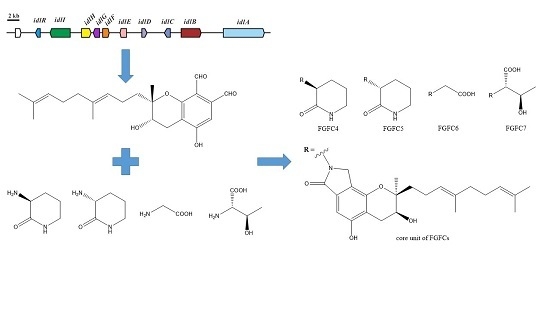
Producing Novel Fibrinolytic Isoindolinone Derivatives in Marine Fungus Stachybotrys longispora FG216 by the Rational Supply of Amino Compounds According to Its Biosynthesis Pathway
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isoindolinone Biosynthesis Pathway in Stachybotrys
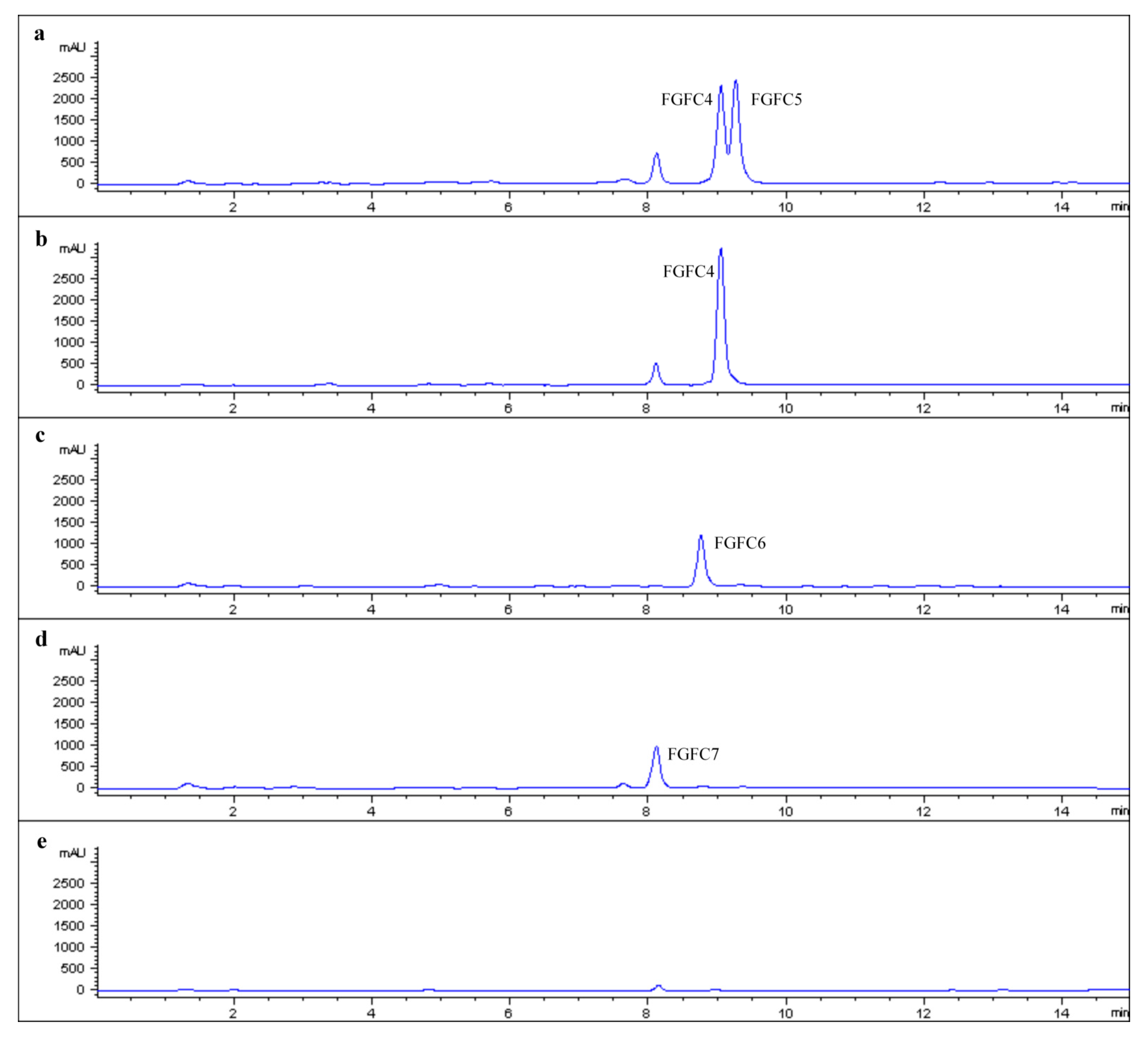
2.2. Production of New FGFCs by Rational Amino Compounds Supply
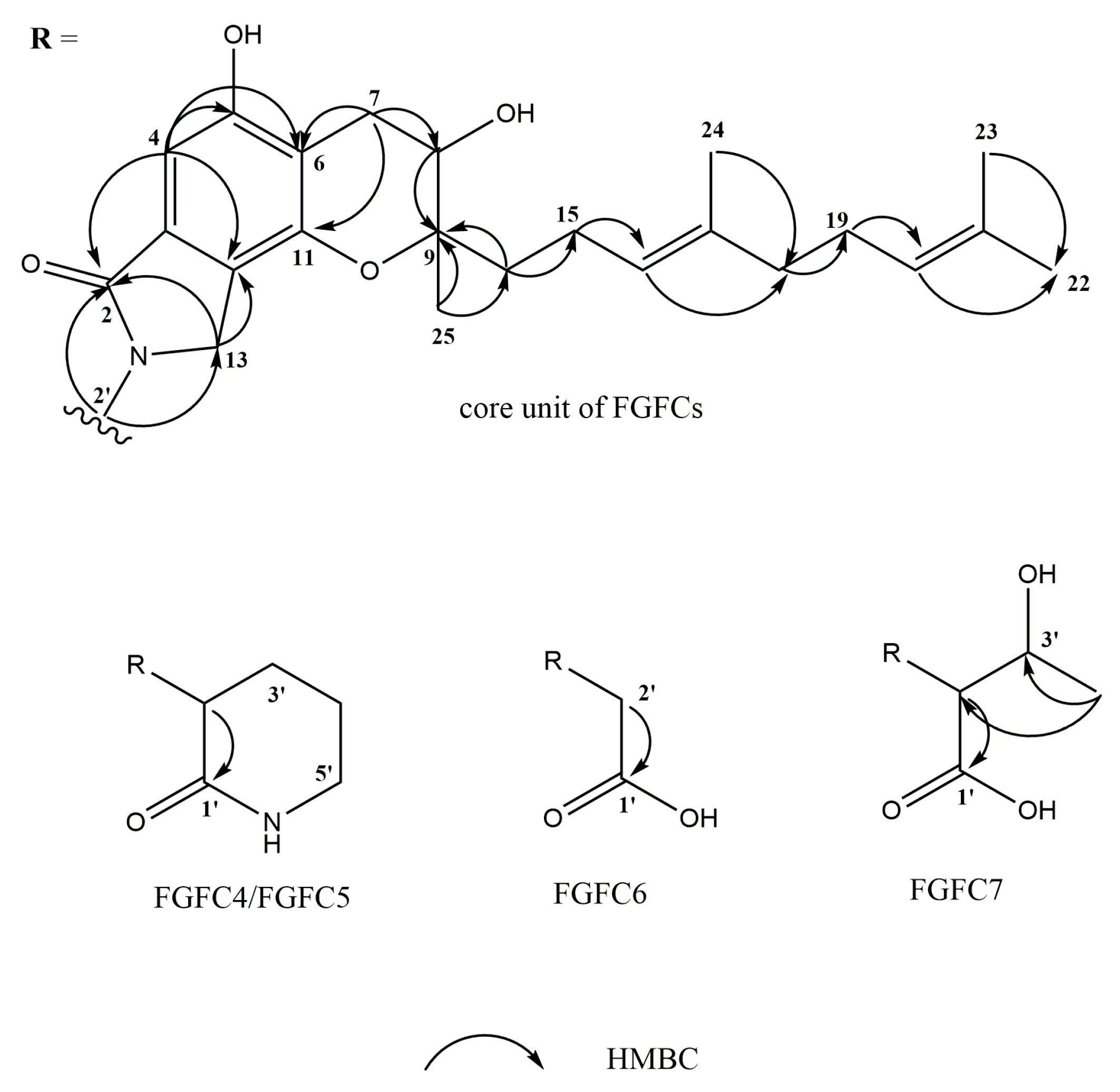
2.3. Structure Determination of New FGFCs
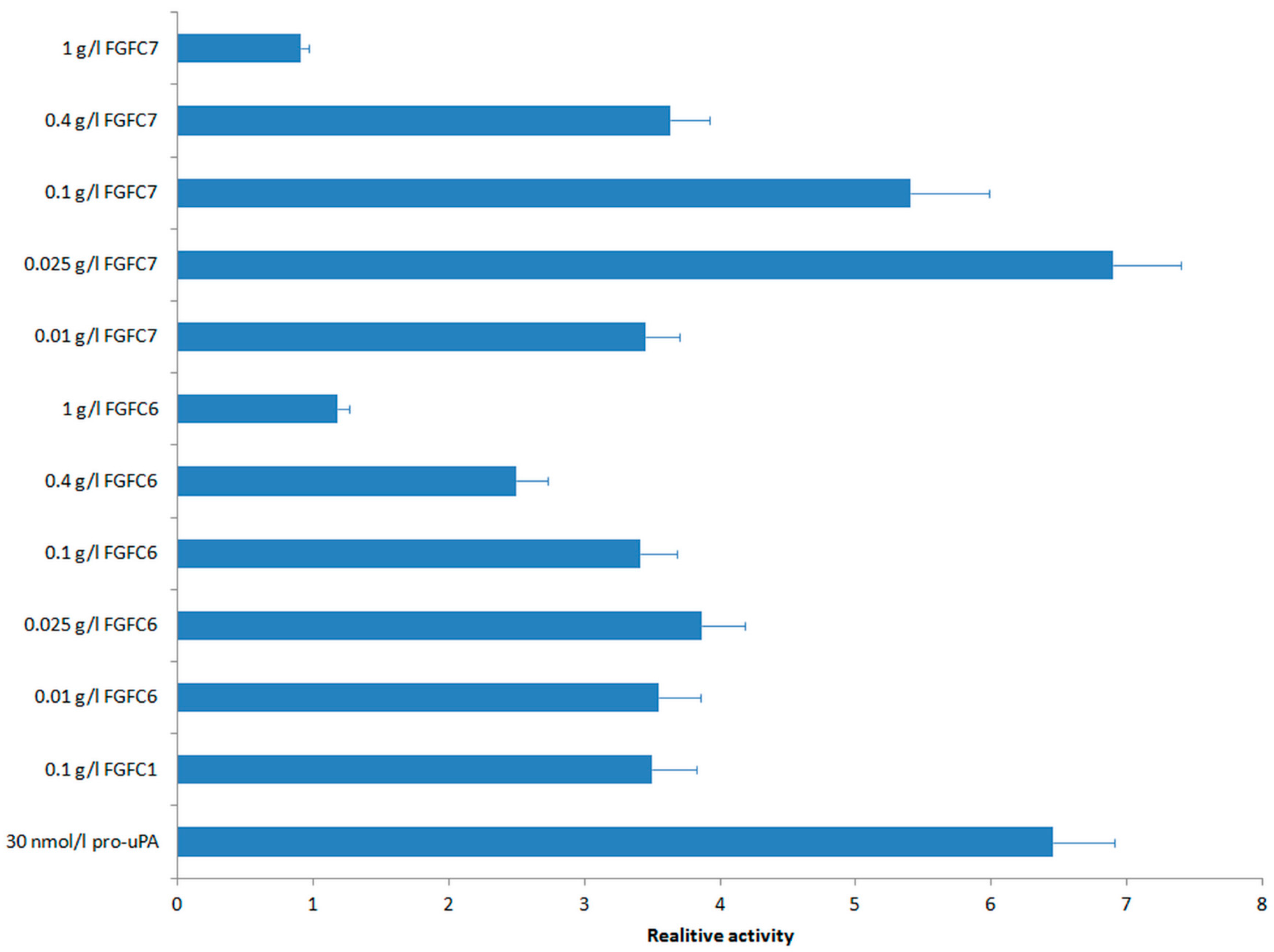
2.4. Fibrinolytic Activities of New FGFCs
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain, Medium, and Cultural Conditions
3.3. Amino Compouds Supply
3.4. Metabolites Detection and Isolation
3.5. Bioactivity Assays
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Leal, M.C.; Sheridan, C.; Osinga, R.; Dioníasio, G.; Rocha, R.J.M.; Silva, B.; Rosa, R.; Calado, R. Marine microorganism-invertebrate assemblages: Perspectives to solve the “supply problem” in the initial steps of drug discovery. Mar. Drugs 2014, 12, 3929–3952. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [PubMed]
- Rijken, D.C.; Lijnen, H.R. New insights into the molecular mechanisms of the fibrinolytic system. J. Thromb. Haemost. 2009, 7, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Hasumi, K.; Yamamichi, S.; Harada, T. Small-molecule modulators of zymogen activation in the fibrinolytic and coagulation systems. FEBS J. 2010, 277, 3675–3687. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, C.; Hasumi, K.; Hatsumi, W.; Endo, A. Staplabin, a novel fungal triprenyl phenol which stimulates the binding of plasminogen to fibrin and U937 cells. J. Antibiot. 