Abstract
Dereplication and chemotaxonomic studies of six marine algae of the Ochrophyta and one of the Rhodophyta phyla resulted in the detection of 22 separate compounds. All 16 secondary metabolites, including four new compounds (16–19), could be rapidly dereplicated using HPLC-NMR and HPLC-MS methodologies in conjunction with the MarinLit database. This study highlights the advantages of using NMR data (acquired via HPLC-NMR) for database searching and for the overall dereplication of natural products.
1. Introduction
The Ochrophyta phylum contains in excess of 1800 species [1]. Recently, 17 new compounds, mostly terpenoids, were reported from the Ochrophyta phylum [2]. In contrast, marine algae belonging to the Rhodophyta phylum, are represented by over 6500 species [1] and produce a large number of halogenated secondary metabolites [3]. For example, nine new compounds were recently reported from the Rhodophyta phylum, a majority representative of bromophenols [2].
The intention of this study was to select a range of marine algae and conduct a dereplication/chemotaxonomical investigation as a means to rapidly differentiate the secondary metabolites present across different genera and between species of the same genera. For instance, marine algae of the Ochrophyta phylum such as Cystophora retroflexa have been reported to produce carotenoids, phlorethols and fucophlorethols [4,5,6] while Cystophora subfarcinata is known to produce tocotrienols and phloroglucinols [7]. The remaining Ochrophyta phylum genera and species investigated included, Sargassum decipiens, Sargassum vestitum, Sargassum cf. fallax, and Halopteris pseudospicata and importantly these have not had any marine secondary metabolites reported. However, the Sargassum genus is known to produce meroditerpenoid, tocotrienol and terpenoid type compounds [8]. The only marine alga of the Rhodophyta phylum studied was of the Laurencia genus which is known to produce very different secondary metabolites from the marine brown algae [9,10,11,12].
In this study, we examined six specimens of marine brown algae belonging to the Ochrophyta phylum (Sargassum cf. fallax, Sargassum decipiens (R.Brown ex Turner) J.Agardh, Sargassum vestitum (R.Brown ex Turner) C.Agardh, Cystophora retroflexa (Labillardière) J.Agardh, Cystophora subfarcinata (Mertens) J.Agardh and Halopteris pseudospicata Sauvageau) and one marine red alga from the Rhodophyta phylum (Laurencia sp.), all of which were collected from Port Phillip Bay, Victoria, Australia. These marine algae were selected for phytochemical evaluation on the basis of three criteria, but in all three instances the intention was to rapidly dereplicate the secondary metabolites present and to avoid lengthy isolations. The marine algae were either selected on the basis of the observed biological activity of the crude extracts, or due to the fact that no previous secondary metabolites had been described from the species of marine alga. The final motivation for the selection was based upon the fact that our research group has previously conducted studies on other closely related Sargassum, Cystophora and Laurencia species and so the intention was to compare the secondary metabolites in closely related species.
Herein, we report the chemical profiling/dereplication conducted using HPLC-NMR and HPLC-MS leading to the identification of seven different structure classes. In total, 22 compounds were detected in the dichloromethane crude extracts of the marine algae studied, of which 16 could be dereplicated.
2. Results and Discussion
The frozen marine algae were extracted with 3:1 methanol/dichloromethane, evaporated under reduced pressure and sequentially solvent partitioned (triturated) into dichloromethane and methanol soluble fractions, respectively. The dichloromethane and methanol crude extracts were initially analysed by off-line analytical HPLC and 1H-NMR analyses and this established that the majority of the secondary metabolites were present in the dichloromethane crude extracts. Based on this, only the dichloromethane extracts were further examined by chemical profiling methodologies (HPLC-NMR & HPLC-MS).
2.1. Chemical Profiling (HPLC-NMR & HPLC-MS)
The dichloromethane crude extracts were subjected to both HPLC-NMR and HPLC-MS chemical profiling and a total of 22 compounds were detected from the seven separate marine algae. Identical HPLC-NMR and HPLC-MS conditions were employed to probe the dichloromethane crude extracts of each alga to allow for comparison between each genera and/or species. Analysis of the stop-flow WET1D proton NMR spectra and extracted UV profiles for each of the compounds concluded the presence of seven distinct chemical structure classes including phenolic acids, phenols, resorcinols, phloroglucinols, xanthophylls, tocotrienols and C15 halogenated acetogenins, which are known to occur in these genera or species of algae [6,7,8,13,14,15,16,17].
Twelve known (1–5, 11–15, 20 and 21) and four new (16–19) compounds (Figure 1) were dereplicated from the dichloromethane crude extracts of the algae. Compounds were dereplicated by analysis of the HPLC-NMR acquired data (WET1D and various combinations of gCOSY, HSQCAD, and gHMBCAD), high resolution HPLC-MS data, and use of the MarinLit database by searching parameters such as the taxonomy (usually genus), UV, MS and NMR data. While some structure classes are unique to certain genera or species, some can be present across various genera or species of algae (Table 1). For instance, in this study, phloroglucinols were detected in S. cf. fallax, C. subfarcinata and C. retroflexa while tocotrienols were exclusive to S. cf. fallax. The xanthophylls, which are known to occur in many marine brown algae [8,18], were found in high abundance in S. vestitum and H. pseudospicata. The specimen of S. decipiens was concluded to produce phenolic acids, phenols, and resorcinols while the specimen of Laurencia sp. could be deduced as containing C15 halogenated acetogenins. Table 2 summarises each of the components detected in the seven marine algae studied together with the search criteria used to dereplicate the structures present in the dichloromethane crude extracts. The amount of compound present in each of the crude extracts was estimated on the basis of the limit of detection (LOD) methodology recently reported for our HPLC-NMR system [19]. In this study, the LOD for five key NMR experiments was established for a given set of parameters. These LODs and the parameters utilized were reviewed, and on this basis, the approximate amount of each compound present in the crude extract for each of the HPLC-NMR analyses undertaken was estimated (see Table 2). In HPLC-NMR analyses, it is imperative to supress signals arising from the HPLC solvents (HDO signal arising from D2O and the CH3CN peak) in order to maximise signal intensity and obtain better quality NMR spectra. Unfortunately, during this process NMR signals of the compound of interest which occur within this suppression region are also inadvertently suppressed.

Table 1.
