Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula
Abstract
:1. Introduction
2. Results
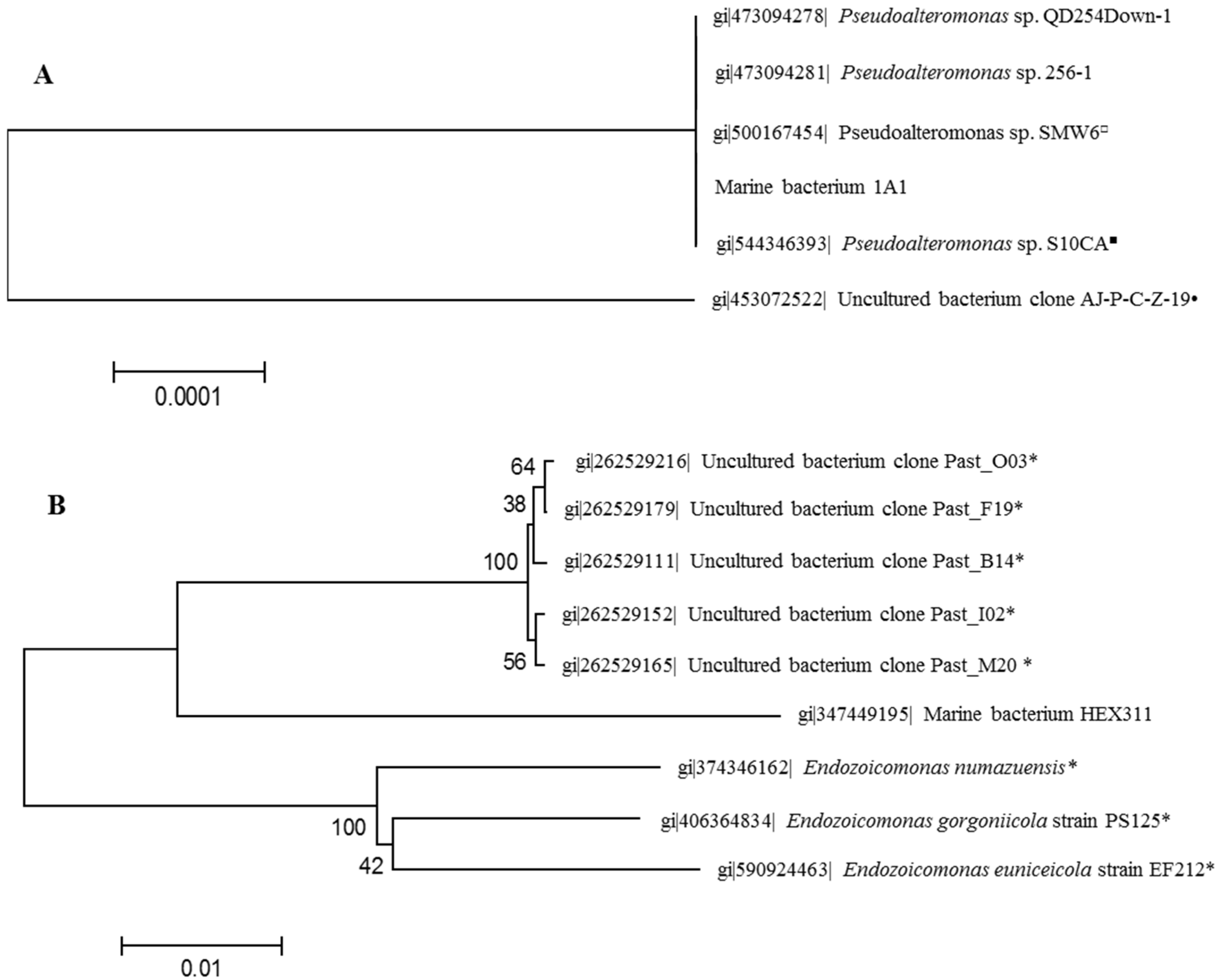
2.1. Identification of Sponge-Associated Bacteria
2.2. Electrophoretic Pattern of Bacterial LPS

2.3. Carbohydrate Composition
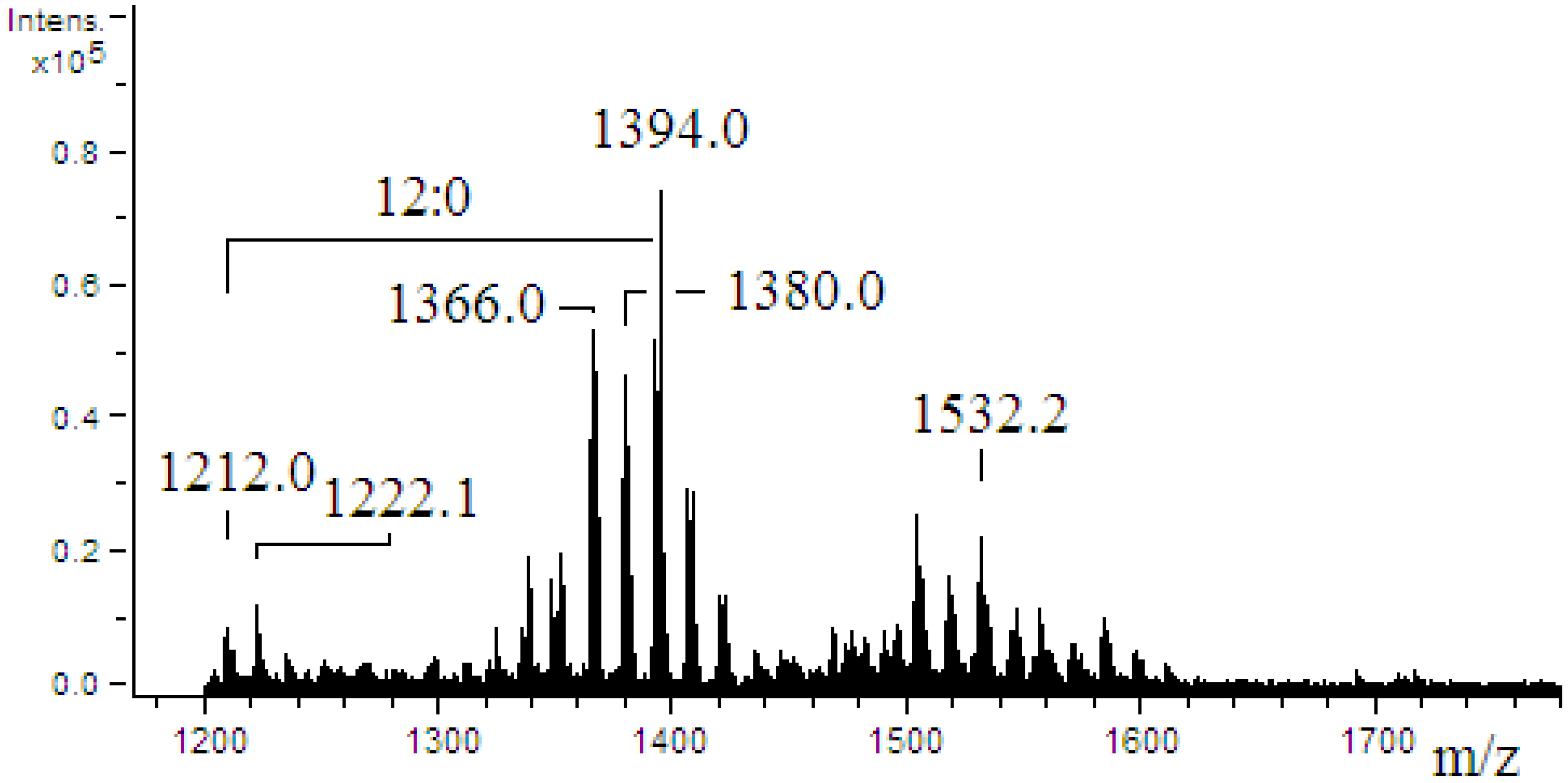
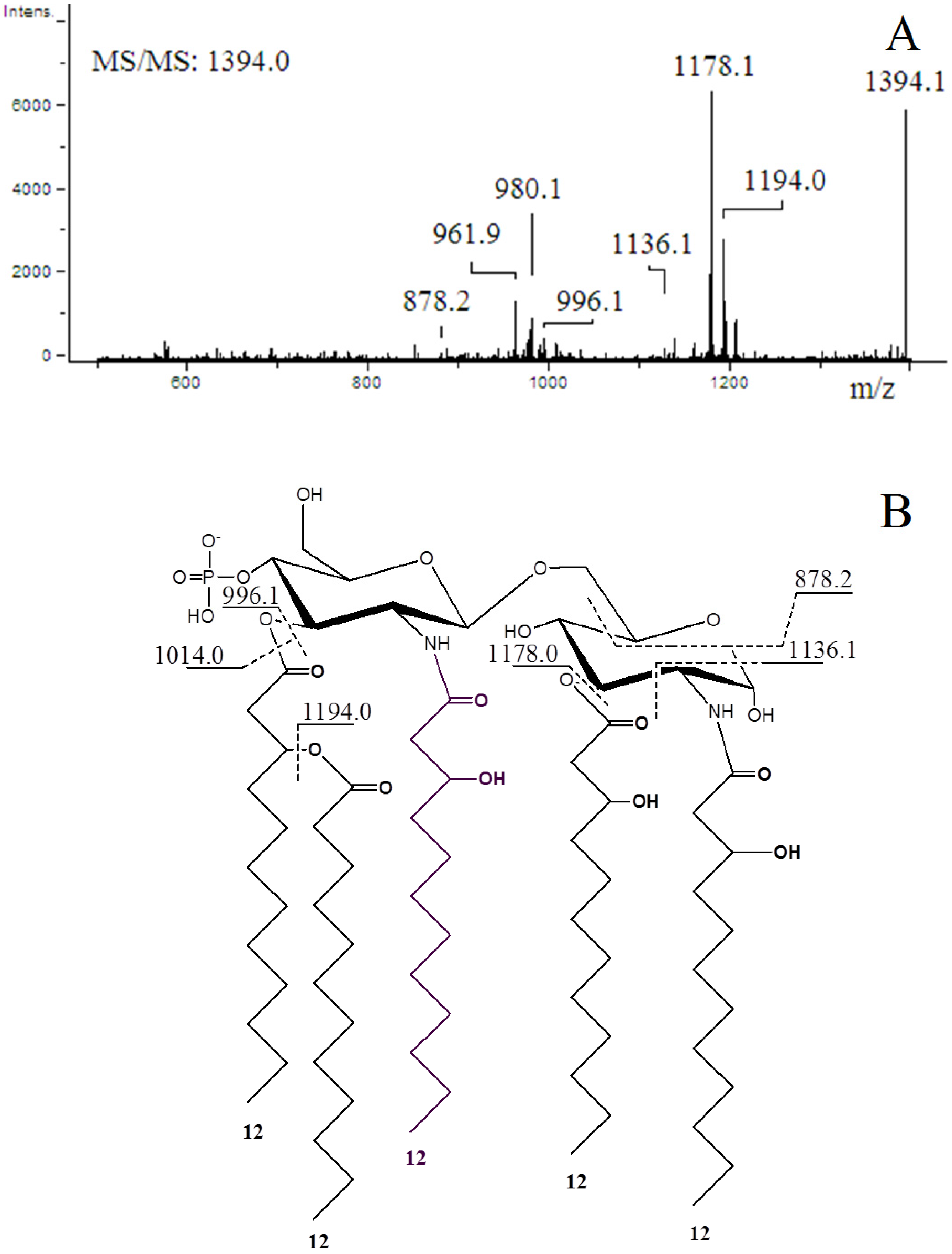
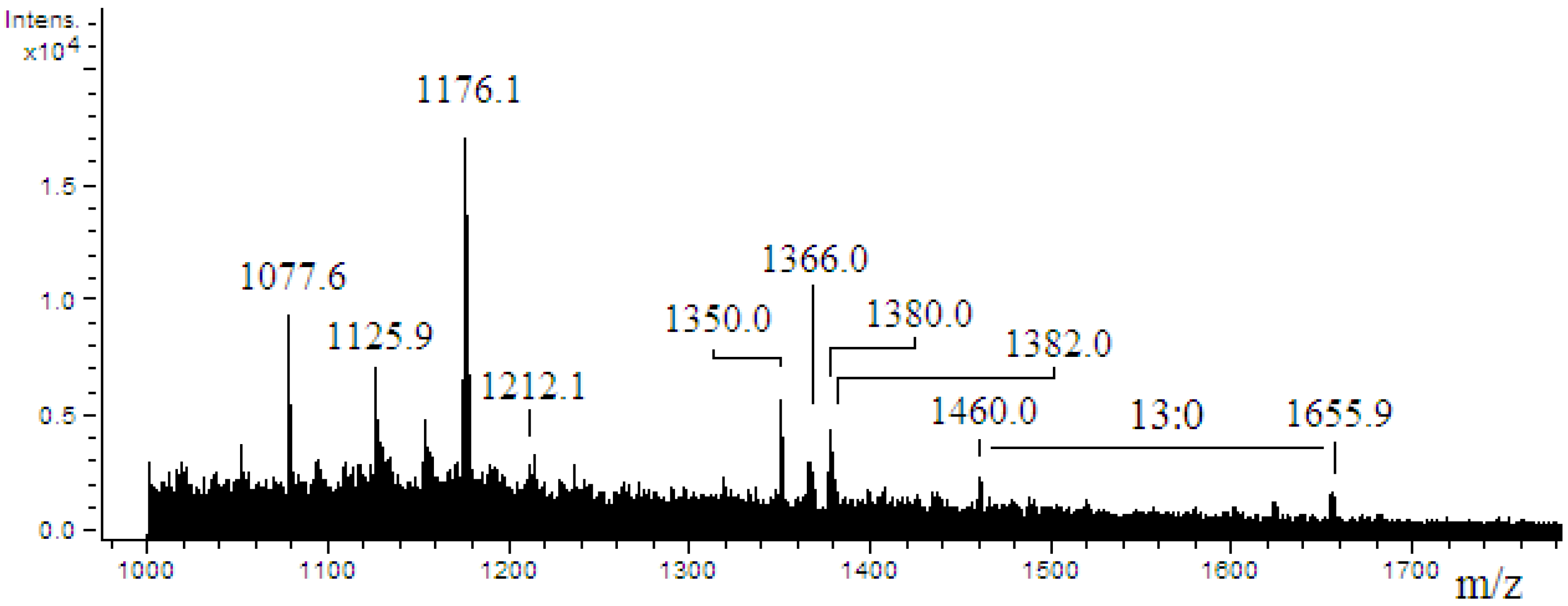
2.4. ESI-MS/MS of Lipid A
2.5. Stimulation of Sponge Primmorphs with LPS
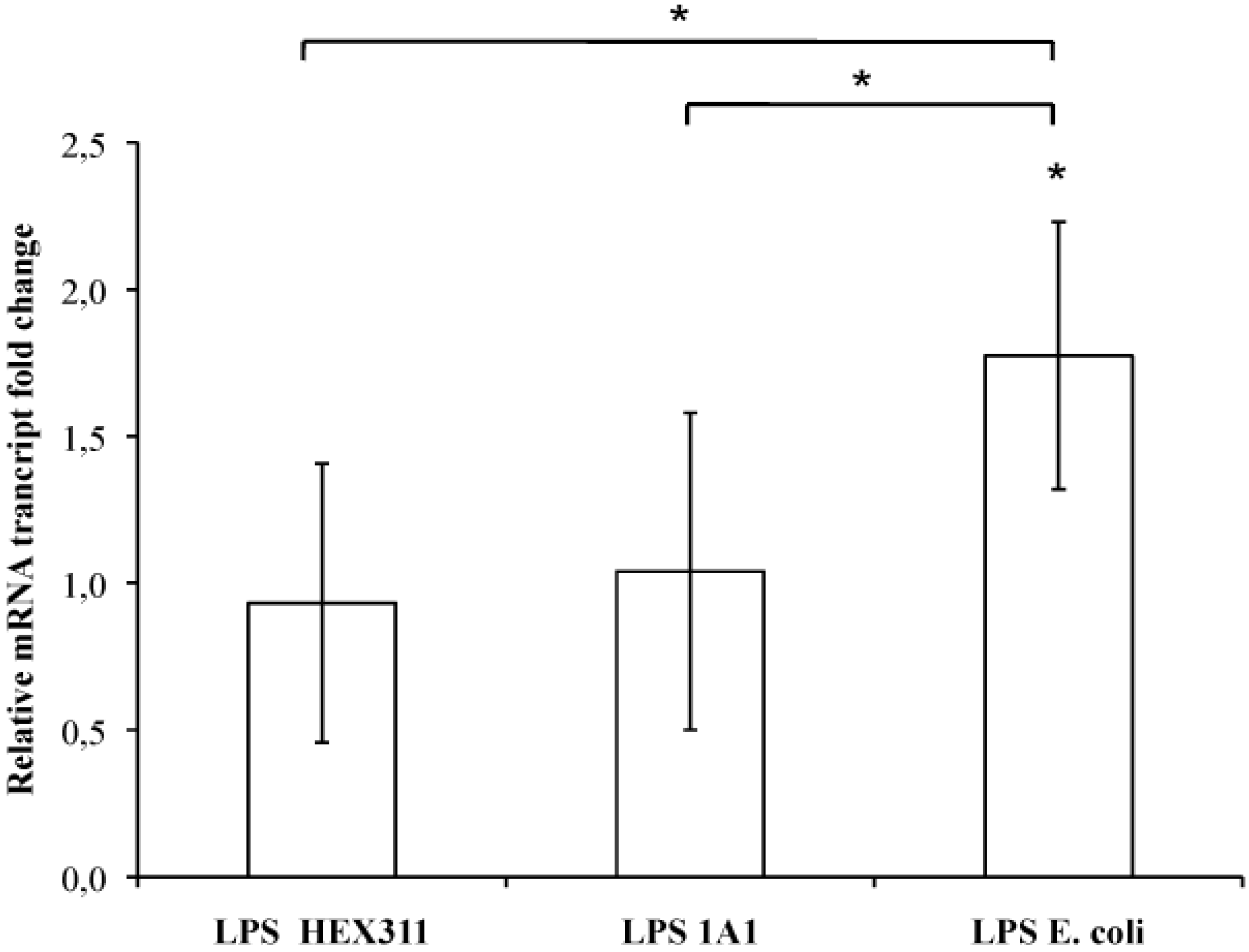
3. Discussion
3.1. Identification of Sponge-Isolated Bacteria
3.2. Characterization of Bacterial LPS
3.3. Effect of LPS on the Expression of the MPEG Gene: Structure–Function Relationship
3.3.1. The Degree of Acylation of the Lipid A
3.3.2. The Position of the Phosphate on the Lipid A
3.3.3. The Carbohydrate Composition
4. Experimental Section
4.1. Sample Collection
4.2. Isolation of Sponge-Associated Bacteria and Culture of Reference Bacteria
4.3. Identification of Sponge-Associated Bacteria
4.4. Lipopolysaccharide Preparation
4.5. SDS-PAGE and Silver Nitrate Coloration
4.6. Carbohydrate Composition
4.7. Lipid A Sample Preparation for Mass Spectrometry
4.8. Mass Spectrometry
4.9. Sponge Cell Culture and Primmorph Stimulation
4.10. Quantitative Reverse Transcription (qRT-PCR) Analysis
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Ethical Statements
References
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, S.; Tsai, P.; Bell, J.; Fromont, J.; Ilan, M.; Lindquist, N.; Perez, T.; Rodrigo, A.; Schupp, P.J.; Vacelet, J.; et al. Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012, 6, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 2012, 10, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.R. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 1978, 49, 161–167. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Vacelet, J.; Donadey, C. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 1977, 30, 301–314. [Google Scholar] [CrossRef]
- Gardères, J.; Henry, J.; Bernay, B.; Ritter, A.; Zatylny-Gaudin, C.; Wiens, M.; Müller, W.E.G.; Le Pennec, G. Cellular effects of bacterial N-3-oxo-dodecanoyl-l-Homoserine lactone on the sponge Suberites domuncula (Olivi, 1792): Insights into an intimate inter-kingdom dialogue. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Hadas, E.; Shpigel, M.; Ilan, M. Particulate organic matter as a food source for a coral reef sponge. J. Exp. Biol. 2009, 212, 3643–3650. [Google Scholar] [CrossRef] [PubMed]
