Comprehensive Expression Profiling and Functional Network Analysis of Porphyra-334, One Mycosporine-Like Amino Acid (MAA), in Human Keratinocyte Exposed with UV-radiation
Abstract
:1. Introduction
2. Results and Discussion
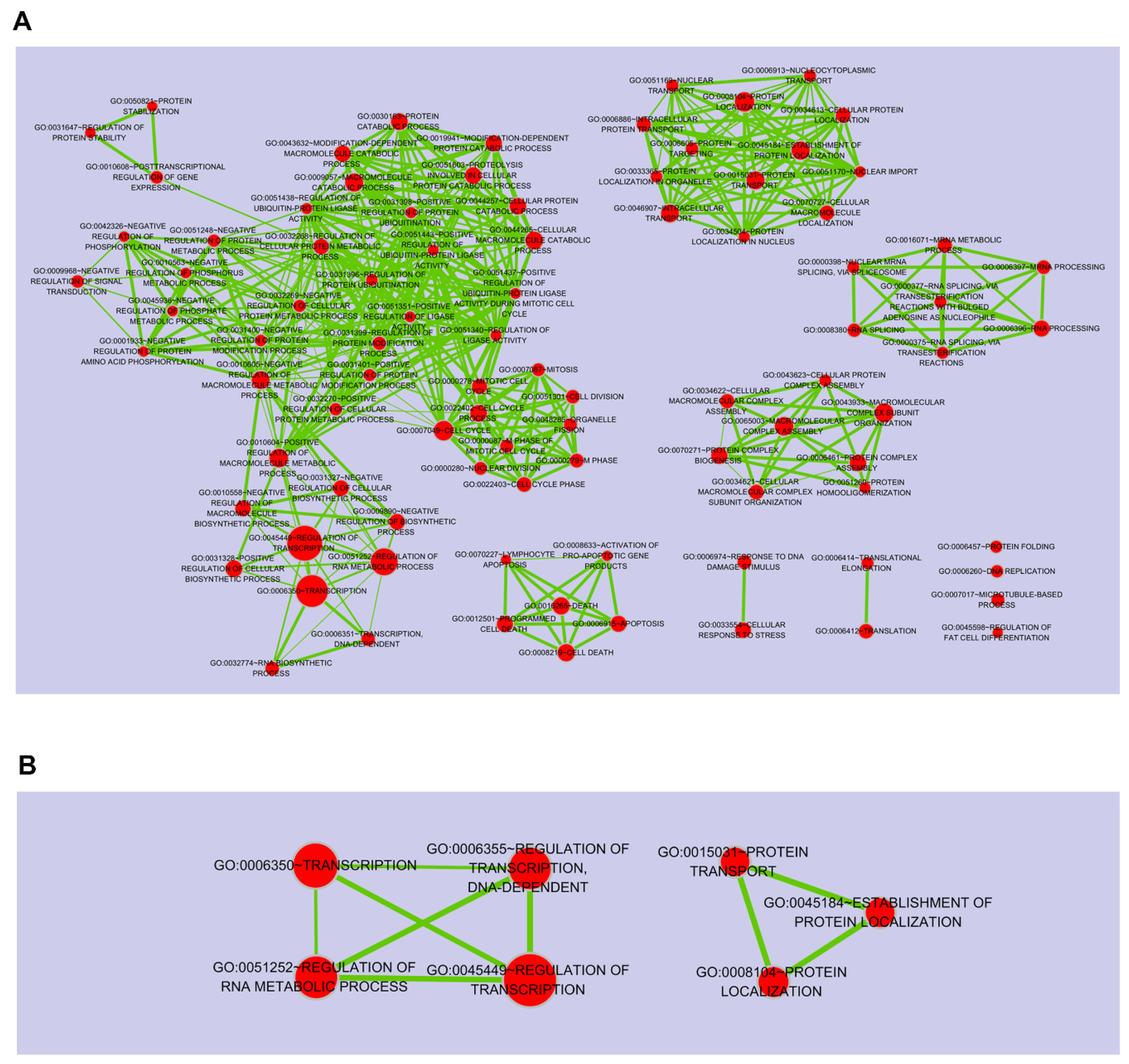
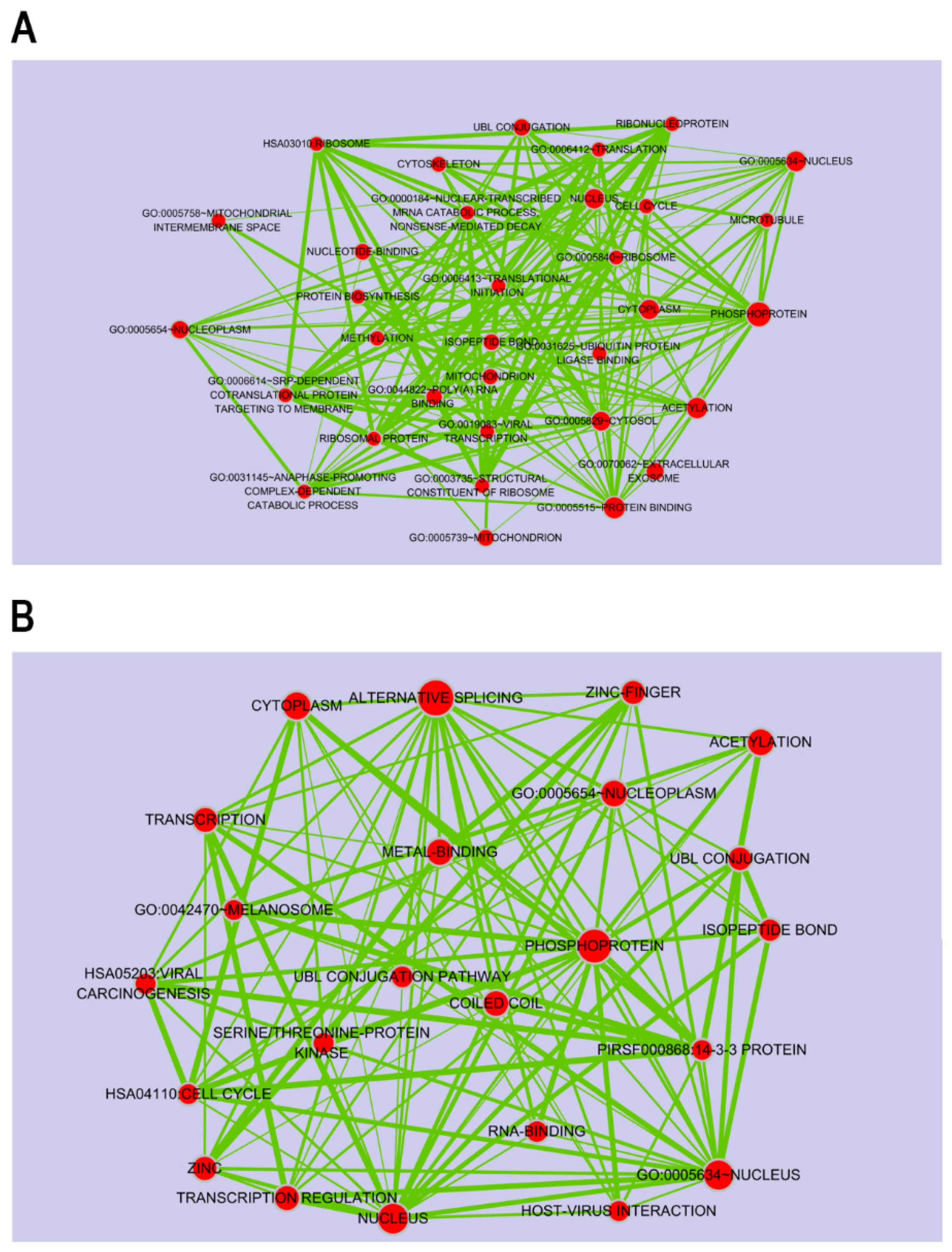
2.1. Porphyra-334-Modulated Differentially Expressed Genes (DEGs)
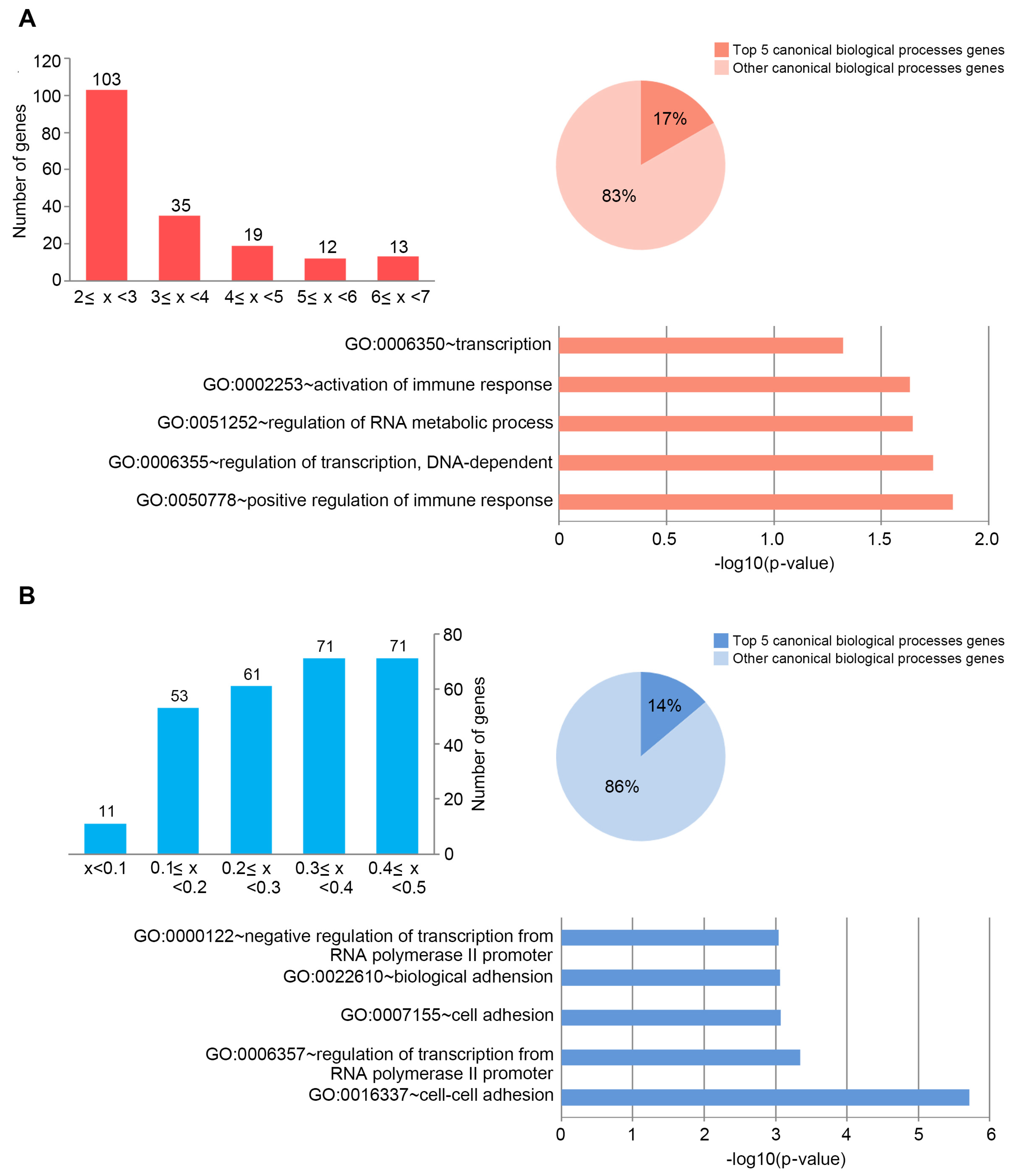
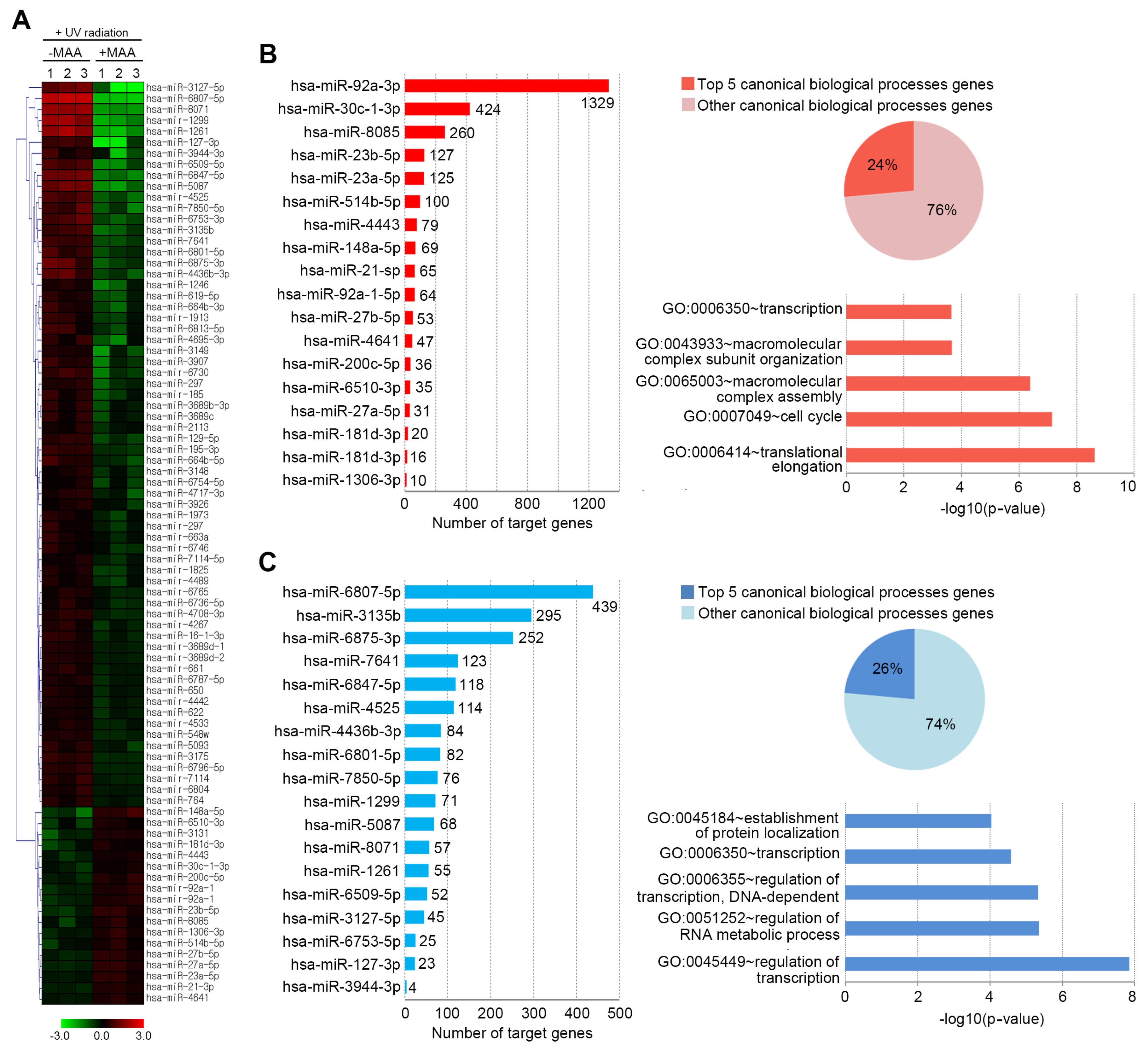
2.2. Porphyra-334-Modulated miRNAs
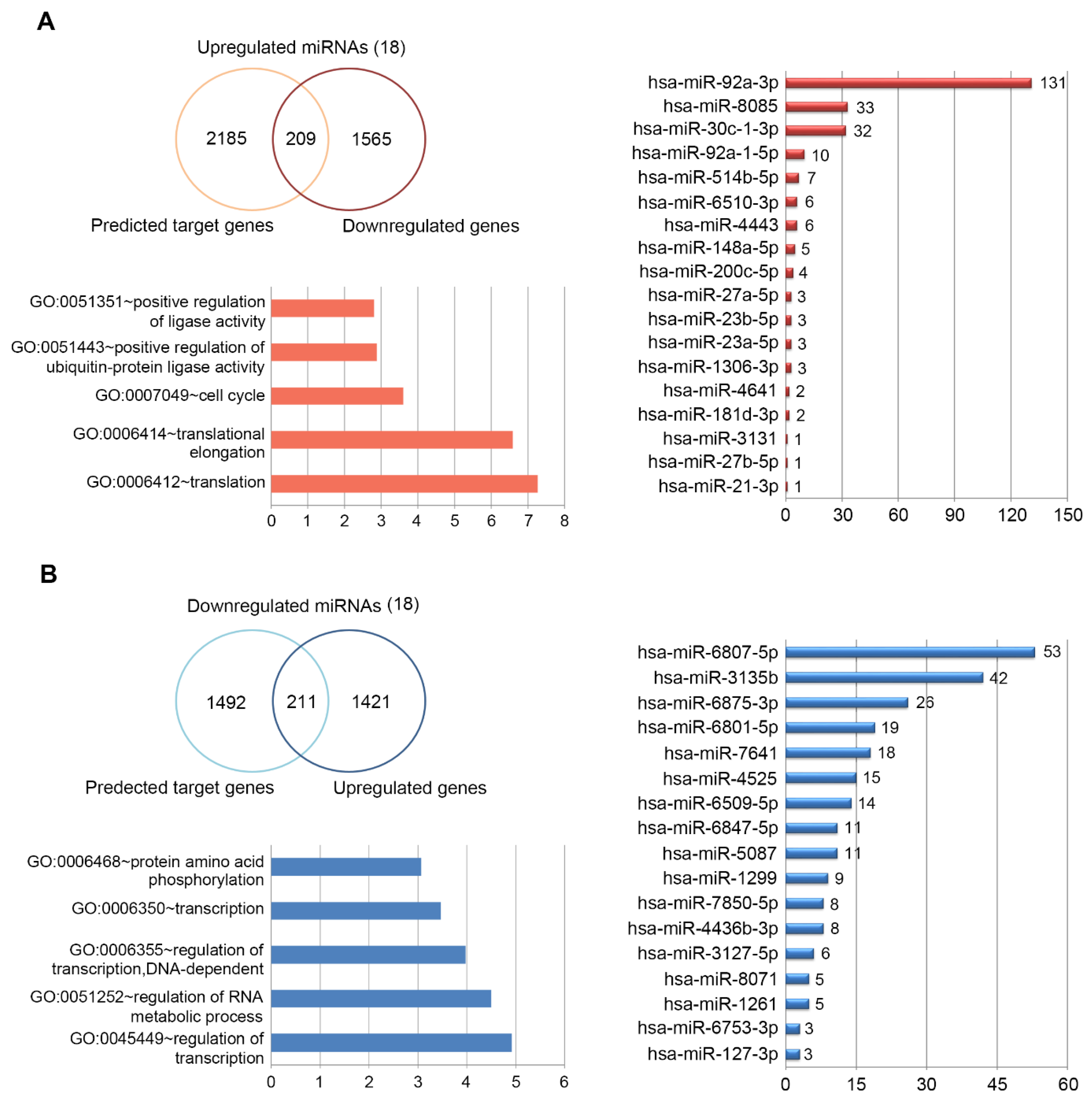
2.3. Target DEGs were Inversely Correlated with Functionally Enriched miRNAs
3. Experimental Section
3.1. Cell Cultures and Irradiation
3.2. RNA extraction
3.3. MicroRNA Microarray Analysis
3.4. Data Acquisition and Analysis
3.5. Library Oreparation and Sequencing
3.6. Data Analysis
3.7. qRT-PCR Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yagura, T.; Makita, K.; Yamamoto, H.; Menck, C.F.M.; Schuch, A.P. Biological sensors for solar ultraviolet radiation. Sensors 2011, 11, 4277–4294. [Google Scholar] [CrossRef] [PubMed]
- Schuch, A.P.; Garcia, C.C.; Makita, K.; Menck, C.F. DNA damage as a biological sensor for environmental sunlight. Photochem. Photobiol. Sci. 2013, 12, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Weatherhead, E.C.; Andersen, S.B. The search for signs of recovery of the ozone layer. Nature 2006, 441, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, R.; Paris, D.; Melck, D.; Fulgentini, L.; Colombetti, G.; Motta, A. In vivo NMR metabolic profiling of Fabrea salina reveals sequential defense mechanisms against ultraviolet radiation. Biophys. J. 2011, 100, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Garg, A.; Sharma, K.; Kumar, S.; Sharma, A.; Puohit, A.P. Mycosporine and mycosporine-like amino acids: A paramount tool against ultra violet irradiation. Pharmacog. Rev. 2011, 5, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Nazifi, E.E.; Wada, N.; Asano, T.; Nishiuchi, T.; Iwamuro, Y.; Chinaka, S.; Matsugo, S.; Sakamoto, T. Characterization of the chemical diversity of glycosylated mycosporine-like amino acids in the terrestrial cyanobacterium Nostoc commune. J. Photochem. Photobiol. B. 2015, 148, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Sakamoto, T.; Matsugo, S. Mycosporine-like amino acids and their derivatives as natural antioxidants. Antioxidants 2015, 4, 603–646. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, C.A.; Airs, R.L. Distribution and abundance of MAAs in 33 species of microalgae across 13 classes. Mar. Drugs 2010, 8, 1273–1291. [Google Scholar] [CrossRef] [PubMed]
- Athukorala, Y.; Trang, S.; Kwok, C.; Yuan, Y.V. Antiproliferative and antioxidant activities and mycosporine-like amino acid profiles of wild harvested and cultivated edible Canadian marine red macroalgae. Molecules 2016, 21, E119. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Enk, C.D.; Hochberg, M.; Srebnik, M. Porphyra-334, a potential natural source for UVA protective sunscreens. Photochm. Photobiol. Sci. 2006, 4, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.S.; Hwang, J.; Park, M.; Seo, H.H.; Kim, H.S.; Lee, J.H.; Moh, S.H.; Lee, T.K. Anti-inflammation activities of mycosporine-like amino acids (MAAs) in response to UV radiation suggest potential anti-skin aging activity. Mar. Drugs 2014, 12, 5174–5187. [Google Scholar] [CrossRef] [PubMed]
