang, Q.; Yuan, C.S.; Zhu, Y.; Zhang, C. Pyrazolofluostatins A–C, pyrazole-fused benzo[a]fluorenes from South China Sea-derived Micromonospora rosaria SCSIO N160. Org. Lett. 2017, 19, 592–595. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.E.; Dalisay, D.S.; Chen, J.; Polishchuck, E.A.; Patrick, B.O.; Narula, G.; Ko, M.; Av-Gay, Y.; Li, H.; Magarvey, N.; et al. Aminorifamycins and sporalactams produced in culture by a Micromonospora sp. isolated from a Northeastern-Pacific marine sediment are potent antibiotics. Org. Lett. 2017, 19, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Huang, R.; Zhu, S.; Li, H.; Zheng, G. Identification and characterization of a novel (+)-γ-lactamase from Microbacterium hydrocarbonoxydans. Appl. Microbiol. Biotechnol. 2016, 100, 9543–9553. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Lin, H.; Proksch, P.; Tang, X.; Shao, Z.; Lin, W. Microbacterins A and B, new peptaibols from the deep sea actinomycete Microbacterium sediminis sp. nov. YLB-01(T). Org. Lett. 2015, 17, 1220–1223. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.L.; Liu, Q.; Xia, J.M.; Gao, Y.; Yang, Q.; Shao, Z.Z.; Liu, G.; Yang, X.W. Anti-allergic compounds from the deep-sea-derived actinomycete Nesterenkonia flava MCCC 1K00610. Mar. Drugs 2017, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Si, L.; Liu, D.; Zhou, A.; Zhang, Z.; Shao, Z.; Wang, S.; Zhang, L.; Zhou, D.; Lin, W. Spiromastilactones: A new class of influenza virus inhibitors from deep-sea fungus. Eur. J. Med. Chem. 2016, 108, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Peng, K.; Liu, Z.; Zhang, G.Y.; Li, J.; Wang, N.; Steinmetz, A.; Liu, Y. Strepsesquitriol, a rearranged zizaane-type sesquiterpenoid from the deep-sea-derived actinomycete Streptomyces sp. SCSIO 10355. J. Nat. Prod. 2013, 76, 2360–2363. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tian, L.; Huang, J.; Ma, H.Y.; Zheng, Z.; Lv, A.L.; Yasukawa, K.; Pei, Y.H. Trichodermatides A–D, novel polyketides from the marine-derived fungus Trichoderma reesei. Org. Lett. 2008, 10, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Kirk, D.N. The chiroptical properties of carbonyl compounds. Tetrahedron 1986, 42, 777–818. [Google Scholar] [CrossRef]
- Netz, N.; Opatz, T. Marine indole alkaloids. Mar. Drugs 2015, 13, 4814–4914. [Google Scholar] [CrossRef] [PubMed]
- Ishikura, M.; Abe, T.; Choshi, T.; Hibino, S. Simple indole alkaloids and those with a non-rearranged monoterpenoid unit. Nat. Prod. Rep. 2013, 30, 694–752. [Google Scholar] [CrossRef] [PubMed]
- Gupta, L.; Talwar, A.; Chauhan, P.M. Bis and tris indole alkaloids from marine organisms: new leads for drug discovery. Curr. Med. Chem. 2007, 14, 1789–1803. [Google Scholar] [CrossRef] [PubMed]
- Henne, P.; Zeeck, A.; Grabley, S.; Thiericke, R. Secondary metabolites by chemical screening.35. 6,7-Dihydroxy-4,5,6,7-tetrahydroindole-4-one, a new type of indole-derivative from Nocardia sp. Nat. Prod. Lett. 1997, 10, 43–47. [Google Scholar] [CrossRef]