- Reiswig, H.M. Bacteria as food for temperate-water marine sponges. Can. J. Zool. 1975, 53, 582–589. [Google Scholar] [CrossRef]
- Reiswig, H.M. Particle feeding in natural populations of three marine demosponges. Biol. Bull. 1971, 141, 568–591. [Google Scholar] [CrossRef]
- Lauckner, G. Diseases of Porifera. In Diseases of Marine Animals: 1. General Aspects, Protozoa to Gastropoda; Kinne, O., Ed.; John Wiley and Sons: Chichester, UK, 1980; Volume 1, pp. 139–165. [Google Scholar]
- Luter, H.M.; Whalan, S.; Webster, N.S. Prevalence of tissue necrosis and brown spot lesions in a common marine sponge. Mar. Freshw. Res. 2010, 61, 484–489. [Google Scholar] [CrossRef]
- Webster, N.; Negri, A.; Webb, R.; Hill, R. A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 2002, 232, 305–309. [Google Scholar] [CrossRef]
- Wilkinson, C.R. Symbiotic interactions between marine sponges and algae. In Algae and symbioses; Reisser, W., Ed.; Biopress: Bristol, UK, 1992; pp. 112–151. [Google Scholar]
- Lee, Y.K.; Lee, J.H.; Lee, H.K. Microbial symbiosis in marine sponges. J. Microbiol. 2001, 39, 254–264. [Google Scholar]
- Schmitt, S.; Angermeier, H.; Schiller, R.; Lindquist, N.; Hentschel, U. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 2008, 74, 7694–7708. [Google Scholar] [CrossRef] [PubMed]
- Proksch, P. Defensive roles for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon. 1994, 32, 639–655. [Google Scholar] [CrossRef]
- Müller, W.E.G.; Müller, I.M. Origin of the metazoan immune system: Identification of the molecules and their functions in sponges. Integr. Comp. Biol. 2003, 43, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Schröder, H.C.; Kljajić, Z.; Weiler, B.E.; Gasić, M.; Uhlenbruck, G.; Kurelec, B.; Müller, W.E.G. The galactose-specific lectin from the sponge Chondrilla nucula displays anti-human immunodeficiency virus activity in vitro via stimulation of the (2′-5′)oligoadenylate metabolism. Antivir. Chem. Chemother. 1990, 1, 99–105. [Google Scholar] [CrossRef]