1996, 49, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-M.; Narasaki, R.; Ohyama, S.; Hasumi, K. Selective production of staplabin and SMTPs in cultures of Stachybotrys microspora fed with precursor amines. J. Antibiot. 2001, 54, 962–966. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Koide, H.; Hu, W.-M.; Nishimura, N.; Narasaki, R.; Kitano, Y.; Hasumi, K. Structure–activity relationships of 11 new congeners of the SMTP plasminogen modulator. J. Antibiot. 2010, 63, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Koide, H.; Hasegawa, K.; Nishimura, N.; Narasaki, R.; Hasumi, K. A new series of the SMTP plasminogen modulators with a phenylamine-based side chain. J. Antibiot. 2012, 65, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-M.; Narasaki, R.; Nishimura, N.; Hasumi, K. SMTP (Stachybotrys microspora triprenyl phenol) enhances clot clearance in a pulmonary embolism model in rats. Thromb. J. 2012, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Nishimura, N.; Suzuki, E.; Zhuang, J.; Hasegawa, K.; Takamatsu, H.; Honda, K.; Hasumi, K. SMTP-7, a novel small-molecule thrombolytic for ischemic stroke: A study in rodents and primates. J. Cereb. Blood Flow Metab. 2014, 34, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wu, W.-H.; Zhu, Q.-G.; Fu, S.-Q.; Wang, X.-Y.; Hong, S.-T.; Guo, R.-H.; Bao, B. Identification and fibrinolytic evaluation of an isoindolone derivative isolated from a rare marine fungus Stachybotrys longispora FG216. Chin. J. Chem. 2015, 33, 1089–1095. [Google Scholar] [CrossRef]
- Wang, M.-X.; He, H.; Na, K.; Cai, M.-H.; Zhou, X.-S.; Zhao, W.-J.; Zhang, Y.-X. Designing novel glucose/ornithine replenishment strategies by biosynthetic and bioprocess analysis to improve fibrinolytic FGFC1 production by the marine fungus Stachybotrys longispora. Process. Biochem. 2015, 50, 2012–2018. [Google Scholar] [CrossRef]