Chemotaxonomic comparison of the seven marine algae studied and the chemical classes present in each.
| Alga | Chemical Class(es) Present | Compounds Present * |
|---|---|---|
| C. subfarcinata | Phloroglucinols | 18, 19 |
| C. retroflexa | Phloroglucinols | 11-13, 16, 18, 20, 21 |
| S. cf. fallax | Phlroglucinols, tocotrienols | 12, 14, 15, 17, 20, 21 |
| S. decipiens | Phenols, phenolic acids, resorcinols | 1–4 |
| S. vestitum | Xanthophylls | 5 |
| H. pseudospicata | Xanthophylls | 5 |
| Laurencia sp. | Polyhalogenated C15 acetogenins | - |
* Retention times for compounds present are provided in Table 2.

Table 2.
Identification of chemical structure classes present in seven marine algae studied (ordered on the basis of HPLC-NMR retention time, Rt).
| Peak # | Rt (min) | Compound | Structure Class | Marine Alga (~Amount Present in μg) | UV (nm) | MarinLit Search Parameters | New/Known |
|---|---|---|---|---|---|---|---|
| 1 | 2.29 | (2) | Phenolic acid | S. decipiens (500–1000) | 240, 302 | Compound not in MarinLit database | Known |
| 2 | 2.44 | (3) | Phenol | S. decipiens (500–1000) | 236, 301 | Compound not in MarinLit database | Known |
| 3 | 3.42 | (11) | Phloroglucinol | C. retroflexa (750–1000) | 235, 285 | Molecular formula, UV ± 5 | Known |
| 4 | 3.55 | (1) | Phenolic acid | S. decipiens (<100) | 235, 301 | Molecular formula, UV ± 10 | Known |
| 5 | 4.45 | (16) | Phloroglucinol | C. retroflexa (750–1000) | 235, 285 | Compound not in MarinLit database | New |
| 6 | 5.00 | n.a. | C15 acetogenin | Laurencia sp. (<25) | 220, 237 | Genus, UV ± 5, 1 triplet methyl group | Not Identified |
| 7 | 6.05 | n.a. | Unknown | Laurencia sp. (500–1000) | 220, 240, 255 | Unable to dereplicate using any parameters | Not Identified |
| 8 | 6.70 | n.a. | C15 acetogenin | Laurencia sp. (<25) | 220, 237 | Genus, UV ± 5, 1 triplet methyl group | Not Identified |
| 9 | 7.87 | (4) | Resorcinol | S. decipiens (<10) | 229, 276, 281 | Compound not in MarinLit database | Known |
| 10 | 9.98 | (12) | Phloroglucinol | C. retroflexa (500–1000), S. cf. fallax (<250) | 230, 285 | Molecular formula, UV ± 5 | Known |
| 11 | 12.95 | (13) | Phloroglucinol | C. retroflexa (250–750) | 230, 285 | Molecular formula, UV ± 5 | Known |
| 12 | 13.65 | (17) | Phloroglucinol | S. cf. fallax (<250) | 215, 228, 285 | Compound not in MarinLit database | New |
| 13 | 14.53 | (5) | Xanthophyll | S. vestitum (<100), H. pseudospicata (<100) | 450 | Molecular formula | Known |
| 14 | 15.50 | (20) | Phloroglucinol | C. retroflexa (<500), S. cf. fallax (<100) | 212, 228, 285 | Compound not in MarinLit database | Known [13] |
| 15 | 20.15 | (21) | Phloroglucinol | C. retroflexa (<250), S. cf. fallax (<50) | 230, 285 | Compound not in MarinLit database | Known [13] |
| 16 | 21.62 | (14) | Phloroglucinol | S. cf. fallax (<100) | 212, 228, 285 | Molecular formula, UV ± 5 | Known |
| 17 | 22.96 | (18) | Phloroglucinol | C. retroflexa (<100), C. subfarcinata (<50) | 228, 285 | Compound not in MarinLit database | New |
| 18 | 23.16 | n.a. | Phloroglucinol | C. retroflexa (<100) | 238, 288 | Genus, UV ± 5 | Not Identified |
| 19 | 26.71 | n.a. | Xanthophyll | S. vestitum (<5), H. pseudospicata (<5) | 450 | Insufficient data to search MarinLit Database | Not Identified |
| 20 | 30.27 | n.a. | Xanthophyll | S. vestitum (<5), H. pseudospicata (<5) | 450 | Insufficient data to search MarinLit Database | Not Identified |
| 21 | 33.40 | (19) | Phloroglucinol | C. subfarcinata (<50) | 213, 228, 285 | Compound not in MarinLit database | New |
| 22 | 60.80 | (15) | Tocotrienol | S. cf. fallax (<10) | 212, 300 | Class, UV ± 5, contains only singlet aromatic/vinyl CH3 groups, aromatic ring | Known |
n.a. indicates that a structure could not be concluded.

Figure 1.
Compounds and range of structure classes as dereplicated by HPLC-NMR and HPLC-MS in conjunction with the MarinLit database.
2.2. Identification of Phenols, Phenolic Acids and Resorcinols
The specimen of S. decipiens was concluded to contain compounds 1–4, which represent three different structure classes (phenols, phenolic acids and resorcinols). While the phenols and phenolic acids displayed similar UV absorbances (300 nm), the resorcinol exhibited unique UV absorbances (276 and 281 nm). The compound eluting at Rt = 3.55 min was dereplicated from the MarinLit database using the molecular formula and UV absorbances as search parameters. This yielded four possible compounds, but analysis of the NMR data obtained from HPLC-NMR supported only one of these structures. In this way, the compound eluting at Rt = 3.55 min could be identified as 6-tridecylsalicylic acid (1) which had been previously reported from the marine brown alga Caulocystis cephalornithos [20].
The remaining compounds eluting at Rt = 2.29, 2.44 and 7.87 min did not yield any matches in the MarinLit database when using various combinations of UV, mass and molecular formulae, and, when only UV data was searched this resulted in far too many possible structure classes. On the basis of the WET1D NMR spectra and UV profiles of the remaining three compounds (Rt = 2.29, 2.44 and 7.87 min) it could be concluded that they were closely related to 6-tridecylsalicylic acid (1). The marine brown alga C. cephalornithos, which is known to produce 6-tridecylsalicylic acid (1), also produces a variety of structurally related analogues [20,21]. Comparison of the data obtained from HPLC-NMR and HPLC-MS to the data for the compounds reported from C. cephalornithos, allowed for the compounds eluting at Rt = 2.29, 2.44 and 7.87 min to be identified as undecylsalicylic acid (2), 3-undecylphenol (3) and 5-undecylresorcinol (4) respectively. It is important to point out that these compounds do not appear separately in the MarinLit database and care must be taken in such dereplication exercises. This is the first study conducted on S. decipiens and compounds 1–4 have not been reported from the Sargassum genus. This highlights the importance of profiling different genera and species as a means of extending knowledge on marine biodiversity. The HPLC-NMR and HPLC-MS data of the compounds identified were consistent with the data reported previously [20,21,22,23].