- Nizifi, E.; Wada, N.; Yamaba, M.; Asano, T.; Nishiuchi, T.; Matsugo, S.; Sakamoto, T. Glycosylated porphyra-334 and palythine-threonine from the terrestrial cyanobacterium Nostoc commune. Mar. Drugs 2013, 11, 3124–3154. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Gunde-Cimerman, N. Mycosporines and mycosporine-like amino acids: UV protectants or multipurpose secondary metabolites? FEMS Microbiol. Lett. 2007, 269, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Steiglitz, B.M.; Kreider, J.M.; Frankenburg, E.P.; Pappano, W.N.; Hoffman, G.G.; Meganck, J.A.; Liang, X.; Höök, M.; Goldstein, S.A.; Greenspan, D.S. Procollagen C proteinase enhancer 1 genes are important determinants of the mechanical properties and geometry of bone and the ultrastructure of connective tissues. Mol. Cell Biol. 2006, 26, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Daniel, S.; Cornelia, S.; Fred, Z. UV-A sunscreen from red algae for protection against premature skin aging. Cosmet. Toilet. Manufact. Worldw. 2004, 139–143. [Google Scholar]
- Lee, C.H.; Wu, S.B.; Hong, C.H.; Yu, H.S.; Wei, Y.H. Molecular mechanisms of UV-induced apoptosis and its effects on skin residential cells: the implication in UV-based phothotherapy. Int. J. Mol. Sci. 2013, 14, 6414–6435. [Google Scholar] [CrossRef] [PubMed]
- Tosato, M.G.; Orallo, D.E.; Ali, S.M.; Churio, M.S.; Martin, A.A.; Dicellio, L. Confocal Raman spectroscopy: In vivo biochemical changes in the human skin by topical formulations under UV radiation. J. Photochem. Photobiol. B. 2015, 153, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Park, S.J.; Kim, I.H.; Choi, Y.H.; Nam, T.J. Protective effect of