- Yan, T.; Wu, W.-H.; Su, T.-W.; Chen, J.-J.; Zhu, Q.-G.; Zhang, C.-Y.; Wang, X.-Y.; Bao, B. Effects of a novel marine natural product: Pyrano indolone alkaloid fibrinolytic compound on thrombolysis and hemorrhagic activities in vitro and in vivo. Arch. Pharm. Res. 2015, 38, 1530–1540. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Suzuki, E.; Hasegawa, K.; Nishimura, N.; Kitano, Y.; Hasumi, K. Pre-SMTP, a key precursor for the biosynthesis of the SMTP plasminogen modulators. J. Antibiot. 2012, 65, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Matsuda, Y.; Gao, H.; Hu, D.; Yao, X.-S.; Abe, I. Biosynthesis of LL-Z1272β: Discovery of a new member of NRPS-like enzymes for aryl-aldehyde formation. ChemBioChem 2016, 17, 904–907. [Google Scholar] [CrossRef] [PubMed]
- Semeiks, J.; Borek, D.; Otwinowski, Z.; Grishin, N.V. Comparative genome sequencing reveals chemotype-specific gene clusters in the toxigenic black mold Stachybotrys. BMC Genom. 2014, 15, 590. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-M.; De Guzman, F.S.; Gloer, J.B.; Shearer, C.A. Stachybotrins A and B: Novel bioactive metabolites from a brackish water isolate of the fungus Stachybotrys sp. J. Org. Chem. 1992, 57, 6700–6703. [Google Scholar] [CrossRef]
- Nozawa, Y.; Ito, M.; Sugawara, K.; Hanada, K.; Mizoue, K. Stachybotrin C and parvisporin, novel neuritogenic compounds. II. Structure determination. J. Antibiot. 1997, 50, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, D.; Cen, S.; Proksch, P.; Lin, W.-H. Isoindolinone-type alkaloids from the sponge-derived fungus Stachybotrys chartarum. Tetrahedron 2014, 70, 7010–7015. [Google Scholar] [CrossRef]
