Hormaomycins B and C: New Antibiotic Cyclic Depsipeptides from a Marine Mudflat-Derived Streptomyces sp.
Abstract
:1. Introduction
2. Results and Discussion
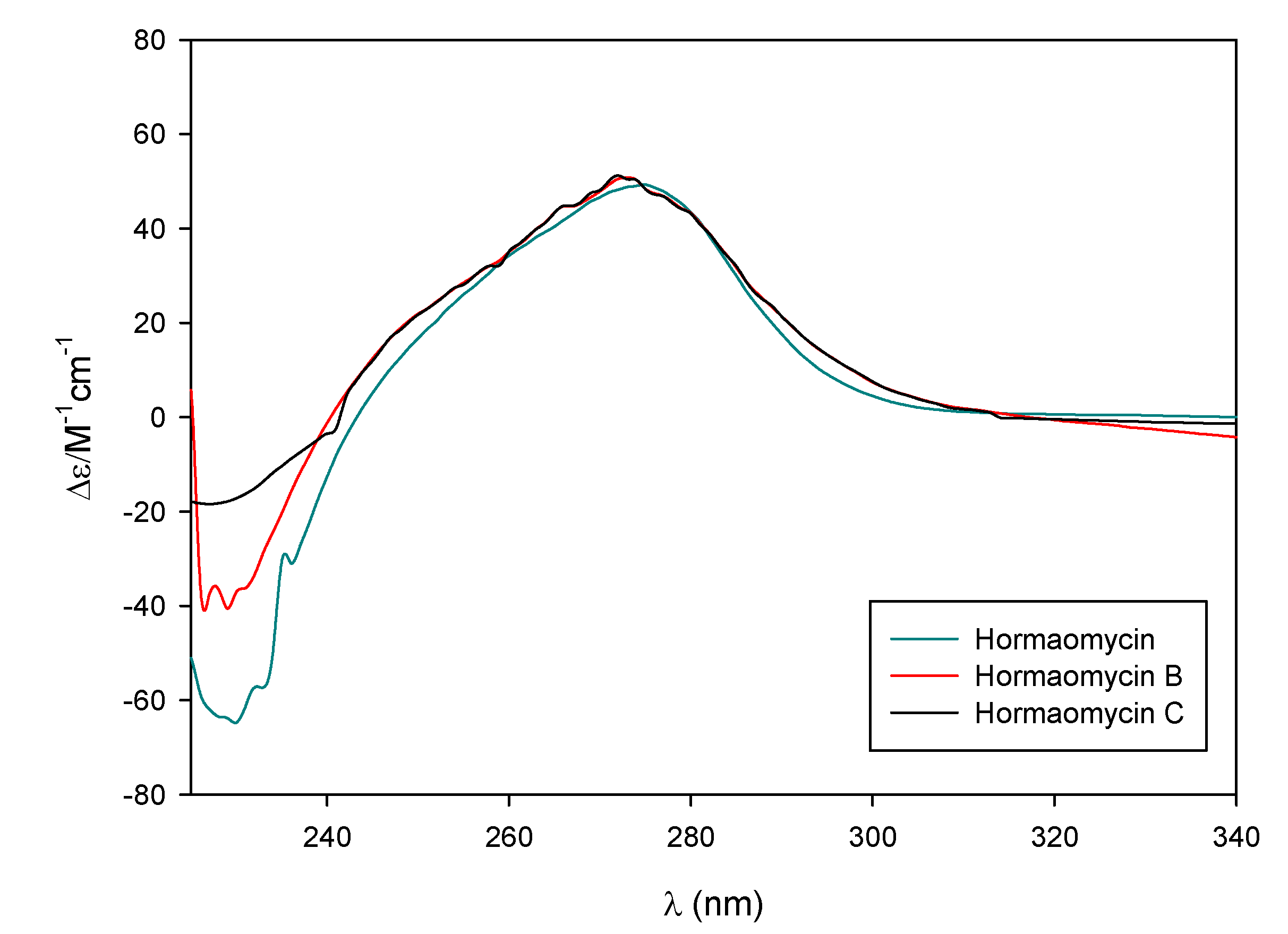
2.1. Structural Elucidation
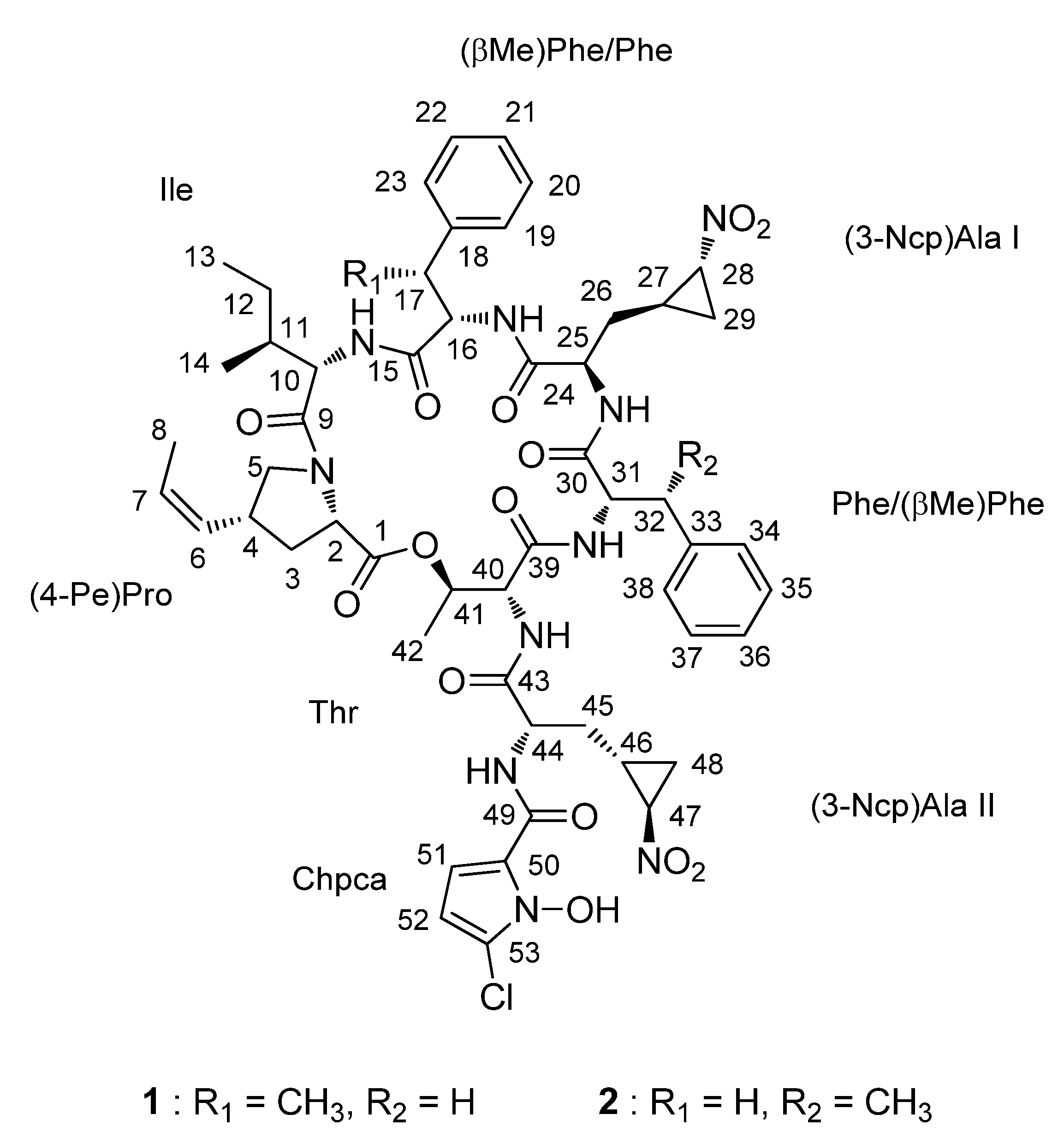
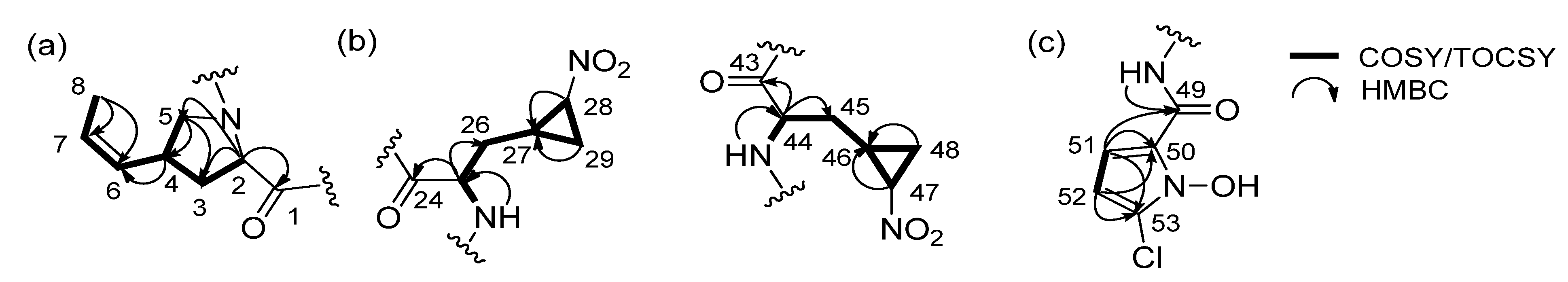
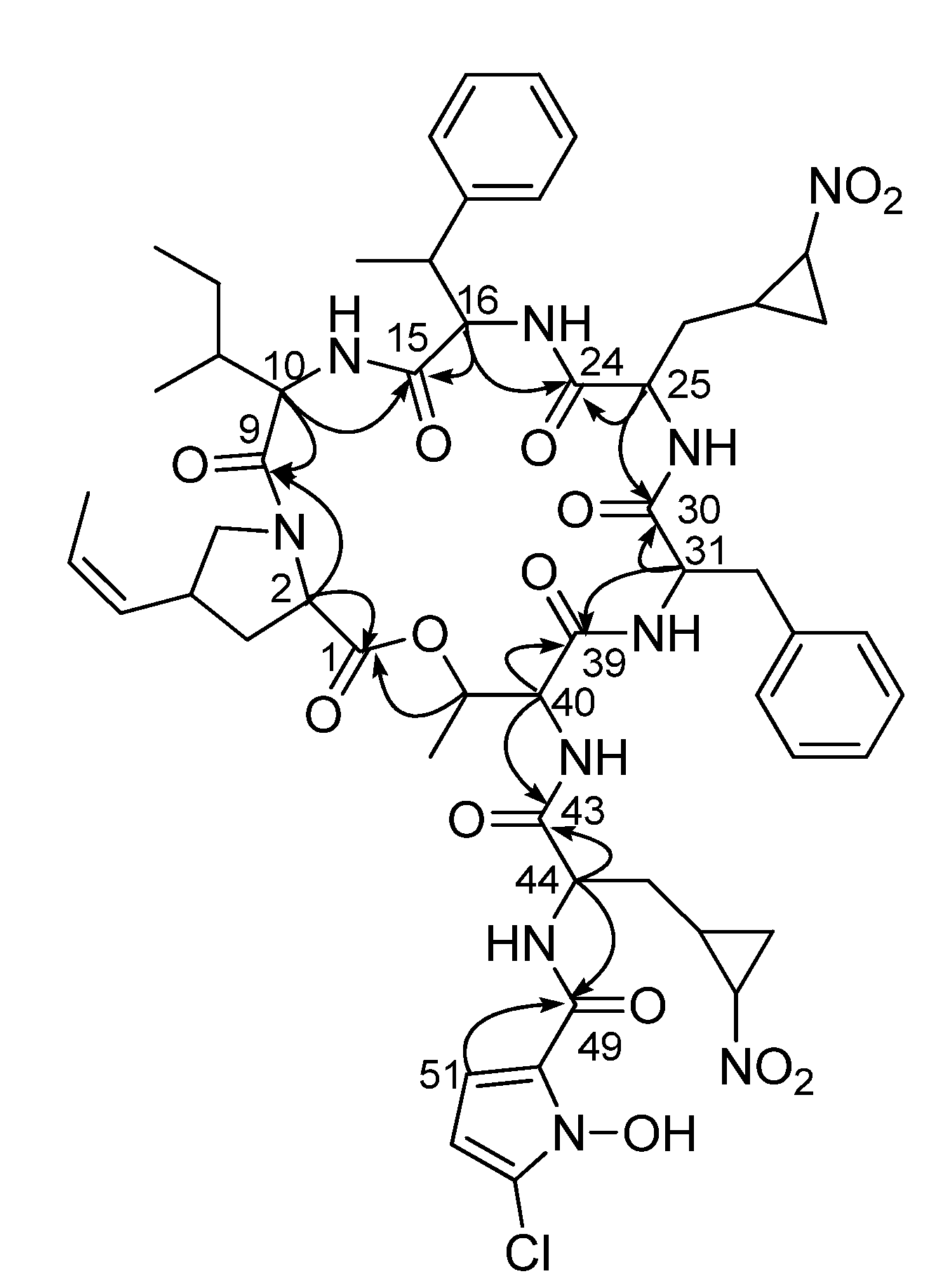
| Unit | C/H | 1 | 2 | ||||
|---|---|---|---|---|---|---|---|
| δH, mult (J in Hz) | δC | Type | δH, mult (J in Hz) | δC | Type | ||
| (4-Pe)Pro | 1 | – | 171.8 | C | – | 171.6 | C |
| 2 | 4.26, 1H, m | 61.7 | CH | 4.26, 1H, dd (11.0, 6.5) | 61.8 | CH | |
| 3 | 2.37, 1H, m | 34.9 | CH2 | 2.36, 1H, m | 35.8 | CH2 | |
| 1.80, 1H, m | 1.81, 1H, m | ||||||
| 4 | 3.27, 1H, m | 36.9 | CH | 3.25, 1H, m | 37.0 | CH | |
| 5 | 3.98, 1H, m | 53.0 | CH2 | 3.98, 1H, m | 53.0 | CH2 | |
| 3.31, 1H, m | 3.27, 1H, m | ||||||
| 6 | 5.26, 1H, dd (9.5, 9.5) | 127.8 | CH | 5.26, 1H, dd (9.0, 9.0) | 127.9 | CH | |
| 7 | 5.64, 1H, dq (9.5, 7.0) | 128.8 | CH | 5.63, 1H, dq (9.0, 7.0) | 128.8 | CH | |
| 8 | 1.68, 1H, d (7.0) | 13.5 | CH3 | 1.66, 1H, d (7.0) | 13.5 | CH3 | |
| Ile | 9 | – | 171.5 | C | – | 171.5 | C |
| 10 | 4.64, 1H, m | 54.5 | CH | 4.64, 1H, m1q | 54.9 | CH | |
| 10-NH | 7.31, 1H, brs | – | 7.23, 1H, brs | – | |||
| 11 | 1.88, 1H, m | 38.6 | CH | 1.90, 1H, m | 38.2 | CH | |
| 12 | 1.54, 1H, m | 25.0 | CH2 | 1.55, 1H, m | 25.0 | CH2 | |