2.3. Identification of Xanthophylls
The dichloromethane crude extracts of S. vestitum and H. pseudospicata were found to contain an abundance of xanthophylls. The main compound in the dichloromethane crude extract of each of these two algae (compound eluting at Rt = 14.53 min) was identified as all-trans-fucoxanthin (5), the main pigment found in marine brown algae. This pigment was identified using the MarinLit database by searching the molecular formula obtained by high resolution HPLC-MS, which yielded only one structure. The UV profile, WET1D, gCOSY and HSQCAD NMR spectra of 5 were found to be in agreement with the structure of all-trans-fucoxanthin [24,25] A ROESYAD NMR experiment conducted in stop-flow HPLC-NMR mode confirmed that the configuration for all of the double bonds was trans (Figure 2).
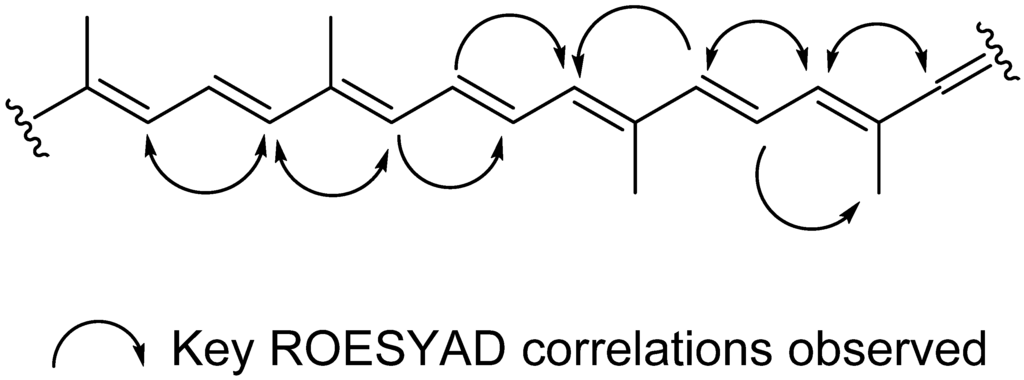
Figure 2.
key ROESYAD correlations confirming the all trans configuration in all-trans-fucoxanthin (5).
A further two structurally related compounds eluting at Rt = 26.71 and 30.27 min were also detected in the dichloromethane crude extract of both S. vestitum and H. pseudospicata, but in much lower quantities. Identification was not possible due to the insufficient quantities present to obtain HPLC-NMR data, and due to the fact that the compounds did not ionize in either positive or negative ESI high resolution HPLC-MS. However, these compounds could be deduced to be two xanthophyll structural analogues based upon comparison of their UV profiles to all-trans-fucoxanthin (5) (Figure 3). This is the first report of all-trans-fucoxanthin (5) and other xanthophylls occurring in the Halopteris genus [8].
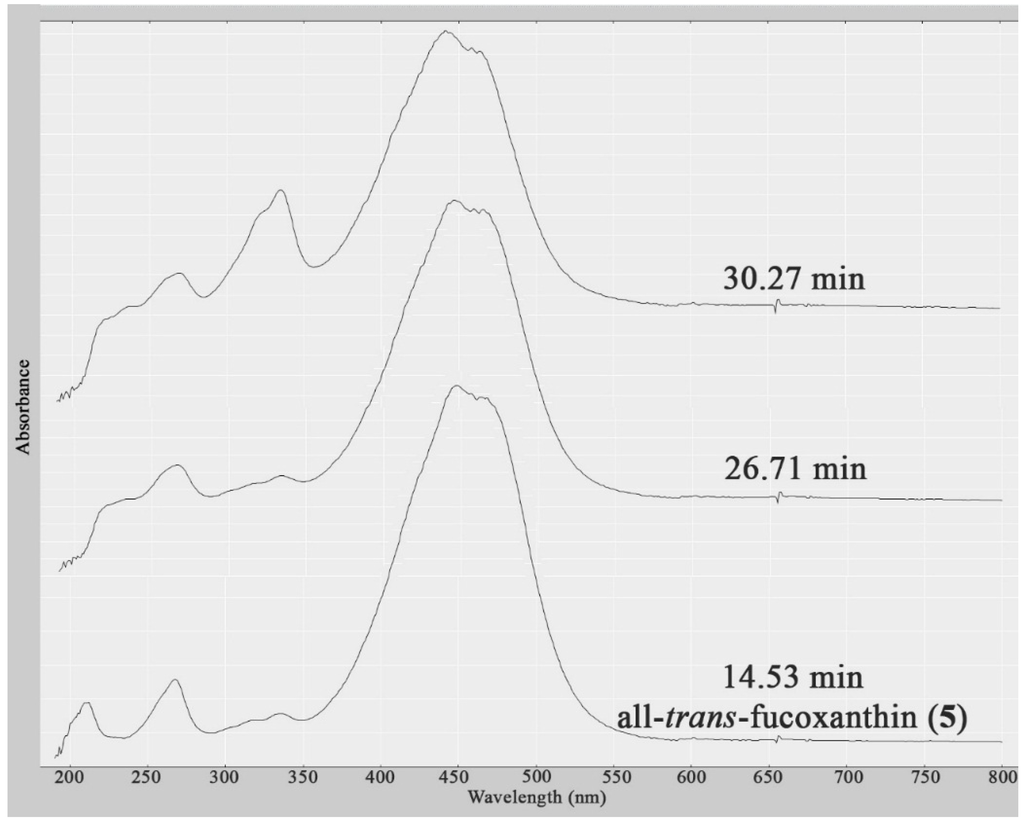
Figure 3.
UV profiles (obtained from HPLC-NMR) of the three related xanthophylls present in the dichloromethane crude extracts of H. pseudospicata and S. vestitum showing their similarity.
