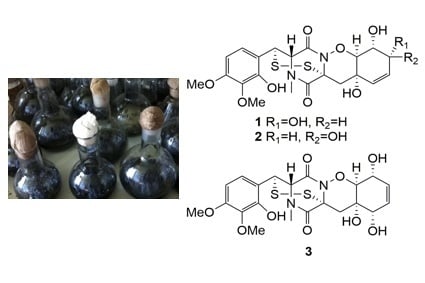
Pretrichodermamides D–F from a Marine Algicolous Fungus Penicillium sp. KMM 4672
Abstract
:1. Introduction
2. Results
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Strain
3.3. Cultivation of Fungus
3.4. Extraction and Isolation
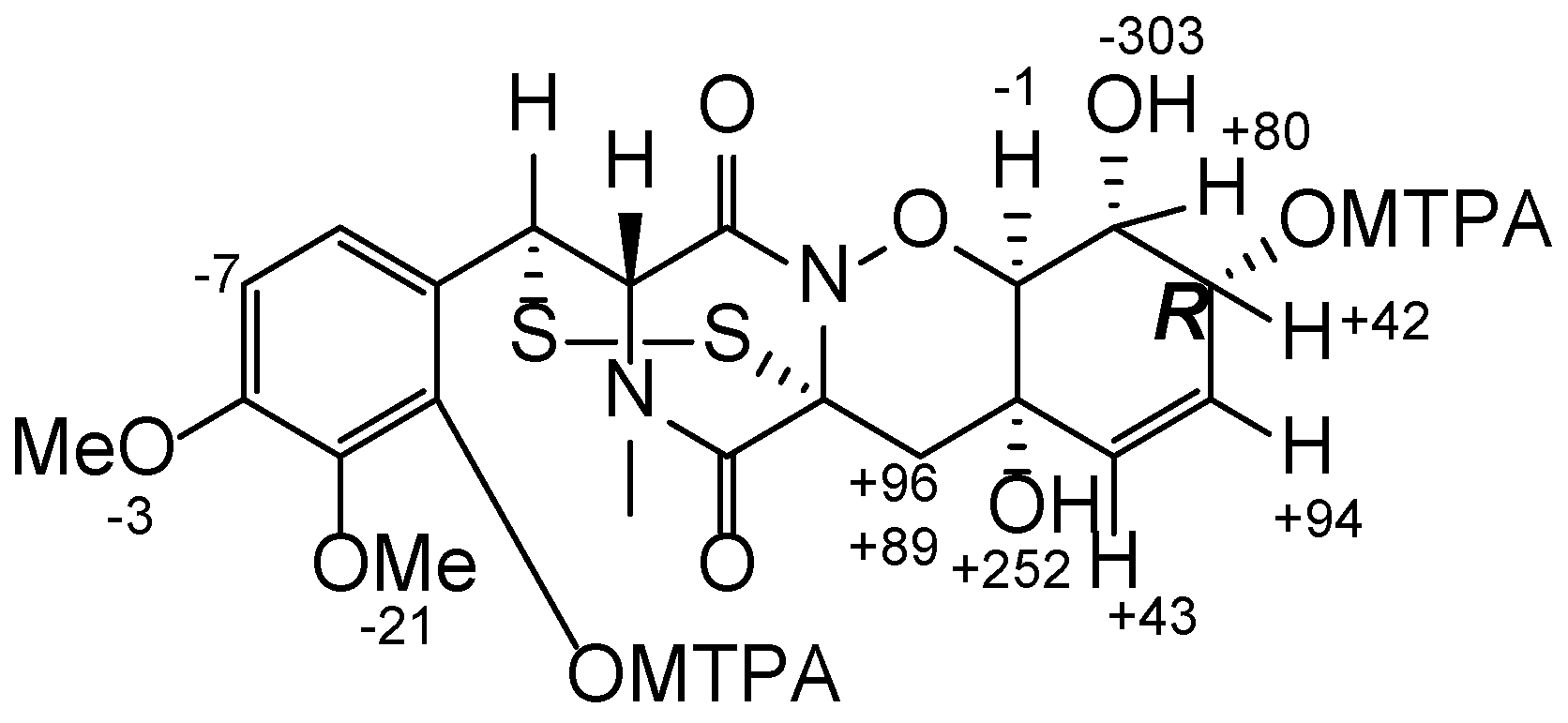
3.5. Preparation of (S)-MTPA and (R)-MTPA Esters of Pretrichodermamide D (1)
3.6. Cell Culture
3.7. Cytotoxicity Assay
3.8. Cell Cycle and Apoptosis Induction Analysis
3.9. Hemolytic Activity
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mfuh, A.M.; Zhang, Y.; Stephens, D.E.; Vo, A.X.T.; Arman, H.D.; Larionov, O.V. Concise total synthesis of trichodermamides A, B, and C enabled by an efficient construction of the 1,2-oxazadecaline core. J. Am. Chem. Soc. 2015, 137, 8050–8053. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Rotinsulu, H.; Narita, R.; Takahashi, R.; Namikoshi, M. Induced production of halogenated epidithiodiketopiperazines by a marine-derived Trichoderma cf. brevicompactum with sodium halides. J. Nat. Prod. 2015, 78, 2319–2321. [Google Scholar] [PubMed]
- Yamazaki, H.; Takahashi, O.; Murakami, K.; Namikoshi, M. Induced production of a new unprecedented epitrithiodiketopiperazine, chlorotrithiobrevamide, by a culture of the marine-derived Trichoderma cf. brevicompactum with dimethyl sulfoxide. Tetrahedron Lett. 2015, 56, 6262–6265. [Google Scholar] [CrossRef]
- Kajula, M.; Ward, J.M.; Turpeinen, A.; Tejesvi, M.V.; Hokkanen, J.; Tolonen, A.; Häkkänen, H.; Picart, P.; Ihalainen, J.; Sahl, H.G.; et al. Bridged epipolythiodiketopiperazines from Penicillium raciborskii, an endophytic fungus of Rhododendron tomentosum Harmaja. J. Nat. Prod. 2016, 79, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, C.; Yokose, K.; Furumai, T.; Maruyama, H.B. A new epidithiodiketopiperazine group antibiotic, FA-2097. J. Antibiot. 1982, 35, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Stipanovic, R.D.; Howell, C.R. The structure of gliovirin, a new antibiotic from Gliocladium virens. J. Antibiot. 1982, 35, 1326–1330. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Debbab, A.; Mándi, A.; Wray, V.; Schulz, B.; Müller, W.E.G.; Kassack, M.; Lin, W.; Kurtán, T.; Proksch, P.; et al. Alkaloids from the sponge-associated fungus Aspergillus sp. Eur. J. Org. Chem. 2013, 894, 894–906. [Google Scholar] [CrossRef]