- Jarvis, B.B.; Salemme, J.; Morals, A. Stachybotrys toxins. 1. Nat. Toxins 1995, 3, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Roggo, B.E.; Hug, P.; Moss, S.; Stämpfli, A.; Kriemler, H.P.; Peter, H.H. Novel spirodihydrobenzofuranlactams as antagonists of endothelin and as inhibitors of HIV-1 protease produced by Stachybotrys sp. II. Structure determination. J. Antibiot. 1996, 49, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-H.; Li, L.-T.; Zhu, T.-J.; Ba, M.-Y.; Li, G.-Q.; Gu, Q.-Q.; Guo, Y.; Li, D.-H. Phenylspirodrimanes with anti-HIV activity from the sponge-derived fungus Stachybotrys chartarum MXH-X73. J. Nat. Prod. 2013, 76, 2298–2306. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Oesker, V.; Wiese, J.; Malien, S.; Schmaljohann, R.; Imhoff, J.F. Spirocyclic drimanes from the marine fungus Stachybotrys sp. strain MF347. Mar. Drugs 2014, 12, 1924–1938. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, C.-M.; Liu, D.; Proksch, P.; Guo, P.; Lin, W.-H. Chartarlactams A-P, phenylspirodrimanes from the sponge-associated fungus Stachybotrys chartarum with antihyperlipidemic activities. J. Nat. Prod. 2014, 77, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.F.; Entwistle, R.; Corcoran, D.; Oakley, B.R.; Wang, C.C.C. Identification and molecular genetic analysis of the cichorine gene cluster in Aspergillus nidulans. Med. Chem. Comm. 2012, 3, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cai, M.-H.; Zhou, X.-S.; Li, Z.-Y.; Zhang, Y.-X. Polyketides in Aspergillus terreus: Biosynthesis pathway discovery and application. Appl. Microbiol. Biotechnol. 2016, 100, 7787–7798. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.-C.; Entwistle, R.; Guo, C.-J.; Ahuja, M.; Szewczyk, E.; Hung, J.-H.; Chiang, Y.-M.; Oakley, B.R.; Wang, C.C.C. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 2012, 134, 4709–4720. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.-J.; Knox, B.P.; Chiang, Y.