- Perović-Ottstadt, S.; Adell, T.; Proksch, P.; Wiens, M.; Korzhev, M.; Gamulin, V.; Müller, I.M.; Müller, W.E.G. A (1→3)-β-d-glucan recognition protein from the sponge Suberites domuncula. Eur. J. Biochem. 2004, 271, 1924–1937. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.; Korzhev, M.; Krasko, A.; Thakur, N.L.; Perović-Ottstadt, S.; Breter, H.J.; Ushijima, H.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E.G. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway. Induction of a perforin-like molecule. J. Biol. Chem. 2005, 280, 27949–27959. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.L.; Perović-Ottstadt, S.; Batel, R.; Korzhev, M.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E.G. Innate immune defense of the sponge Suberites domuncula against Gram-positive bacteria: Induction of lysozyme and AdaPTin. Mar. Biol. 2004, 146, 271–282. [Google Scholar] [CrossRef]
- Wiens, M.; Korzhev, M.; Perović-Ottstadt, S.; Luthringer, B.; Brandt, D.; Klein, S.; Müller, W.E.G. Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 2007, 24, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Reveillaud, J.; Maignien, L.; Eren, A.M.; Huber, J.A.; Apprill, A.; Sogin, M.L.; Vanreusel, A. Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 2014, 8, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Fieseler, L.; Horn, M.; Wagner, M.; Hentschel, U. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 2004, 70, 3724–3732. [Google Scholar] [CrossRef] [PubMed]
- Simister, R.L.; Deines, P.; Botté, E.S.; Webster, N.S.; Taylor, M.W. Sponge-specific clusters revisited: A comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 2012, 14, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Gloeckner, V.; Lindquist, N.; Schmitt, S.; Hentschel, U. Ectyoplasia ferox, an experimentally tractable model for vertical microbial transmission in marine sponges. Microb. Ecol. 2013, 65, 462–474. [Google Scholar] [CrossRef] [PubMed]
- De Bary, A. De la symbiose. Rev. Int. Sci. 1879, 3, 301–309. [Google Scholar]
- Gardères, J.; Taupin, L.; Saïdin, J.B.; Dufour, A.; Le Pennec, G. N-acyl homoserine lactone production by bacteria within the sponge Suberites domuncula (Olivi, 1792) (Porifera, Demospongiae). Mar. Biol. 2012, 159, 1685–1692. [Google Scholar] [CrossRef]