porphyra-334 on UVA-induced photoaging in human skin fibroblasts. Int. J. Mol. Med. 2014, 34, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Gostner, J.; Fuchs, J.M.; Chaita, E.; Aligiannis, N.; Skaltsounis, J.; Ganzera, M. Inhibition of collagenase by mycosporine-like amino acids from marine source. Plants Med. 2015, 81, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Hartmann, A.; Ganzera, M.; Fuchs, D.; Gostner, J.M. Immunomodulatory effects of the mycosporine-like amino acids shinorine and porphyra-334. Mar. Drugs 2016, 14, 119. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Yang, D.J.; Kulkarni, A.; Moh, S.H.; Kim, K.W. Mycosporine-like amino acids promote wound healing through focal adhesion kinase (FAK) and mitogen-activated protein kinase (MAP kinases) signaling pathway in keratinocytes. Mar. Drugs 2015, 13, 7055–7066. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.S.; Oh, S.K.; Lee, S.G.; Kim, I.C.; Kim, S. Porphyra-334, a mycosporine-like amino acid, attenuates UV-induced apoptosis in HaCaT cells. Acta Pharm. 2017, 67, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Roy, S.S. Wnt/β-catenin pathway is regulated by PITX2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, SKOV-3. J. Biol. Chem. 2013, 288, 4355–4367. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Reichert, E.C.; Nakano, T.; Lohse, M.; Gardner, A.A.; Revelo, M.P.; Topham, M.K.; Stafforini, D.M. Deficiency of phospholipase A2 group 7 decreases intestinal polyposis and colon tumorigenesis in Apc(Min/+) mice. Cancer Res. 2013, 73, 2806–2816. [Google Scholar] [CrossRef] [PubMed]