2.4. Identification of Halogenated C15 Acetogenins
Profiling of a specimen of Laurencia sp. yielded three compounds (eluting at Rt = 5.00, 6.05 and 6.70 min), which displayed similar WET1D NMR spectra (Figure 4) confirming that they were structurally related. WET1D and gCOSY NMR data were obtained for the compounds eluting at Rt = 5.00 and 6.70 min, while WET1D, gCOSY, HSQCAD and gHMBCAD NMR data was obtained for the compound eluting at Rt = 6.05 min.
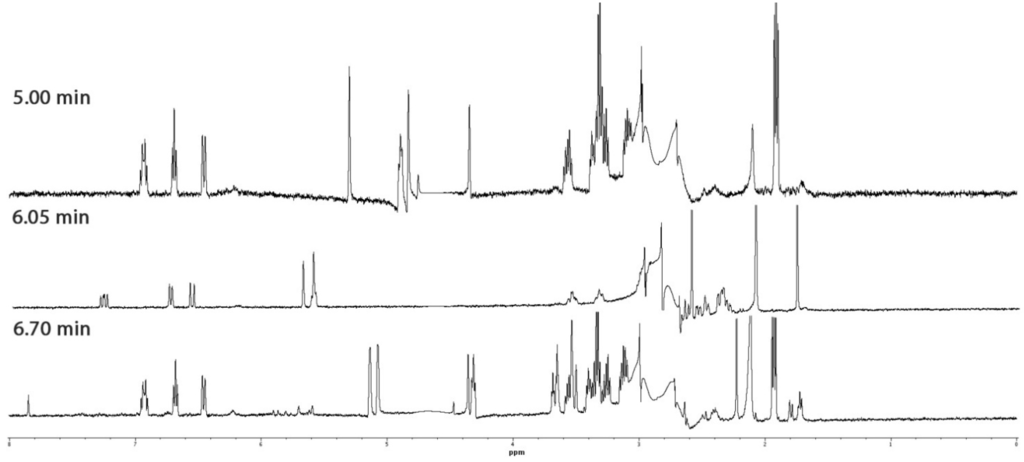
Figure 4.
WET1D NMR spectra (75% CH3CN/D2O, 500 MHz) of compounds eluting at Rt = 5.00, 6.05 and 6.70 min from the dichloromethane crude extract of Laurencia sp.
Based upon the HPLC-NMR WET1D spectra (Figure 4), it was clear that the compounds eluting at Rt = 5.00 and 6.70 min were structural analogues, whereas the compound eluting at Rt = 6.05 min was a closely related structural derivative. A MarinLit search was conducted for the compound eluting at Rt = 5.00 min using the genus, UV and the presence of a single triplet methyl in the WET1D proton NMR spectrum obtained from stop-flow HPLC-NMR. This yielded nine possible structures including five polyhalogenated C15 acetogenins (6–10) (Figure 1). The WET1D NMR data for the compound eluting at Rt = 5.00 min supported the presence of olefinic protons (δ 6.93, dt, J = 7.5, 10.0 Hz; 6.69, t, J = 8.0 Hz; 6.46, d, J = 10.0 Hz), and the presence of halogens was deduced based on the presence of several deshielded methines (δ 5.30, s; 4.90, m; 4.84, s; 4.76, s; 4.35, s). Unfortunately, due to suppression of the methine signals in the D2O suppression region, it was not possible to confirm the total number of halogens present. Furthermore, the compound eluting at Rt = 5.00 min did not ionize in either positive or negative ESI high resolution HPLC-MS, making it impossible to determine a molecular formula. It could only be proposed that the compounds eluting at Rt = 5.00 min and Rt = 6.70 min are of the polyhalogenated C15 acetogenin structure class, but the exact nature of the ring size could not be concluded.
The WET1D NMR spectrum for the compound eluting at Rt = 6.05 min showed slight differences suggesting it was a structural variant, further supported by the UV profile (220, 237 nm for the compounds eluting at Rt = 5.00 and 6.70 min), which also differed, and the absence of any absorbances in the UV profile suggested very little conjugation (220, 240 and 255 nm). Only partial HSQCAD and gHMBCAD NMR data could be obtained for this component and this hindered the ability to dereplicate the compound. Similar to the compounds eluting at Rt = 5.00 and 6.70 min, the compound eluting at Rt = 6.05 min did not ionize in ether positive or negative ESI high resolution HPLC-MS. Various searches conducted of the MarinLit database using different combinations of data (taxonomy, UV and NMR) did not aid in dereplicating a general structure class, and so the compound eluting at Rt = 6.05 min remains unidentified. Off-line isolation would be necessary to unequivocally deduce the structures of these three compounds.
2.5. Identification of Phloroglucinols and Tocotrienols
The dichloromethane crude extracts of S. cf. fallax, C. subfarcinata and C. retroflexa yielded a total of eleven phloroglucinols (11–14, 16–21) and one tocotrienol (15). These two structure classes display similar UV absorbances, with the phloroglucinols typically displaying an absorbance at 285 nm and the tocotrienols at 300 nm, however, differences are observed in the WET1D NMR spectra. This represents the first instance of phloroglucinols and tocotrienols being reported from C. retroflexa and S. cf. fallax. The known phloroglucinols (11–14) and tocotrienol (15) were all dereplicated with use of the MarinLit database utilising the search parameters as indicated in Table 2, and by comparison of the NMR data obtained by HPLC-NMR. One of the phloroglucinols (compound eluting at Rt = 23.16 min) could not be identified due to insufficient quantities present to permit sufficient HPLC-NMR data acquisition and also due to there being no observed molecular ion in the high resolution HPLC-MS. However, based on the similarities of the UV absorbances and WET1D NMR data to 11, it was concluded to represent a phloroglucinol containing an alcohol substituent in the terpene side chain. δ-tocotrienol (15) did not ionize in the high resolution HPLC-MS analysis, but a search of the MarinLit database together with consideration of the WET1D NMR data, resulted in δ-tocotrienol (15) being identified. The HPLC-NMR and HPLC-MS data of the known compounds was consistent with the data previously reported [26,27,28,29].
As well as compounds 11–15, a series of new phloroglucinols were also determined. A MarinLit search conducted on these phloroglucinols using combinations of taxonomy, UV, molecular formulae and NMR data did not result in any confirmed structures, supporting the fact that these represented new phloroglucinols.