| 1.29, 1H, m | 1.30, 1H, m | ||||||
| 13 | 0.90, 3H, d (7.0) | 10.8 | CH3 | 0.89, 3H, d (7.0) | 10.8 | CH3 | |
| 14 | 1.04, 3H, d (7.0) | 15.2 | CH3 | 1.02, 3H, d (7.0) | 15.3 | CH3 | |
| (βMe)Phe/Phe | 15 | – | 167.9 | C | – | 171.3 | C |
| 16 | 4.33, 1H, m | 60.8 | CH | 4.58, 1H, m | 55.2 | CH | |
| 16-NH | 6.85, 1H, brs | – | 6.91, 1H, brs | – | |||
| 17 | 3.02, 1H, m | 44.6 | CH | 2.91, 1H, m | 38.0 | CH2 | |
| 17-Me | 1.29, 3H, d (6.5) | 18.0 | CH3 | 2.97, 1H, m | |||
| 18 | – | 141.9 | C | – | 136.5 | C | |
| 19 | 7.11, 1H, d (8.5) | 129.0 | CH | 7.11, 1H, d (8.0) | 129.5 | CH | |
| 20 | 7.13, 1H, t (8.5) | 127.5 | CH | 7.15, 1H, t (8.0) | 129.0 | CH | |
| 21 | 7.03, 1H, t (8.5) | 127.6 | CH | 7.09, 1H, t (8.0) | 127.7 | CH | |
| 22 | 7.13, 1H, t (8.5) | 127.5 | CH | 7.15, 1H, t (8.0) | 129.0 | CH | |
| 23 | 7.11, 1H, d (8.5) | 129.0 | CH | 7.11, 1H, d (8.0) | 129.5 | CH | |
| (3-Ncp)Ala I | 24 | – | 169.3 | C | – | 168.2 | C |
| 25 | 3.52, 1H, m | 52.2 | CH | 3.55, 1H, m | 52.8 | CH | |
| 25-NH | 6.26, 1H, brs | – | 6.37, 1H, brs | – | |||
| 26 | 0.56, 1H, m | 33.4 | CH2 | 0.78, 1H, m | 33.0 | CH2 | |
| −0.26, 1H, m | 0.13, 1H, m | ||||||
| 27 | 0.28, 1H, m | 20.2 | CH | 0.58, 1H, m | 20.4 | CH | |
| 28 | 2.92, 1H, m | 58.7 | CH | 3.04, 1H, m | 58.6 | CH | |
| 29 | −0.66, 1H, m | 17.7 | CH2 | −0.34, 1H, m | 17.4 | CH2 | |
| 1.02, 1H, m | 1.13, 1H, m | ||||||