- Lamont, L.B.; Crittenden, S.L.; Bernstein, D.; Wickens, M.; Kimble, J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 2004, 7, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.B.; Kim, J.Y.; Cho, S.D.; Park, K.S.; Jung, J.Y.; Lee, H.Y.; Hong, I.S.; Nam, J.S. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 2015, 5, 12465. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, N.N.; Shao, S.; Hoang, B.H.; Mercola, D.; Zi, X. Wnt signaling in castration-resistant prostate cancer: implications for therapy. Am. J. Clin. Exp. Urol. 2014, 2, 27–44. [Google Scholar] [PubMed]
- Tan, J.J.; Ong, S.A.; Chen, K.S. Rasd1 interacts with Ear2 (Nr2f6) to regulate renin tractription. BMC Mol. Biol. 2011, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Ichim, C.V.; Dervović, D.D.; Zúñiga-Pflücker, J.C.; Wells, R.A. The orphan nuclear receptor Ear-2 (Nr2f6) is a novel negative regulator of T cell development. Exp. Hematol. 2014, 42, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Hermann-kleiter, N.; Baier, G. Orphan nuclear receptor NR2F6 acts as an essential gatekeeper of Th17CD4+ T cell effector functions. Cell Commun. Signal. 2014, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Akai, J.; Halley, P.A.; Storey, K.Q. FGF-dependent Notch signaling maintains the spinal cord stem zone. Genes Dev. 2005, 19, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Yanez, D.A.; Touma, M.; Nakano, H.; Jaroszewicz, A.; Jordan, M.C.; Pellegrini, M.; Roos, K.P.; Nakano, A. Nkx-5 suppresses the proliferation of atrial myocytes and conduction system. Circ. Res. 2014, 114, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Park, G.; Lee, J.E.; Park, J.Y.; Kim, T.H.; Kim, Y.J.; Lee, S.H.; Yoo, H.; Kim, J.H.; Park, J.B. CPEB1 modulates differentiation of glioma stem cells via downregulation of HES1 and SIRT1 expression. Oncotarget 2014, 5, 6756–6769. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.; Osipo, C.; Foreman, K.; Golde, T.; Osborne, B.; Miele, L. Rational targeting of Notch signaling in cancer. Oncogene 2008, 27, 5124–5131. [Google Scholar] [CrossRef] [PubMed]
- Pancewicz, J.; Taylor, J.M.; Datta, A.; Baydoun, H.H.; Waldmann, T.A.; Hermine, O.; Nicot, C. Notch signaling contributes to proliferation and tumor formation of human T-cell leukemia virus type 1-associated adult T-cell leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 16619–16624. [Google Scholar] [CrossRef] [PubMed]
- Fre, S.; Pallavi, S.K.; Huyghe, M.; Laé, M.; Janssen, K.P.; Robine, S.; Artavanis-Taskonas, S.; Louvard, D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc. Natl. Acad. Sci. USA 2009, 106, 6309–6314. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. NUMB is a break of WNT-Notch signaling cycle. Int. J. Mol. Med. 2006, 18, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kato, A.; Angelocci, C.; Watanabe, M.; Kato, Y. A potential molecular pathogenesis of cardiac/laterality defects in Oculo-Facio-Cardio-Dental syndrome. Dev. Biol. 2014, 387, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Nickoloff, B.J.; Osborne, B.A.; Miele, L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene 2003, 22, 6598–6608. [Google Scholar] [CrossRef] [PubMed]