High resolution HPLC-MS yielded a molecular formula of C24H34O5 for 16 (observed 401.2337 [M − H]−, calcd. for C24H33O5, 401.2328) and stop-flow HPLC-NMR enabled for WET1D proton, gCOSY, HSQCAD and gHMBCAD NMR data to be obtained (Table 3). Three double bonds were concluded to be present based on the degrees of unsaturation and inspection of the WET1D NMR spectrum (δH 7.30, 1H; 6.80, 1H; 6.48, 1H; 6.32, 1H; 6.28, 1H; 6.23, 1H). A diagnostic methine at δH 4.96 was observed which suggested the presence of an alcohol substituent, together with a diagnostic methylene at δH 3.89, which is typical of the methylene group attached adjacent to the ketone functionality. The gCOSY and gHMBCAD NMR data was able to secure the spin system from this diagnostic methylene (δH 3.89, H-2) to the first olefinic proton (δH 6.32, H-6). Further interpretation of the gCOSY NMR data secured the spin system from H-6 (δH 6.32) to H-14 (δH 3.00). The proton at position H-14 lies on the edge of the suppression region of the acetonitrile signal, which prevented any gCOSY NMR correlations from the proton being observed. Fortunately, in the gHMBCAD NMR spectrum, key correlations were observed from this methylene which allowed for the carbon at position C-15 to be assigned (δC 29.9). The terminal methyl group (δH 1.72, H-18) displayed a key gHMBCAD NMR correlation to the carbon at position C-16 (δC 32.0), confirming its assignment. The protons at positions H-5, H-15 and H-16 could not be assigned as these signals resided in the acetonitrile suppression region. The configuration of the double bonds was established by consideration of the NMR coupling constants and on biosynthetic grounds. Compound 16 represents a structure analogue of the known phloroglucinol 11, which is known to have a 6Z configuration [28].The co-occurrence of 11 and 16 led to the conclusion that their double bond configuration about position 6/7 would be the same when considering their biosynthesis. As such, the structure for 16 was proposed as 9-hydroxy-1-(2,4,6-trihydroxyphenyl)octadeca-6Z,10E,12Z-trien-1-one, attributed the trivial name retroflexanone (16). The configuration of the single stereogenic center at position C-9 remains undefined.

Table 3.
NMR data (500 MHz, 75% CH3CN/D2O, suppression of HDO and CH3CN at δH 4.64 and 2.82 ppm, respectively) for retroflexanone (16) obtained via stop-flow HPLC-NMR.
| Retroflexanone (16) | ||||
|---|---|---|---|---|
| Position | δH (J in Hz) | δC a, Type | gCOSY | gHMBCAD |
| 1 | 207.2, s | |||
| 2 | 3.89, t (7.0) | 44.2, t | 3 | 1, 3, 4 |
| 3 | 2.48, p (7.0) | 25.1, t | 2, 4 | 1, 2, 4, 5 |
| 4 | 2.23, m | 29.9, t | 3 | 2, 3, 5, 6 |
| 5 | SS | 27.8, t | ||
| 6 | 6.32, m | 132.6, d | 7 | |
| 7 | 6.23, m | 126.2, d | 6, 8 | |
| 8 | 3.12, m | 35.8, t | 7 w, 9 | 6, 7, 9 |
| 9 | 4.96, dt (6.5, 7.0) | 72.6, d | 8, 10 | 11 |
| 10 | 6.48, dd (7.0, 15.0) | 136.6, d | 9, 11 | 12 |
| 11 | 7.30, dd (15.0, 11.0) | 126.4, d | 10, 12 | |
| 12 | 6.80, t (11.0) | 128.8, d | 11, 13 w | |
| 13 | 6.28, m | 133.3, d | 12, 14 | |
| 14 | 3.00, m b | ND | 12, 13, 15 | |
| 15 | SS | 29.9, t | ||
| 16 | SS | 32.0, t | ||
| 17 | 2.13, m | 23.0, t | 18 | 16 |
| 18 | 1.72, t (6.5) | 14.3, q | 17 | 16, 17 |
| 1′ | 105.2, s | |||
| 2′ | 165.0, s * | |||
| 3′ | 6.72, s | 95.7, d | 1′, 2′, 5′ | |
| 4′ | ND * | |||
| 5′ | 6.72, s | 95.7, d | 1′, 3′, 6′ | |
| 6′ | 165.0, s * | |||
| 2′-OH | ND | |||
| 4′-OH | ND | |||
| 6′-OH | ND | |||
a carbon assignments based on HSQCAD and gHMBCAD NMR experiments; b signal assigned based on correlations in gCoSY experiment; * signals for C-2′ and C-6′are interchangeable with C4′; w indicates weak or long range correlation; SS Signal suppressed; ND Not Detected.
Based upon the molecular formula of the phloroglucinols eluting at Rt = 13.65, 22.96 and 33.40 min, the number of double bonds present in each compound could be concluded. Unfortunately, NMR and MS data alone is not sufficient to determine the placement of the double bond within the structures. This meant that only tentative structures could be proposed, which are represented as 17–19, respectively. To establish the position of the double bonds in the terpene side chain, off-line approaches such as chemical derivatization or degradation are required. The double bonds were assigned on the basis that olefinic protons that correspond to a cis geometry appear as a narrow multiplet in the proton NMR spectrum [28] which was also observed for the known compounds (12–14) during this study.
Phloroglucinols 20 and 21 (compounds eluting at Rt = 20.15 and 15.50 min) do not appear in the MarinLit database, however, a recent study conducted by the research group describes the first report of these phloroglucinols occurring in C. monilifera and C. subfarcinata [13].
2.6. Anti-Microbial Activity
The crude extracts of S. cf. fallax, S. vestitum, C. subfarcinata, H. pseudospicata and C. retroflexa were assessed for their anti-microbial activity against six bacteria and three fungi (Table 4). The crude extracts of the algae showed varying selectivity and activity across the micro-organisms tested, with S. vestitum, C. subfarcinata and C. retroflexa showing the most promising activity. The crude extracts of the two marine algae specimens containing the xanthophylls (S. vestitum and H. pseudospicata) displayed different biological activity suggesting that the active component(s) was unlikely to be the xanthophylls but other chemical component(s). However, all-trans-fucoxanthin (5) has been reported to display anti-inflammatory, anti-oxidant and anti-tumor activity [30,31,32,33]. The crude extracts of S. cf. fallax, C. subfarcinata, C. retroflexa all showed similar anti-microbial activities which can potentially be attributed to the presence of the phloroglucinols. Phloroglucinols have been demonstrated to possess various anti-microbial activities [34,35,36]. The crude extracts of S. decipiens and Laurencia sp. were not evaluated for their biological activity.