| Phe/(βMe)Phe | 30 | – | 168.8 | C | – | 168.3 | C |
| 31 | 4.48, 1H, m | 56.4 | CH | 4.48, 1H, m | 60.6 | CH | |
| 31-NH | 6.79, 1H, brs | – | 6.79, 1H, brs | – | |||
| 32 | 2.79, 1H, m | 38.7 | CH2 | 3.67, 1H, m | 37.1 | CH | |
| 3.38, 1H, m | 1.40, 3H, d (6.5) | 13.7 | CH3 | ||||
| 33 | – | 137.3 | C | – | 142.6 | C | |
| 34 | 7.22, 1H, m | 129.3 | CH | 7.24, 1H, m | 127.9 | CH | |
| 35 | 7.23, 1H, m | 127.8 | CH | 7.25, 1H, m | 128.8 | CH | |
| 36 | 7.15, 1H, t (8.5) | 127.2 | CH | 7.17, 1H, t (8.5) | 127.3 | CH | |
| 37 | 7.23, 1H, m | 127.8 | CH | 7.25, 1H, m | 128.8 | CH | |
| 38 | 7.22, 1H, m | 129.3 | CH | 7.24, 1H, m | 127.9 | CH | |
| Thr | 39 | – | 171.1 | C | – | 171.1 | C |
| 40 | 4.55, 1H, m | 58.4 | CH | 4.52, 1H, m | 55.8 | CH | |
| 40-NH | 8.98, 1H, brs | – | 8.95, 1H, brs | – | |||
| 41 | 5.40, 1H, m | 69.4 | CH | 5.38, 1H, m | 69.6 | CH | |
| 42 | 1.52, 3H, m | 17.2 | CH3 | 1.48, 3H, m | 17.3 | CH3 | |
| (3-Ncp)Ala II | 43 | – | 172.2 | – | 172.1 | ||
| 44 | 5.14, 1H, m | 51.2 | CH | 5.09, 1H, m | 51.5 | CH | |
| 44-NH | 8.16, 1H, brs | – | 7.96, 1H, brs | – | |||
| 45 | 1.80, 1H, m | 35.5 | CH2 | 1.83, 1H, m | 35.2 | CH2 | |
| 1.60, 1H, m | 1.61, 1H, m | ||||||
| 46 | 1.93, 1H, m | 21.7 | CH | 1.91, 1H, m | 21.7 | CH | |
| 47 | 4.04, 1H, m | 59.8 | CH | 4.05, 1H, m | 59.6 | CH | |
| 48 | 1.95, 1H, m | 17.8 | CH2 | 1.95, 1H, m | 17.9 | CH2 | |