-M.; Lo, H.-C.; Sanchez, J.F.; Lee, K.-H.; Oakley, B.R.; Bruno, K.S.; Wang, C.C.C. Molecular genetic characterization of a cluster in A. terreus for biosynthesis of the meroterpenoid terretonin. Org. Lett. 2012, 14, 5684–5687. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-S.; Chiang, Y.-M.; Wang, C.C.C. Biosynthetic pathway of the reduced polyketide product citreoviridin in Aspergillus terreus var. aureus revealed by heterologous expression in Aspergillus nidulans. Org. Lett. 2016, 18, 1366–1369. [Google Scholar] [CrossRef] [PubMed]
- Koide, H.; Narasaki, R.; Hasegawa, K.; Nishimura, N.; Hasumi, K. A new series of the SMTP plasminogen modulator with a phenylglycine-based side chain. J. Antibiot. 2012, 65, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Suzuki, E.; Tsujihara, K.; Nishimura, Y.; Hasumi, K. Structure–activity relationships of the plasminogen modulator SMTP with respect to the inhibition of soluble epoxide hydrolase. J. Antibiot. 2015, 68, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-M.; Ohyama, S.; Hasumi, K. Activation of fibrinolysis by SMTP-7 and -8, novel staplabin analogs with a pseudosymmetric structure. J. Antibiot. 2000, 53, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Pannel, R.; Gurewich, V. Activation of plasminogen by single-chain urokinase or by tow-chain urokinase—A demonstration that single-chain urokinase has a low catalytic activity. Blood 1987, 69, 22–26. [Google Scholar]
- Petersen, L.C.; Lund, L.R.; Nielsen, L.S.; Dano, K.; Skriver, L. One-chain urokinase-type plasminogen activator from human sarcoma cell is a proenzyme with little or no intrinsic activity. J. Biol. Chem. 1988, 263, 11189–11195. [Google Scholar] [PubMed]