3. Experimental Section
3.1. Marine Alga Material
S. cf. fallax was collected at low tide on January 3, 2003 from St Paul’s Beach, Sorrento, Port Phillip Bay, Victoria, Australia. C. subfarcinata, S. vestitum and H. pseudospicata were collected at low tide on January 22, 2006 from the Borough of Queenscliff (near Point Lonsdale), Port Phillip Bay, Victoria, Australia. Laurencia sp., S. decipiens and C. retroflexa were collected by SCUBA on 21 April 2010 from Governor Reef (near Indented Head), Port Phillip Bay, Victoria, Australia. The marine algae were identified by Dr. Gerald Kraft (The University of Melbourne) and voucher specimens (designated the code numbers 2003-06, 2006-11, 2006-12, 2006-13, 2010-04, 2010-08 and 2010-09, respectively) are deposited at the School of Applied Sciences (Discipline of Applied Chemistry), RMIT University, Melbourne, Australia.
3.2. Extraction
Each of the frozen marine algae were extracted with 3:1 methanol/dichloromethane (1 L). The crude extracts were then decanted and concentrated under reduced pressure and sequentially solvent partitioned (triturated) into dichloromethane and methanol soluble extracts, respectively.

Table 4.
Anti-microbial activity of the crude extracts obtained from S. cf. fallax, C. subfarcinata, C. retroflexa, S. vestitum and H. pseudospicata showing zones of inhibition (mm).
| Crude | Microorganism Concentration (mg/mL) | E. coli ATCC 25922 | S. aureus ATCC 25923 | S. aureus MRSA 344/2-32 | P. aeruginosa ATCC 27853 | S. pyogenes 345/1 | B. subtilis ATCC 19659 | C. albicans ATCC 10231 or 14053* | T. mentagrophytes ATCC 28185 | C. resinae |
|---|---|---|---|---|---|---|---|---|---|---|
| S. vestitum (3:1 MeOH/DCM) | 50 | ND a | NT b | NT b | 3 | NT b | ND a | 3 * | 2 | ND a |
| H. pseudospicata (3:1 MeOH/DCM) | 50 | ND a | NT b | NT b | ND a | NT b | ND a | ND a | ND a | ND a |
| S. cf. fallax (3:1 MeOH/DCM) | 50 | ND a | NT b | NT b | ND a | NT b | 2 | ND a* | ND a | ND a |
| C. subfarcinata (3:1 MeOH/DCM) | 50 | ND a | NT b | NT b | 1 | NT b | ND a | 3* | ND a | ND a |
| C. retroflexa (DCM) | 50 | ND a | 2 | 3 | 5 | ND a | NT b | ND a | NT b | NT b |
| C. retroflexa (MeOH) | 50 | 1 | 4 | 6 | 1 | ND a | NT b | ND a | NT b | NT b |
a indicates no zone of inhibition detected; b indicates not tested; * indicates tested against ATCC 14053. Please note: S. decipiens and Laurencia sp. were not evaluated for biological activity.
3.3. Biological Evaluation
For details on the biological evaluation of the dichloromethane and methanol crude extracts of C. retroflexa please refer to [37]. The 3:1 MeOH/DCM crude extracts of S. cf. fallax, C. subfarcinata, S. vestitum and H. pseudospicata were assessed for anti-microbial activity at the University of Canterbury, Christchurch, New Zealand. A standardized inoculum was prepared by transferring a loop of bacterial/fungal cells, from a freshly grown stock slant culture, into a 10 mL vial of sterile water. This was vortexed and compared to a 5% BaCl2 in water standard to standardize the cell density. This gave a cell density of 108 colony-forming units per millilitre. Ten millilitres of the standardized inoculum was then added to 100 mL of Mueller Hinton or potato dextrose agar (at between 40 and 50 °C) and mixed by swirling, giving a final cell density of 107 colony forming units per millilitre. Five millilitres of this was poured into sterile 85 mm petri dishes. The suspensions were allowed to cool and solidify on a level surface to give a “lawn” of bacteria/fungi over the dish. The crude extract was pipetted onto 6 millimeter diameter filter paper disks and the solvent evaporated. These disks were then placed onto the prepared seeded agar dishes (with appropriate solvent and positive controls) and incubated. Active anti-microbial samples displayed a zone of inhibition outside the disk, which was measured in mm as the radius of inhibition for each bacteria/fungi. The six organisms were Eschericha coli (ATCC 25922), Bacillus subtilis (ATCC 19659) and Pseudomonas aeruginosa (ATCC27853) for the bacteria and, Candida albicans (ATCC 14053), Trichophyton mentagrophytes (ATCC 28185) and Cladosporium resinae for the fungi.
3.4. Chemical Profiling
Chemical profiling was carried out on the dichloromethane soluble extracts of the marine algae employing HPLC-NMR and HPLC-MS methodologies. The dichloromethane extracts were dissolved in HPLC-NMR grade CH3CN and filtered through a 0.45 PTFE membrane filter (Grace Davison Discovery Sciences, Columbia, MD, USA). S. decipiens: 109.1 mg dissolved in 1090 μL (5005 μg per HPLC-NMR injection); C. retroflexa: 185.6 mg dissolved in 2230 μL (4161 μg per HPLC-NMR injection); S. cf. fallax: 166.5 mg dissolved in 1665 μL (5000 μg per HPLC-NMR injection); S. vestitum: 342.8 mg dissolved in 3430 μL (4997 μg per HPLC-NMR injection); H. pseudospicata: 61.6 mg dissolved in 615 μL (5008 μg per HPLC-NMR injection); Laurencia sp.: 216.2 mg dissolved in 2160 μL (5005 μg per HPLC-NMR injection); C. subfarcinata: 741.9 mg dissolved in 7420 μL (4999 μg per HPLC-NMR injection).