- Thomas, T.; Rusch, D.; DeMaere, M.Z.; Yung, P.Y.; Lewis, M.; Halpern, A.; Heidelberg, K.B.; Egan, S.; Steinberg, P.D.; Kjelleberg, S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010, 4, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Siegl, A.; Kamke, J.; Hochmuth, T.; Piel, J.; Richter, M.; Liang, C.; Dandekar, T.; Hentschel, U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011, 5, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C. The Toll receptor family and microbial recognition. Trends Microbiol. 2000, 8, 452–456. [Google Scholar] [CrossRef]
- Janeway, C.A.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.; Krasko, A.; Perovic, S.; Müller, W.E.G. Caspase-mediated apoptosis in sponges: Cloning and function of the phylogenetic oldest apoptotic proteases from Metazoa. Biochim. Biophys. Acta 2003, 1593, 179–189. [Google Scholar] [CrossRef]
- Wiens, M.; Müller, W.E.G. Cell death in Porifera: Molecular players in the game of apoptotic cell death in living fossils. Can. J. Zool. 2006, 84, 307–321. [Google Scholar] [CrossRef]
- Hritz, I.; Mandrekar, P.; Velayudham, A.; Catalano, D.; Dolganiuc, A.; Kodys, K.; Kurt-Jones, E.; Szabo, G. The critical role of Toll-like receptor (TLR) 4 in alcoholic liver disease is independent of the common TLR adapter MyD88. Hepatology. 2008, 48, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.S.; Apicella, M.A.; McFall-Ngai, M.J. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 2000, 226, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Cheesman, S.E.; Guillemin, K. We know you are in there: Conversing with the indigenous gut microbiota. Res. Microbiol. 2007, 158, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Böhm, M.; Hentschel, U.; Friedrich, A.; Fieseler, L.; Steffen, R.; Gamulin, V.; Müller, I.; Müller, W. Molecular response of the sponge Suberites domuncula to bacterial infection. Mar. Biol. 2001, 139, 1037–1045. [Google Scholar]
- Grebenjuk, V.A.; Kuusksalu, A.; Kelve, M.; Schütze, J.; Schröder, H.C.; Müller, W.E.G. Induction of (2′-5′)oligoadenylate synthetase in the marine sponges Suberites domuncula and Geodia cydonium by the bacterial endotoxin lipopolysaccharide. Eur. J. Biochem. 2002, 269, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Schröder, H.C.; Ushijima, H.; Krasko, A.; Gamulin, V.; Thakur, N.L.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E.G. Emergence and disappearance of an immune molecule, an antimicrobial lectin, in basal Metazoa. A tachylectin-related protein in the sponge Suberites domuncula. J. Biol. Chem. 2003, 278, 32810–32817. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.G.; Klemt, M.; Thakur, N.L.; Schröder, H.C.; Aiello, A.; D’Esposito, M.; Menna, M.; Fattorusso, E. Molecular/chemical ecology in sponges: Evidence for an adaptive antibacterial response in Suberites domuncula. Mar. Biol. 2003, 144, 19–29. [Google Scholar] [CrossRef]