| 1.02, 1H, m | 1.01, 1H, m | ||||||
| Chpca | 49 | – | 159.7 | C | – | 160.1 | C |
| 50 | – | 119.8 | C | – | 119.8 | C | |
| 51 | 6.68, 1H, d (4.5) | 109.8 | CH | 6.81, 1H, d (4.5) | 109.8 | CH | |
| 52 | 6.08, 1H, d (4.5) | 103.5 | CH | 6.13, 1H, d (4.5) | 104.0 | CH | |
| 53 | – | 121.5 | C | – | 120.5 | C | |
| NOH | 10.8, 1H, brs | – | 10.8, 1H, brs | – | |||
2.2. Bioactivities of the Hormaomycins
| Compound | MICs (μM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram-Positive | Gram-Negative | |||||||
| S. aureus | B. subtilis | K. rhizophila | S. pyogenes | K. pneumoniae | S. enterica | P. hauseri | E. coli | |
| Hormaomycin | 0.4 | 1.8 | 0.03 | 1.8 | >113 | >113 | 0.9 | >113 |
| Hormaomycin B | 7 | 14 | 0.4 | 14 | >115 | 29 | 29 | >115 |
| Hormaomycin C | 7 | 56 | 0.23 | 8 | >114 | 114 | 14 | >114 |
| Ampicillin | <0.17 | 0.17 | <0.17 | 0.17 | 45.9 | 3.7 | <0.17 | 11.5 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Isolation, Cultivation and Extraction of Bacteria
3.3. Isolation of the Hormaomycins
3.4. Antibacterial Activity Assay
3.5. Antifungal Activity Assay
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clardy, J.; Walsh, C. Lessons from natural molecules. Nature 2004, 432, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.-P.; Yuan, J.; Sun, L.; She, Z.-G.; Wu, J.-H.; Lan, X.-J.; Zhu, X.; Lin, Y.-C.; Chen, S.