3.5. HPLC-NMR & HPLC-MS Conditions
For details of the HPLC-NMR conditions please refer to [37]. For both on-flow and stop-flow HPLC-NMR modes, 50 μL injections of the dichloromethane extracts were injected onto an Agilent Eclipse Plus C18 (150 × 4.6) 5 μ column using a solvent composition of 75% CH3CN/D2O at a flow rate of 1 mL/min. During HPLC-NMR analyses, the HDO and CH3CN signals are suppressed at 4.64 and 2.82 ppm, respectively. In the stop-flow HPLC-NMR mode, WET1D, gCOSY, HSQCAD and gHMBCAD NMR experiments were acquired. HRESILCMS was carried out on an Agilent 6540 Series TOF system in either the positive or negative ionization mode (ESI operation conditions of 12 L/min N2, 325 °C drying gas temperature, and 3500 V capillary voltage) equipped with an Agilent 1260 Infinity Binary Pump, Agilent 1290 Infinity Autosampler and Agilent 1260 DAD detector (Agilent, Santa Clara, CA, USA). The instrument was calibrated using the “Agilent Tuning Mix” with purine as the reference compound and the Hewlett-Packard standard HP0921. The separations were carried out using an Agilent Eclipse Plus C18 (4.6 × 150) 5 μ column using a solvent composition of 75% CH3CN/H2O at a flow rate of 1 mL/min.
3.6. On-Line (HPLC-NMR & HPLC-MS) Partial Characterization of Compounds
1-(2,4,6-trihydroxyphenyl)-hexadecadien-1-one (unknown double bond positions) (17): HPLC-NMR WET1D NMR (500 MHz, 75% CH3CN/D2O, suppression of HDO and CH3CN at t δH 4.64 and 2.82 ppm, respectively) obtained from stop-flow mode δ 6.71 (2H, s, H-3′/H-5′), 6.22 (4H, m, CHCH), 3.89 (2H, t, J = 8.0, H-2), 3.62 (2H, m, CHCH2CH), 2.11 (m, CH2)*, 1.77 (3H, t, J = 8.0; HRESILCMS; m/z 359.2226 (calcd for C22H31O4, 359.2222).
1-(2,4,6-trihydroxyphenyl)-octadecadien-1-one (unknown double bond positions) (18): HPLC-NMR WET1D NMR (500 MHz, 75% CH3CN/D2O, suppression of HDO and CH3CN at t δH 4.64 and 2.82 ppm, respectively) obtained from stop-flow mode δ 6.70 (2H, s, H-3’/H-5’), 6.20 (4H, m, CHCH), 3.87 (2H, t, J = 7.5, H-2), 3.61 (2H, dd, J = 6.0, 7 Hz, CHCH2CH), 2.46 (2H, m, H-3), 2.11 (m, CH2)*, 1.71 (3H, t, J = 7.0; HRESILCMS; m/z 387.2539 (calcd for C24H35O4, 387.2535).
1-(2,6-dihydroxy-4-methoxyphenyl)-octadecatrien-1-one (unknown double bond positions) (19): HPLC-NMR WET1D NMR (500 MHz, 75% CH3CN/D2O, suppression of HDO and CH3CN at t δH 4.64 and 2.82 ppm, respectively) obtained from stop-flow mode δ 6.82 (2H, s, H-3’/H-5’), 6.21 (6H, m, CHCH), 3.91 (2H, t, J = 7.0, H-2), 3.66 (4H, m, CHCH2CH), 2.49 (2H, m, H-3), 2.25 (2H, m, H-4), 2.10, (m, CH2)*, 1.70 (3H, t, J = 6.5; HRESILCMS; m/z 399.2539 (calcd for C25H35O4, 399.2535).
Please refer to the supplementary information file containing UV profiles, HPLC-NMR data including tabulated NMR data and the high resolution HPLC-MS chromatograms for all remaining compounds (these are listed in order of retention time).
4. Conclusions
This study has demonstrated the importance and ability of HPLC-NMR in conjunction with high resolution HPLC-MS and databases such as MarinLit to rapidly dereplicate the identity of secondary metabolites from a range of marine algae. The chemical diversity between, and within, marine algae genera was highlighted (See Table 1 and Table 2). The phloroglucinols (11–14, 16–21) are an example of compounds that can be found occurring across different species of the same genus (Cystophora), while the xanthophylls (15) provide an example of compounds that can extend over different genera (Halopteris and Sargassum). The application of HPLC-NMR to investigate either genera or species of marine algae which were previously not studied (S. decipiens, S. vestitum, S. cf. fallax, and H. pseudospicata) and to rapidly identify structure classes present, without the need for traditional and lengthy isolation procedures is noteworthy. Finally, this study also highlights the advantage of using HPLC-NMR over HPLC-MS data for dereplication, especially when compounds do not easily ionize during HPLC-MS analyses. In such cases, crucial NMR data, which can be acquired via HPLC-NMR, provides important structural features and connectivity, which can enable a compound to be dereplicated. Unlike HPLC-MS, HPLC-NMR also allows for the differentiation of structural isomers.
Acknowledgments
The Marine And Terrestrial Natural Product (MATNAP) research group would like to thank Roderick Watson (MAFRI), Rick Tinker, Daniel Dias and Priyanka Reddy for their assistance with the collection of the marine algae; Gerald Kraft (The University of Melbourne) for the taxonomic identification of the marine algae; Nerida Thurbon (School of Applied Sciences (Discipline of Biotechnology and Biological Sciences), Science Engineering and Health, RMIT University) for providing access to the micro-organisms to conduct the anti-microbial assays and for her invaluable technical support; Gill Ellis (University of Canterbury, Christchurch, New Zealand) for conducting some of the anti-microbial biological testing; Sally Duck (School of Chemistry, Faculty of Science, Monash University) for conducting the high resolution mass spectrometric analyses (HPLC-MS). Robert Brkljača would like to acknowledge his Australian Postgraduate Award (APA) scholarship that has supported his studies. Emrehan Gӧker was a recipient of the School of Applied Sciences Vacation scholarship that supported his BSc (AppSc) (honours) studies on C. retroflexa.
Author Contributions
Experimental work for S. cf. fallax, C. subfarcinata, S. vestitum, H. pseudospicata, Laurencia sp., and S. decipiens was conducted by R. Brkljača. Experimental work for C. retroflexa was conducted by E. S. Göker. Manuscript was prepared by R. Brkljača and S. Urban.
Supplementary Information
HPLC-NMR and HPLC-MS data and UV profiles for the crude extracts of the seven marine algae are included (these are listed in order of retention time). This material is available free of charge via MDPI Website.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guiry, M.D.; Guiry, G.M. AlgaeBase. Available online: http://www.algaebase.org (accessed on 6 March 2015).