- Gauthier, M.E.A.; Du Pasquier, L.; Degnan, B.M. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol. Dev. 2010, 12, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Woodley, C.M.; Medina, M. Threatened Corals Provide Underexplored Microbial Habitats. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Costello, C.E.; Vath, J.E. Tandem mass spectrometry of glycolipids. In Methods in Enzymology; McCloskey, J.A., Ed.; Academic Press: Waltham, MA, USA, 1990; Volume 193, pp. 738–768. [Google Scholar]
- Angermeier, H.; Glckner, V.; Pawlik, J.R.; Lindquist, N.L.; Hentschel, U. Sponge white patch disease affecting the Caribbean sponge Amphimedon compressa. Dis. Aquat. Organ. 2012, 99, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Angermeier, H.; Kamke, J.; Abdelmohsen, U.R.; Krohne, G.; Pawlik, J.R.; Lindquist, N.L.; Hentschel, U. The pathology of sponge orange band disease affecting the Caribbean barrel sponge Xestospongia muta. FEMS Microbiol. Ecol. 2011, 75, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.B.; Thacker, R.W.; Gochfeld, D.J. Molecular community profiling reveals impacts of time, space, and disease status on the bacterial community associated with the Caribbean sponge Aplysina cauliformis. FEMS Microbiol. Ecol. 2014, 87, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, J.D.; Pramanik, A.; Webster, N.S.; Llewellyn, L.E.; Gachhui, R.; Mukherjee, J. The pathogen of the great barrier reef sponge Rhopaloeides odorabile is a new strain of Pseudoalteromonas agarivorans containing abundant and diverse virulence-related genes. Mar. Biotechnol. 2015, 17, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Carre-Mlouka, A.; Domart-Coulon, I.; Vacelet, J.; Bourguet-Kondracki, M.-L. Exploring cultivable bacteria from the prokaryotic community associated with the carnivorous sponge Asbestopluma hypogea. FEMS Microbiol. Ecol. 2014, 88, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Sadovskaya, I.; Brisson, J.R.; Thibault, P.; Richards, J.C.; Lam, J.S.; Altman, E. Structural characterization of the outer core and the O-chain linkage region of lipopolysaccharide from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. FEBS 2000, 267, 1640–1650. [Google Scholar] [CrossRef]