- Orfali, R.S.; Aly, A.H.; Ebrahim, W.; Abdel-Aziz, M.S.; Müller, W.E.G.; Lin, W.; Daletos, G.; Proksch, P. Pretrichodermamide C and N-methylpretrichodermamide B, two new cytotoxic epidithiodiketopiperazines from hyper saline lake derived Penicillium sp. Phytochem. Lett. 2015, 11, 168–172. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.M.; Meng, L.H.; Jiang, W.L.; Xu, G.M.; Huang, C.G.; Wang, B.G. Bisthiodiketopiperazines and acorane sesquiterpenes produced by the marine-derived fungus Penicillium adametzioides AS-53 on different culture media. J. Nat. Prod. 2015, 78, 1294–1299. [Google Scholar] [CrossRef] [PubMed]
- Yokose, K.; Nakayama, N.; Miyamoto, C.; Furumai, T.; Maruyama, H.B.; Stipanovic, R.D.; Howell, C.R. Structure of Fa-2097, a new member of the dioxopiperazine antibiotics. J. Antibiot. 1984, 37, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, T.; Ooi, T.; Ohkubo, Y.; Yabuuchi, T. The modified Mosher’s method and the sulfoximine method. Bull. Chem. Soc. Jpn. 2006, 79, 965–980. [Google Scholar] [CrossRef]
- Liu, C.; Lou, W.; Zhu, Y.; Nadiminty, N.; Schwartz, C.T.; Evans, C.P.; Gao, A.C. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin. Cancer Res. 2014, 20, 3198–3210. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplificaion and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Hong, S.B.; Go, S.J.; Shin, H.D.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 2005, 97, 1316–1329. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, S.A.; Menchinskaya, E.S.; Venz, S.; Rast, S.; Amann, K.; Hauschild, J.; Otte, K.; Kalinin, V.I.; Silchenko, A.S.; Avilov, S.A.; et al. The marine triterpene glycoside frondoside A exhibits activity in vitro and in vivo in prostate cancer. Int. J. Cancer 2016, 138, 2450–2465. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, S.A.; Venz, S.; Shubina, L.K.; Fedorov, S.N.; Walther, R.; Jacobsen, C.; Stonik, V.A.; Bokemeyer, C.; Balabanov, S.; Honecker, F. Activity of aaptamine and two derivatives, demethyloxyaaptamine and isoaaptamine, in cisplatin-resistant germ cell cancer. J. Proteom. 2014, 96, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Freshney, R.I. Culture of Animal Cells: A Manual of Basic Technique; Wiley-Liss: New York, NY, USA, 1994; p. 486. [Google Scholar]