- Nozawa, Y.; Yamamoto, K.; Ito, M.; Sakai, N.; Mizoue, K.; Mizobe, F.; Hanada, K. Stachybotrin C and parvisporin, novel neuritogenic compounds. I. Taxonomy, isolation, physico-chemical and biological properties. J. Antibiot. 1997, 50, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Roggo, B.E.; Petersen, F.; Sills, M.; Roesel, J.L.; Moerker, T.; Peter, H.H. Novel spirodihydrobenzofuranlactams as antagonists of endothelin and as inhibitors of HIV-1 protease produced by Stachybotrys sp. I. Fermentation, isolation and biological activity. J. Antibiot. 1996, 49, 13–19. [Google Scholar] [CrossRef] [PubMed]

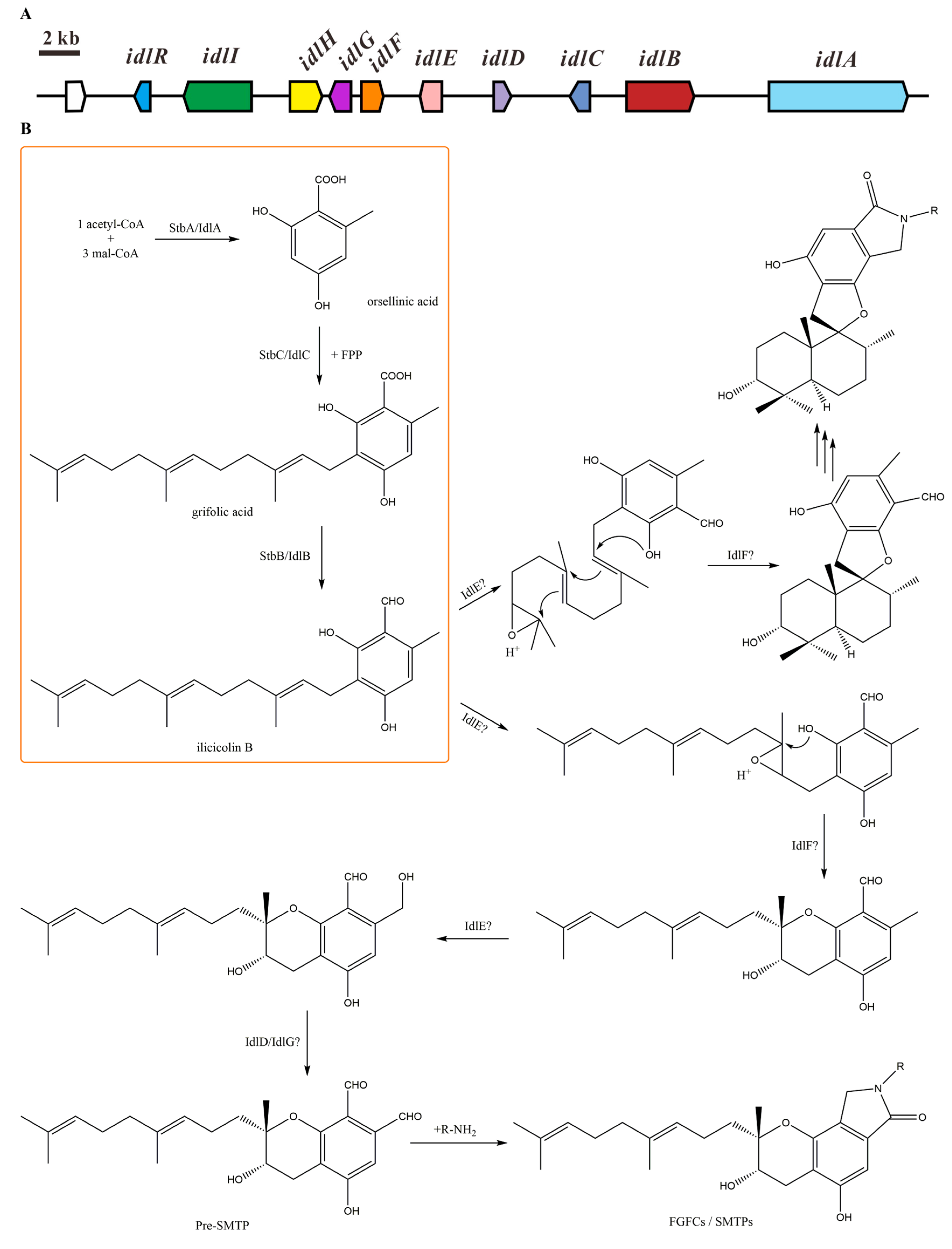
| Gene | Locus Tag | S. chartarum IBT 7711 Homologue | Protein Identity (%) | S. bisbyi PYH05-7 Homologue | Protein Identity (%) | Putative Function |
|---|---|---|---|---|---|---|
| idlR | S40285_07604 | S7711_05923 | 98 | Transcriptional regulator | ||
| idlI | S40285_07605 | S7711_05924 | 78 (34% coverage) | Nitrate reductase (partial in S. chartarum IBT 7711) | ||
| S7711_05925 | Carboxylesterase | |||||
| idlH | S40285_07606 | S7711_05926 | 97 | Esterase | ||
| idlG | S40285_07607 | S7711_05927 | 100 | Short-chain dehydrogenase | ||
| idlF | S40285_07608 | S7711_05928 | 94 | Isomerase/epimerase | ||
| idlE | S40285_07609 | S7711_05929 | 97 | Copper dependent oxidase | ||
| idlD | S40285_07610 | S7711_05930 | 96 | Short-chain dehydrogenase | ||
| idlC | S40285_10521 | S7711_10996 | 98 | stbC | 76 | PT |
| idlB | S40285_07611 | S7711_05931 | 98 | stbB | 73 | NRPS-like |
| idlA | S40285_07612 | S7711_05932 | 98 | stbA | 75 | NR-PKS |
| FGFC4 | FGFC5 | FGFC6 | FGFC7 | |||||
|---|---|---|---|---|---|---|---|---|
| No. | δC | δH | δC | δH | δC | δH | δC | δH |
| 2 | 171.6, C | 171.5, C | 171.7, C | 172.6, C | ||||
| 3 | 132.4, C | 132.4, C | 132.2, C | 131.7, C | ||||