-P. Statistical research on marine natural products based on data obtained between 1985 and 2008. Mar. Drugs 2011, 9, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. A new antibiotic and the evolution of resistance. N. Engl. J. Med. 2015, 372, 1168–1170. [Google Scholar] [PubMed]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem.-Int. Edit. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Perez Baz, J.; Canedo, L.M.; Fernandez Puentes, J.L.; Silva Elipe, M.V. Thiocoraline, a novel depsipeptide with antitumor activity produced by a marine Micromonospora. II. Physico-chemical properties and structure determination. J. Antibiot. 1997, 50, 738–741. [Google Scholar]
- Kim, S.-H.; Shin, Y.; Lee, S.-H.; Oh, K.-B.; Lee, S.K.; Shin, J.; Oh, D.-C. Salternamides A−D from a halophilic Streptomyces sp. actinobacterium. J. Nat. Prod. 2015, 78, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.; Ahn, C.-H.; Shin, Y.; Won, T.H.; Ko, K.; Lee, S.K.; Oh, K.-B.; Shin, J.; Nam, S.-I.; Oh, D.-C. New benzoxazine secondary metabolites from an Arctic actinomycete. Mar. Drugs 2014, 12, 2526–2538. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.; Chung, B.; Shin, Y.; Rheingold, A.L.; Moore, C.E.; Park, S.J.; Park, S.; Lee, S.K.; Oh, K.