- Canguilhem, B.; Pradines, A.; Baudouin, C.; Boby, C.; Lajoie-Mazenc, I.; Charveron, M.; Favre, G. RhoB protects human keratinocytes from UVB-induced apoptosis through epidermal growth factor receptor signaling. J. Biol. Chem. 2005, 280, 43257–43263. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.S.; Yoo, J.Y.; Nuovo, G.J.; Jeon, Y.J.; Kim, S.; Lee, T.J.; Kim, T.; Bakàcs, A.; Alder, H.; Kaur, B.; et al. MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc. Natl. Acad. Sci. USA 2012, 109, 5316–5321. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell. Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.J.; Kim, O.Y.; Lee, G.T.; Lee, K.S.; Park, I.C.; Lee, S.J.; Kim, Y.R.; Ahn, K.J.; An, I.S.; An, S.; et al. Identification of ultraviolet B radiation-induced microRNAs in normal human dermal papilla cells. Mol. Med. Rep. 2014, 10, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Syed, D.N.; Khan, M.I.; Shabbir, M.; Mukhtar, H. MicroRNAs in skin response to UV radiation. Curr. Drug Targets 2013, 14, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, W.; Ding, R.; Xiong, L.; Dou, R.; Zhang, Y.; Guo, Z. Comprehensisve expression profiling and functional network analysis of p53-regulated micrRNAs in HepG2 cells treated with doxorubicin. PLoS ONE 2016, 11, e0149227. [Google Scholar]
- Sinha, A.U.; Kaimal, V.; Chen, J.; Jegga, A.G. Dissecting microreulation of a master regulatory network. BMC Genom. 2008, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Meek, D.W. The p53 response to DNA damage. DNA Repair 2004, 3, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Verschoor, M.L.; Verschoor, C.P.; Singh, G. Est-1 global gene expression profile reveals associations with metabolism and oxidative stress in ovarian and breast cancers. Cancer Metab. 2013, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Powley, I.R.; Kondrashov, A.; Young, L.A.; Dobbyn, H.C.; Hill, K.; Cannell, I.G.; Stoneley, M.; Kong, Y.W.; Cotes, J.A.; Smith, G.C.; et al. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 2009, 23, 1207–1220. [Google Scholar] [CrossRef] [PubMed]