- Dietera, A.; Hamm, A.; Fiedler, H.P.; Goodfellow, M.; Muller, W.E.; Brun, R.; Beil, W.; Bringmann, G. Pyrocoll, an antibiotic, antiparasitic and antitumor compound produced by a novel alkaliphilic Streptomyces strain. J. Antibiot. 2003, 56, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Deslauriers, R.; Grzonka, Z.; Schaumburg, K.; Shiba, T.; Walter, R. Carbon-13 nuclear magnetic resonance studies of the conformations of cyclic dipeptides. J. Am. Chem. Soc. 1975, 97, 5093–5100. [Google Scholar] [CrossRef] [PubMed]
- Thajudeen, H.; Park, K.; Moon, S.-S.; Hong, I.S. An efficient green synthesis of proline-based cyclic dipeptides under water-mediated catalyst-free conditions. Tetrahedron Lett. 2010, 51, 1303–1305. [Google Scholar] [CrossRef]
- Mollica, A.; Costante, R.; Fiorito, S.; Genovese, S.; Stefanucci, A.; Mathieu, V.; Kiss, R.; Epifano, F. Synthesis and anti-cancer activity of naturally occurring 2,5-diketopiperazines. Fitoterapia 2014, 98, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; Toriuchi, Y.; Fukayama, S.; Sano, S. Synthesis and conformational characterization of diketopiperazines bearing a benzyl moiety. Chem. Lett. 2014, 43, 340–342. [Google Scholar] [CrossRef]
- Ienaga, K.; Nakamura, K.; Goto, T. Bioactive compounds produced in animal tissues. I. Two diketopiperidine plant-growth regulators containing hydroxyproline isolated from rabbit skin tissue extract. Tetrahedron Lett. 1987, 28, 1285–1286. [Google Scholar] [CrossRef]
- Cronan, J.M., Jr.; Davidson, T.R.; Singleton, F.L.; Colwell, R.R.; Cardellina, J.H., II. Plant growth promoters isolated from a marine bacterium associated with Palythoa sp. Nat. Prod. Lett. 1998, 11, 271–278. [Google Scholar] [CrossRef]
- Maurer, G.; Kiechel, J.R. Ergopeptide Alkaloids. Patent DE2805977A1, 1978. [Google Scholar]
- Hendea, D.; Laschat, S.; Baro, A.; Frey, W. Diastereoselective alkylation of a proline-derived bicyclic lactim ether. Helv. Chim. Acta 2006, 89, 1894–1909. [Google Scholar] [CrossRef]
- Ding, Z.G.; Zhao, J.Y.; Yang, P.W.; Li, M.G.; Huang, R.; Cui, X.L.; Wen, M.L. 1H and 13C NMR assignments of eight nitrogen containing compounds from Nocardia alba sp.nov (YIM 30243T). Magn. Reson. Chem. 2009, 47, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Lankiewicz, L.; Nyasse, B.; Fransson, B.; Grehn, L.; Ragnarsson, U. Synthesis of amino acid derivatives substituted in the backbone with stable isotopes for application in peptide synthesis. J. Chem. Soc. Perkin Trans. 1 1994, 17, 2503–2510. [Google Scholar] [CrossRef]
- McNulty, J.; Nair, J.J.; Cheekoori, S.; Larichev, V.; Capretta, A.; Robertson, A.J. Scope and mechanistic insights into the use of tetradecyl(trihexyl)phosphonium bistriflimide: A remarkably selective ionic liquid solvent for substitution reactions. Chem. A Eur. J. 2006, 12, 9314–9322. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.E.; Storz, T.; Colyer, J.T.; Thiel, O.R.; Dilmeghani Seran, M.; Larsen, R.D.; Murry, J.A. Iodide-catalyzed reductions: Development of a synthesis of phenylacetic acids. J. Org. Chem. 2011, 76, 9519–9524. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.J.; Lu, X.M.; Zhu, T.J.; Fang, Y.C.; Gu, Q.Q.; Zhu, W. GPR12 Selections of the metabolites from an endophytic Streptomyces sp. asociated with Cistanches deserticola. Arch. Pharmacal Res. 2008, 31, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.P.; Wang, Y.; Liu, P.P.; Hong, K.; Chen, H.; Yin, X.; Zhu, W.M. Aromatic compounds from the halotolerant fungal strain of Wallemia sebi PXP-89 in a hypersaline medium. Arch. Pharmacal Res. 2011, 34, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.L. Phytochemical profile of the stems of Aeonium lindleyi. Rev. Bras. Farmacogn. 2012, 22, 676–679. [Google Scholar] [CrossRef]
- Goswami, S.; Dey, S.; Jana, S. Design and synthesis of a unique ditopic macrocyclic fluorescent receptor containing furan ring as a spacer for the recognition of dicarboxylic acids. Tetrahedron 2008, 64, 6358–6363. [Google Scholar] [CrossRef]
- Yang, X.W.; Zeng, H.W.; Liu, X.H.; Li, S.M.; Xu, W.; Shen, Y.H.; Zhang, C.; Zhang, W.D. Anti-inflammatory and anti-tumour effects of Abies georgei extracts. J. Pharm. Pharmacol. 2008, 60, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, Y.; Cao, M.; Pan, T.; Yang, Y.; Mao, H.; Sun, L.; Liu, G. Anti-allergic activity of R-phycocyanin from Porphyra haitanensis in antigen-sensitized mice and mast cells. Int. Immunopharmacol. 2015, 25, 465–473. [Google Scholar] [CrossRef] [PubMed]
| Position | δC, Type | δH (J in Hz) |
|---|---|---|
| 1 | 11.3, brs | |
| 2 | 120.3, CH | 6.74, dd (2.9, 2.4) |
| 3 | 105.2, CH | 6.25, dd (2.9, 2.2) |
| 3a | 118.4, C | |
| 4 | 194.1, C | |
| 5 | 72.6, CH | 4.05, ddd (11.6, 4.5, 3.8) |
| 6 | 33.0, CH2 | 1.87, m; 2.20, m |
| 7 | 21.3, CH3 | 2.83, m |
| 7a | 143.4, C | |
| 5-OH | 4.98, d (3.8) |
| Concentrations (µg/mL) | Cell Viability (%) |
|---|---|
| 20 | 91 ± 10 |
| 10 | 93 ± 1.4 |
| 5 | 90 ± 10 |
| 2.5 | 93 ± 12 |
| 1.25 | 94 ± 12 |
| 0.625 | 99 ± 14 |
| Compound | Concentration (μg/mL) | Inhibition Rate (%) |
|---|---|---|
| 1 | 20 | −1.4 ± 0.8 |
| Loratadine | 20 | 37 ± 5.3 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, S.; Zhou, T.-T.; Xie, C.-L.; Zhang, G.-Y.; Yang, X.-W. Microindolinone A, a Novel 4,5,6,7-Tetrahydroindole, from the Deep-Sea-Derived Actinomycete Microbacterium sp. MCCC 1A11207. Mar. Drugs 2017, 15, 230. https://doi.org/10.3390/md15070230
Niu S, Zhou T-T, Xie C-L, Zhang G-Y, Yang X-W. Microindolinone A, a Novel 4,5,6,7-Tetrahydroindole, from the Deep-Sea-Derived Actinomycete Microbacterium sp. MCCC 1A11207. Marine Drugs. 2017; 15(7):230. https://doi.org/10.3390/md15070230
Chicago/Turabian StyleNiu, Siwen, Ting-Ting Zhou, Chun-Lan Xie, Gai-Yun Zhang, and Xian-Wen Yang. 2017. "Microindolinone A, a Novel 4,5,6,7-Tetrahydroindole, from the Deep-Sea-Derived Actinomycete Microbacterium sp. MCCC 1A11207" Marine Drugs 15, no. 7: 230. https://doi.org/10.3390/md15070230
APA StyleNiu, S., Zhou, T.-T., Xie, C.-L., Zhang, G.-Y., & Yang, X.-W. (2017). Microindolinone A, a Novel 4,5,6,7-Tetrahydroindole, from the Deep-Sea-Derived Actinomycete Microbacterium sp. MCCC 1A11207. Marine Drugs, 15(7), 230. https://doi.org/10.3390/md15070230