| 4 | 100.9, CH | 6.77 s | 101.0, CH | 6.77 s | 101.1, CH | 6.78 s | 101.0, CH | 6.80 s |
| 5 | 158.0, C | 158.2, C | 158.0, C | 157.8, C | ||||
| 6 | 113.5, C | 113.6, C | 113.6, C | 113.6, C | ||||
| 7 | 27.7, CH2 | 3.00 dd (17.6, 5.4) | 27.8, CH2 | 3.01 dd (17.7, 5.5) | 27.8, CH2 | 3.00 dd (17.7, 5.4) | 27.8, CH2 | 3.01 dd (17.7, 5.3) |
| 2.67 dd (17.6, 7.1) | 2.66 dd (17.7, 7.4) | 2.68 dd (17.7, 7.0) | 2.70 dd (17.7, 6.8) | |||||
| 8 | 68.3, CH | 3.90 dd (7.0, 5.6) | 68.5, CH | 3.89 dd (7.2, 5.7) | 68.4, CH | 3.90 t (6.2) | 68.4, CH | 3.91 t (6.0) |
| 9 | 80.2, C | 80.2, C | 80.3, C | 80.2, C | ||||
| 11 | 150.0, C | 150.0, C | 150.0, C | 150.0, C | ||||
| 12 | 121.9, C | 121.9, C | 122.2, C | 123.1, C | ||||
| 13 | 46.8, CH2 | 4.36 d (16.7) | 46.9, CH2 | 4.34 d (16.7) | 49.6, CH2 | 4.39 s | 48.6, CH2 | 4.65 m |
| 4.24 d (16.7) | 4.25 d (16.7) | |||||||
| 14 | 38.6, CH2 | 1.69 m | 38.6, CH2 | 1.71 m | 38.6, CH2 | 1.69 m | 38.5, CH2 | 1.70 m |
| 15 | 22.6, CH2 | 2.20 m | 22.6, CH2 | 2.21 m | 22.6, CH2 | 2.20 m | 22.6, CH2 | 2.19 m |
| 16 | 125.5, CH | 5.16 t (6.8) | 125.6, CH | 5.17 t (6.7) | 125.5, CH | 5.16 t (6.9) | 125.4, CH | 5.16 t (6.9) |
| 17 | 136.3, C | 136.2, C | 136.3, C | 136.3, C | ||||
| 18 | 40.8, CH2 | 1.99 m | 40.9, CH2 | 1.99 m | 40.8, CH2 | 1.98 m | 40.8, CH2 | 1.97 m |
| 19 | 27.7, CH2 | 2.07 m | 27.7, CH2 | 2.07 m | 27.7, CH2 | 2.07 m | 27.7, CH2 | 2.07 m |
| 20 | 125.4, CH | 5.10 t (7.0) | 125.4, CH | 5.09 t (6.9) | 125.4, CH | 5.08 t (6.9) | 125.4, CH | 5.07 t (6.9) |
| 21 | 132.2, C | 132.2, C | 132.2, C | 132.2, C | ||||
| 22 | 25.9, CH3 | 1.67 s | 25.9, CH3 | 1.67 s | 25.9, CH3 | 1.66 s | 25.9, CH3 | 1.65 s |
| 23 | 17.8, CH3 | 1.59 s | 17.8, CH3 | 1.59 s | 17.8, CH3 | 1.58 s | 17.8, CH3 | 1.57 s |
| 24 | 16.0, CH3 | 1.60 s | 16.0, CH3 | 1.61 s | 16.0, CH3 | 1.60 s | 16.0, CH3 | 1.58 s |