- Holst, O. Chemical Structure of the core region of liposaccharides. In Endotoxin in Health and Disease; Helmut, B., Opal, S.M., Vogel, S.N., Morrison, D.C., Eds.; Marcel Dekker Inc: New York, NY, USA, 1999; pp. 115–154. [Google Scholar]
- Kooistra, O.; Bedoux, G.; Brecker, L.; Lindner, B.; Carballo, P.S.; Haras, D.; Zähringer, U. Structure of a highly phosphorylated lipopolysaccharide core in the ΔalgC mutants derived from Pseudomonas aeruginosa wild-type strains PAO1 (serogroup O5) and PAC1R (serogroup O3). Carbohydr. Res. 2003, 338, 2667–2677. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Lanzetta, R.; Parrilli, M.; Sturiale, L.; Garozzo, D.; Nazarenko, E.L.; Gorshkova, R.P.; Ivanova, E.P.; Molinaro, A. The complete structure of the core carbohydrate backbone from the LPS of marine halophilic bacterium Pseudoalteromonas carrageenovora type strain IAM 12662T. Carbohydr. Res. 2005, 340, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Carillo, S.; Pieretti, G.; Parrilli, E.; Tutino, M.L.; Gemma, S.; Molteni, M.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Structural investigation and biological activity of the lipooligosaccharide from the psychrophilic bacterium Pseudoalteromonas haloplanktis TAB 23. Chem. Eur. J. 2011, 17, 7053–7060. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, M.M.; Piaz, F.D.; Lanzetta, R.; Parrilli, M. Lipid A structure of Pseudoalteromonas haloplanktis TAC 125: Use of electrospray ionization tandem mass spectrometry for the determination of fatty acid distribution. J. Mass Spectrom. 2002, 37, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Kussak, A.; Weintraub, A. Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from Gram-negative bacteria. Anal. Biochem. 2002, 307, 131–137. [Google Scholar] [CrossRef]
- Bedoux, G.; Vallée-Réhel, K.; Kooistra, O.; Zähringer, U.; Haras, D. Lipid A components from Pseudomonas aeruginosa PAO1 (serotype O5) and mutant strains investigated by electrospray ionization ion-trap mass spectrometry. J. Mass Spectrom. 