- Dyshlovoy, S.A.; Hauschild, J.; Amann, K.; Tabakmakher, K.M.; Venz, S.; Walther, R.; Guzii, A.G.; Makarieva, T.N.; Shubina, L.K.; Fedorov, S.N.; et al. Marine alkaloid monanchocidin a overcomes drug resistance by induction of autophagy and lysosomal membrane permeabilization. Oncotarget 2015, 6, 17328–17341. [Google Scholar] [CrossRef] [PubMed]
- Malagoli, D. A full-length protocol to test hemolytic activity of palytoxin on human erythrocytes. Invertebr. Surviv. J. 2007, 4, 92–94. [Google Scholar]
- Taniyama, S.; Arakawa, O.; Terada, M.; Nishio, S.; Takatani, T.; Mahmud, Y.; Noguchi, T. Ostreopsis sp., a possible origin of palytoxin (PTX) in parrotfish Scarus ovifrons. Toxicon 2003, 42, 29–33. [Google Scholar] [CrossRef]


| Position | 1 a | 2 b | 3 b | |||
|---|---|---|---|---|---|---|
| δС, mult | δH (J in Hz) | δС, mult | δH (J in Hz) | δС, mult | δH (J in Hz) | |
| 1 | 165.4, C | - | 165.5, C | - | 165.4, C | - |
| 2 | 68.0, C | - | 67.7, C | - | 69.1, C | - |
| 3 | 38.4, CH2 | α: 2.17, d (15.3) β: 2.33, d (15.3) | 39.0, CH2 | α: 2.22, d (15.7) β: 2.27, d (15.5) | 35.6, CH2 | α: 1.93, brd (15.5) β: 2.06, dd (15.4, 1.5) |
| 4 | 66.9, C | - | 67.0, C | - | 67.4, C | - |
| 5 | 133.8, CH | 5.56, d (10.1) | 131.8, CH | 5.54, dd (10.1, 2.2) | 69.2, CH | 3.69, d (5.5) |
| 6 | 127.4, CH | 5.60, dd (10.0, 4.4) | 129.7, CH | 5.43, dd (10.2, 2.2) | 126.9, CH | 5.69, ddd (10.0, 5.1, 2.3) |
| 7 | 65.8, CH | 4.03, q (4.5) | 72.2, CH | 3.96, tt (7.7, 2.2) | 131.2, CH | 5.56, dd (9.9, 2.5) |
| 8 | 66.2, CH | 3.74, ddd (9.4, 6.6, 4.6) | 71.0, CH | 3.56, ddd (10.7, 7.7, 5.7) | 64.6, CH | 4.16, m |
| 9 | 81.9, CH | 4.12, d (9.4) | 83.4, CH | 3.83, d (10.7) | 83.5, CH | 3.97, dd (7.1, 1.5) |
| 1′ | 164.2, C | - | 164.4, C | - | 163.8, C | - |
| 2′ | 66.0, CH | 4.56, d (2.6) | 65.7, CH | 4.57, d (2.6) | 65.4, CH | 4.57, d (2.6) |
| 3′ | 41.4, CH | 4.55, d (2.5) | 41.5, CH | 4.58, d (2.5) | 41.0, CH | 4.59, d (2.5) |
| 4′ | 116.3, C | - | 116.3, C | - | 116.3, C | - |