| 25 | 18.8, CH3 | 1.29 s | 18.6, CH3 | 1.28 s | 18.8, CH3 | 1.30 s | 19.0, CH3 | 1.31 s |
| 1’ | 171.7, C | 171.8, C | 171.7, C | 173.0, C | ||||
| 2’ | 53.6, CH | 4.80 dd (10.6, 7.1) | 53.6, CH | 4.78 dd (10.6, 6.9) | 45.0, CH2 | 4.34 s | 61.4, CH | 4.89 d (3.8) |
| 3’ | 27.3, CH2 | 2.13 m | 27.3, CH2 | 2.14 m | 68.2, CH | 4.62 m | ||
| 4’ | 22.9, CH2 | 2.03 m | 22.9, CH2 | 2.03 m | 20.7, CH3 | 1.20 d (6.3) | ||
| 5’ | 43.0, CH2 | 3.37 brs | 43.0, CH2 | 3.37 brs | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Y.; Fu, Q.; Wu, W.; Cai, M.; Zhou, X.; Zhang, Y. Producing Novel Fibrinolytic Isoindolinone Derivatives in Marine Fungus Stachybotrys longispora FG216 by the Rational Supply of Amino Compounds According to Its Biosynthesis Pathway. Mar. Drugs 2017, 15, 214. https://doi.org/10.3390/md15070214
Yin Y, Fu Q, Wu W, Cai M, Zhou X, Zhang Y. Producing Novel Fibrinolytic Isoindolinone Derivatives in Marine Fungus Stachybotrys longispora FG216 by the Rational Supply of Amino Compounds According to Its Biosynthesis Pathway. Marine Drugs. 2017; 15(7):214. https://doi.org/10.3390/md15070214
Chicago/Turabian StyleYin, Ying, Qiang Fu, Wenhui Wu, Menghao Cai, Xiangshan Zhou, and Yuanxing Zhang. 2017. "Producing Novel Fibrinolytic Isoindolinone Derivatives in Marine Fungus Stachybotrys longispora FG216 by the Rational Supply of Amino Compounds According to Its Biosynthesis Pathway" Marine Drugs 15, no. 7: 214. https://doi.org/10.3390/md15070214
APA StyleYin, Y., Fu, Q., Wu, W., Cai, M., Zhou, X., & Zhang, Y. (2017). Producing Novel Fibrinolytic Isoindolinone Derivatives in Marine Fungus Stachybotrys longispora FG216 by the Rational Supply of Amino Compounds According to Its Biosynthesis Pathway. Marine Drugs, 15(7), 214. https://doi.org/10.3390/md15070214