-B.; Shin, J.; et al. Pentacyclic antibiotics from a tidal mud flat-derived actinomycete. J. Nat. Prod. 2015, 78, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sandiford, S. K.; van Wezel, G.P. Triggers and cues that activate antibiotic production by actinomycetes. J. Ind. Microbiol. Biotechnol. 2014, 41, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-J.; Li, D.-Y.; Li, Y.-C.; Hua, H.-M.; Ma, E.-L.; Li, Z.-L. Caryophyllene sesquiterpenes from the marine-derived fungus Ascotricha sp. ZJ-M-5 by the one strain-many compounds strategy. J. Nat. Prod. 2014, 77, 1367–1371. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Kim, H.; Moon, K.; Nam, S.-J.; Shin, J.; Oh, K.-B.; Oh, D.-C. Mohangamides A and B, new dilactone-tethered pseudo-dimeric peptides inhibiting Candida albicans isocitrate lyase. Org. Lett. 2015, 17, 712–715. [Google Scholar] [CrossRef] [PubMed]
- Roessner, E.; Zeeck, A.; Koenig, W.A. Structure determination of hormaomycin. Angew. Chem. 1990, 102, 84–85. [Google Scholar]
- Pretsch, E.; Bȕhlmann, P.; Affolter, C. Structure Determination of Organic Compounds-Tables of Spectral Data; Springer: New York, NY, USA, 2000; p. 104. [Google Scholar]
- Zlatopolskiy, B.D.; de Meijere, A. First total synthesis of hormaomycin, a naturally occurring depsipeptide with interesting biological activities. Chem. Eur. J. 2004, 10, 4718–4727. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Teta, R.; Kohlhaas, C.; Cruesemann, M.; Ueoka, R.; Mangoni, A.; Freeman, M.F.; Piel, J. Manipulation of regulatory genes reveals complexity and fidelity in hormaomycin biosynthesis. Chem. Biol. 2013, 20, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Andres, N.; Wolf, H.; Zähner, H. Hormaomycin, a new peptide lactone antibiotic effective in inducing cytodifferentiation and antibiotic biosynthesis in some Streptomyces species. Z. Naturforsch. 1990, 45, 851–855. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, M.; Chung, B.; Oh, K.-B.; Shin, J.; Oh, D.-C. Hormaomycins B and C: New Antibiotic Cyclic Depsipeptides from a Marine Mudflat-Derived Streptomyces sp. Mar. Drugs 2015, 13, 5187-5200. https://doi.org/10.3390/md13085187
Bae M, Chung B, Oh K-B, Shin J, Oh D-C. Hormaomycins B and C: New Antibiotic Cyclic Depsipeptides from a Marine Mudflat-Derived Streptomyces sp. Marine Drugs. 2015; 13(8):5187-5200. https://doi.org/10.3390/md13085187
Chicago/Turabian StyleBae, Munhyung, Beomkoo Chung, Ki-Bong Oh, Jongheon Shin, and Dong-Chan Oh. 2015. "Hormaomycins B and C: New Antibiotic Cyclic Depsipeptides from a Marine Mudflat-Derived Streptomyces sp." Marine Drugs 13, no. 8: 5187-5200. https://doi.org/10.3390/md13085187
APA StyleBae, M., Chung, B., Oh, K.-B., Shin, J., & Oh, D.-C. (2015). Hormaomycins B and C: New Antibiotic Cyclic Depsipeptides from a Marine Mudflat-Derived Streptomyces sp. Marine Drugs, 13(8), 5187-5200. https://doi.org/10.3390/md13085187