| 5′ | 122.6, CH | 7.32, d (8.8) | 122.6, CH | 7.32, d (8.8) | 122.7, CH | 7.35, d (8.8) |
| 6′ | 103.3, CH | 6.55, d (8.8) | 103.3, CH | 6.55, d (8.8) | 103.2, CH | 6.54, d (8.8) |
| 7′ | 153.0, C | - | 153.0, C | - | 152.8, C | - |
| 8′ | 135.9, C | - | 135.9, C | - | 135.8, C | - |
| 9′ | 147.6, C | - | 147.5, C | - | 147.5, C | - |
| 10′ | 32.6, CH3 | 2.96, s | 32.6, CH3 | 2.96, s | 32.5, CH3 | 2.96, s |
| 7′-OMe | 55.7, CH3 | 3.78, s | 55.6, CH3 | 3.78, s | 55.6, CH3 | 3.78, s |
| 8′-OMe | 60.2, CH3 | 3.68, s | 60.2, CH3 | 3.68, s | 60.2, CH3 | 3.68, s |
| 9′-OH | - | 9.43, s | - | 9.42, s | - | 9.38, s |
| 4-OH | - | 5.26, s | - | 5.29, s | - | 4.96, brs |
| 5-OH | - | - | - | - | - | 5.19, d (5.6) |
| 7-OH | - | 4.89, d (5.4) | - | 5.02, d (6.9) | - | |
| 8-OH | - | 4.35, d (6.6) | - | 4.64, d (5.7) | - | 5.15, d (6.7) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yurchenko, A.N.; Smetanina, O.F.; Ivanets, E.V.; Kalinovsky, A.I.; Khudyakova, Y.V.; Kirichuk, N.N.; Popov, R.S.; Bokemeyer, C.; Von Amsberg, G.; Chingizova, E.A.; et al. Pretrichodermamides D–F from a Marine Algicolous Fungus Penicillium sp. KMM 4672. Mar. Drugs 2016, 14, 122. https://doi.org/10.3390/md14070122
Yurchenko AN, Smetanina OF, Ivanets EV, Kalinovsky AI, Khudyakova YV, Kirichuk NN, Popov RS, Bokemeyer C, Von Amsberg G, Chingizova EA, et al. Pretrichodermamides D–F from a Marine Algicolous Fungus Penicillium sp. KMM 4672. Marine Drugs. 2016; 14(7):122. https://doi.org/10.3390/md14070122
Chicago/Turabian StyleYurchenko, Anton N., Olga F. Smetanina, Elena V. Ivanets, Anatoly I. Kalinovsky, Yuliya V. Khudyakova, Natalya N. Kirichuk, Roman S. Popov, Carsten Bokemeyer, Gunhild Von Amsberg, Ekaterina A. Chingizova, and et al. 2016. "Pretrichodermamides D–F from a Marine Algicolous Fungus Penicillium sp. KMM 4672" Marine Drugs 14, no. 7: 122. https://doi.org/10.3390/md14070122
APA StyleYurchenko, A. N., Smetanina, O. F., Ivanets, E. V., Kalinovsky, A. I., Khudyakova, Y. V., Kirichuk, N. N., Popov, R. S., Bokemeyer, C., Von Amsberg, G., Chingizova, E. A., Afiyatullov, S. S., & Dyshlovoy, S. A. (2016). Pretrichodermamides D–F from a Marine Algicolous Fungus Penicillium sp. KMM 4672. Marine Drugs, 14(7), 122. https://doi.org/10.3390/md14070122