2004, 39, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Zubova, S.V.; Ivanov, A.I.; Prokhorenko, I.R. The effect of composition of the core region of Escherichia coli K-12 lipopolysaccharides on the surface properties of cells. Mikrobiologiia 2008, 77, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Galanos, C.; Lüderitz, O.; Rietschel, E.T.; Westphal, O.; Brade, H.; Brade, L.; Freudenberg, M.; Schade, U.; Imoto, M.; Yoshimura, H. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 1985, 148, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, I.N.; Kapustina, N.V.; Isakov, V.V.; Gorshkova, N.M.; Solov’eva, T.F. Elucidation of structure of lipid A from the marine Gram-negative bacterium Pseudoalteromonas haloplanktis ATCC 14393T. Bioorg. Khim. 2004, 30, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Volk, A.S.; Krasikova, I.N.; Anastyuk, S.D.; Dmitrenok, P.S.; Solov’eva, T.F. Structure of lipid A from the marine Gram-negative bacterium Pseudoalteromonas nigrifaciens IAM 13010T. Chem. Nat. Compd. 2007, 43, 519–524. [Google Scholar] [CrossRef]
- Leone, S.; Silipo, A.; L.Nazarenko, E.; Lanzetta, R.; Parrilli, M.; Molinaro, A. Molecular structure of endotoxins from Gram-negative marine bacteria: An update. Mar. Drugs 2007, 5, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, M.; Takahashi, H.; Watanabe, H.; Saito, S.; Kawahara, K. Immunomodulatory effects of Yersinia pestis lipopolysaccharides on human macrophages. Clin. Vaccine Immunol. 2010, 17, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Coats, S.R.; Berezow, A.B.; To, T.T.; Jain, S.; Bainbridge, B.W.; Banani, K.P.; Darveau, R.P. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect. Immun. 2011, 79, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Coats, S.R.; Jones, J.W.; Do, C.T.; Braham, P.H.; Bainbridge, B.W.; To, T.T.; Goodlett, D.R.; Ernst, R.K.; Darveau, R.P. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell. Microbiol. 2009, 11, 1587–1599. [Google Scholar] [CrossRef] [PubMed]
- Phillips, N.J.; Adin, D.M.; Stabb, E.V.; McFall-Ngai, M.J.; Apicella, M.A.; Gibson, B.W. The lipid A from Vibrio fischeri lipopolysaccharide. A unique structure bearing a phosphoglycerol moiety. J. Biol. Chem. 2011, 286, 21203–21219. [Google Scholar] [CrossRef] [PubMed]
- Post, D.M.B.; Yu, L.; Krasity, B.C.; Choudhury, B.; Mandel, M.J.; Brennan, C.A.; Ruby, E.G.; McFall-Ngai, M.J.; Gibson, B.W.; Apicella, M.A. O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide. Composition and analysis of their role in Euprymna scolopes light organ colonization. J. Biol. Chem. 2012, 287, 8515–8530. [Google Scholar] [CrossRef] [PubMed]