- Enujiugha, V.N.; Talabi, J.Y.; Malomo, S.A.; Olagunju, A.I. DPPH radical scavenging capacity of phenolic extracts from African yam bean (Sphenostylis stenocarpa). Food Nutr. Sci. 2012, 3, 7–13. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suh, S.-S.; Lee, S.G.; Youn, U.J.; Han, S.J.; Kim, I.-C.; Kim, S. Comprehensive Expression Profiling and Functional Network Analysis of Porphyra-334, One Mycosporine-Like Amino Acid (MAA), in Human Keratinocyte Exposed with UV-radiation. Mar. Drugs 2017, 15, 196. https://doi.org/10.3390/md15070196
Suh S-S, Lee SG, Youn UJ, Han SJ, Kim I-C, Kim S. Comprehensive Expression Profiling and Functional Network Analysis of Porphyra-334, One Mycosporine-Like Amino Acid (MAA), in Human Keratinocyte Exposed with UV-radiation. Marine Drugs. 2017; 15(7):196. https://doi.org/10.3390/md15070196
Chicago/Turabian StyleSuh, Sung-Suk, Sung Gu Lee, Ui Joung Youn, Se Jong Han, Il-Chan Kim, and Sanghee Kim. 2017. "Comprehensive Expression Profiling and Functional Network Analysis of Porphyra-334, One Mycosporine-Like Amino Acid (MAA), in Human Keratinocyte Exposed with UV-radiation" Marine Drugs 15, no. 7: 196. https://doi.org/10.3390/md15070196
APA StyleSuh, S.-S., Lee, S. G., Youn, U. J., Han, S. J., Kim, I.-C., & Kim, S. (2017). Comprehensive Expression Profiling and Functional Network Analysis of Porphyra-334, One Mycosporine-Like Amino Acid (MAA), in Human Keratinocyte Exposed with UV-radiation. Marine Drugs, 15(7), 196. https://doi.org/10.3390/md15070196