- Rader, B.A.; Kremer, N.; Apicella, M.A.; Goldman, W.E.; McFall-Ngai, M.J. Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis. mBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Altura, M.A.; Stabb, E.; Goldman, W.; Apicella, M.; McFall-Ngai, M.J. Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. Cell. Microbiol. 2011, 13, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Maaetoft-Udsen, K.; Vynne, N.; Heegaard, P.M.; Gram, L.; Frøkiær, H. Pseudoalteromonas strains are potent immunomodulators owing to low-stimulatory LPS. Innate Immun. 2013, 19, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Westphal, O. Bacterial lipopolysaccharide-extraction with phenol water and further application of procedure. Methods Carbohydr. Chem. 1965, 1, 83–91. [Google Scholar]
- Karibian, D.; Deprun, C.; Caroff, M. Use of plasma desorption mass spectrometry in structural analysis of endotoxins: Effects on lipid A of different acid treatments. Prog. Clin. Biol. Res. 1995, 392, 103–111. [Google Scholar] [PubMed]
- Le Pennec, G.; Perovic, S.; Ammar, M.S.A.; Grebenjuk, V.A.; Steffen, R.; Brümmer, F.; Müller, W.E.G. Cultivation of primmorphs from the marine sponge Suberites domuncula: Morphogenetic potential of silicon and iron. J. Biotechnol. 2003, 100, 93–108. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardères, J.; Bedoux, G.; Koutsouveli, V.; Crequer, S.; Desriac, F.; Pennec, G.L. Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula. Mar. Drugs 2015, 13, 4985-5006. https://doi.org/10.3390/md13084985
Gardères J, Bedoux G, Koutsouveli V, Crequer S, Desriac F, Pennec GL. Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula. Marine Drugs. 2015; 13(8):4985-5006. https://doi.org/10.3390/md13084985
Chicago/Turabian StyleGardères, Johan, Gilles Bedoux, Vasiliki Koutsouveli, Sterenn Crequer, Florie Desriac, and Gaël Le Pennec. 2015. "Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula" Marine Drugs 13, no. 8: 4985-5006. https://doi.org/10.3390/md13084985
APA StyleGardères, J., Bedoux, G., Koutsouveli, V., Crequer, S., Desriac, F., & Pennec, G. L. (2015). Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula. Marine Drugs, 13(8), 4985-5006. https://doi.org/10.3390/md13084985