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Pereira, A.R.; Gerwick, W.H. The Chemistry of Marine Algae and Cyanobacteria. In Handbook of Marine Natural Products; Fattorusso, E., Gerwick, W.H., Taglialatela-Scafati, O., Eds.; Springer: London, England, UK, 2012. [Google Scholar]
- Sailler, B.; Glombitza, K.-W. Halogenated phlorethols and fucophlorethols from the brown alga Cystophora retroflexa. Nat. Toxins 1999, 7, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Sailler, B.; Glombitza, K.-W. Phlorethols and fucophlorethols from the brown alga Cystohpora retroflexa. Phytochemistry 1999, 50, 869–881. [Google Scholar] [CrossRef]
- Czeczuga, B.; Taylor, F.J. Carotenoid content in some species of the brown and red algae from the coastal area of New Zealand. Biochem. Syst. Ecol. 1986, 15, 5–8. [Google Scholar] [CrossRef]
- Laird, D.W.; Bennett, S.; Bian, B.; Sauer, B.; Wright, K.; Hughes, V.; van Altena, I.A. Chemical investigation of seven Australasian Cystophora species: New chemistry and taxonomic insights. Biochem. Syst. Ecol. 2010, 38, 187–194. [Google Scholar] [CrossRef]
- MarinLit Database. Available online: http://pubs.rsc.org/marinlit/ (accessed on 6 March 2015).
- Kazlauskas, R.; Murphy, P.T.; Wells, R.J.; Daly, J.J.; Oberhansli, W.E. Heterocladol, a halogenated selinane sesquiterpene of biosynthetic significance from the red alga Laurencia filiformis: Its isolation, crystal structure and absolute configuration. Aust. J. Chem. 1977, 30, 2679–2687. [Google Scholar] [CrossRef]
- Capon, R.; Ghisalberti, E.L.; Jefferies, P.R.; Skelton, B.W.; White, A.H. Sesquiterpene metabolites from Laurencia filiformis. Tetrahedron 1981, 37, 1613–1621. [Google Scholar] [CrossRef]
- Capon, R.J.; Ghisalberti, E.L.; Mori, T.A.; Jefferies, P.R. Sesquiterpenes from Laurencia spp. J. Nat. Prod. 1988, 51, 1302–1304. [Google Scholar] [CrossRef]
- Dias, D.A.; White, J.M.; Urban, S. Laurencia filiformis: Phytochemical profiling by conventional and HPLC-NMR approaches. Nat. Prod. Commun. 2009, 4, 157–172. [Google Scholar] [PubMed]
- Brkljača, R.; Urban, S. Dereplication (HPLC-NMR & HPLC-MS) and structural identification studies of marine brown algae of the genus Cystophora. Phytochemistry 2015. submitted for publication. [Google Scholar]
- Suzuki, M.; Takahashi, Y.; Matsuo, Y.; Guiry, M.D.; Masuda, M. Scanlonenyne, a novel halogenated C15 acetogenin from the red alga Laurencia obtusa in Irish waters. Tetrahedron 1997, 53, 4271–4278. [Google Scholar] [CrossRef]
- San-Martin, A.; Darias, J.; Soto, H.; Contreras, C.; Herrera, J.S.; Rovirosa, J. A new C15 acetogenin from the marine alga Laurencia claviformis. Nat. Prod. Lett. 1997, 10, 303–311. [Google Scholar] [CrossRef]
- Suzuki, M.; Nakano, S.; Takahashi, Y.; Abe, T.; Masuda, M. Bisezakyne-A and -B, halogenated C15 acetogenins from a Japanese Laurencia species. Phytochemistry 1999, 51, 657–662. [Google Scholar] [CrossRef]
- Aydogmus, Z.; Imre, S.; Ersoy, L.; Wray, V. Halogenated secondary metabolites from Laurencia obtusa. Nat. Prod. Res. 2004, 18, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Takaichi, S. Carotenoids in Algae: Distributions, Biosyntheses and Functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef] [PubMed]
- Brkljača, R.; Urban, S. Limit of detection studies for application to natural product identification using high performance liquid chromatography coupled to nuclear magnetic resonance spectroscopy. J. Chromatogr. A 2015, 1375, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, R.; Mulder, J.; Murphy, P.T.; Wells, R.J. New metabolites from the brown alga Caulocystis cephalornithos. Aust. J. Chem. 1980, 33, 2097–2101. [Google Scholar] [CrossRef]
- Narkowicz, C.K.; Blackman, A.J. Further acetogenins from Tasmanian collections of Caulocystis cephalornithos demonstrating chemical variability. Biochem. Syst. Ecol. 2006, 34, 635–641. [Google Scholar] [CrossRef]
- Spencer, G.F.; Tjarks, L.W.; Kleiman, R. Alkyl and phenylalkyl anacardic acids from Knema elegans seed oil. J. Nat. Prod. 1980, 43, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, Y.; Masuya, T.; Tori, M.; Campbell, E.O. Long chain alkyl phenols from the liverwort Schistochila appendiculata. Phytochemistry 1987, 26, 735–738. [Google Scholar] [CrossRef]
- Englert, G.; Bjornland, T.; Liaaen-Jensen, S. 1D and 2D NMR studies of some allenic carotenoids of the fucoxanthin series. Magn. Reson. Chem. 1990, 28, 519–528. [Google Scholar] [CrossRef]
- Haugan, J.A.; Englert, G.; Glinz, E.; Liaaen-Jensen, S. Algal carotenoids. 48. Structural assignments of geometrical isomers of fucoxanthin. Acta Chem. Scand. 1992, 46, 389–395. [Google Scholar] [CrossRef]
- Ohnmacht, S.; West, R.; Simionescu, R.; Atkinson, J. Assignment of the 1H and 13C NMR of tocotrienols. Magn. Reson. Chem. 2008, 46, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Gregson, R.P.; Kazlauskas, R.; Murphy, P.T.; Wells, R.J. New metabolites from the brown alga Cystophora torulosa. Aust. J. Chem. 1977, 30, 2527–2532. [Google Scholar] [CrossRef]
- Kazlauskas, R.; King, L.; Murphy, P.T.; Warren, R.G.; Wells, R.J. New metabolites from the Brown Algal Genus Cystophora. Aust. J. Chem. 1981, 34, 439–447. [Google Scholar] [CrossRef]
- Amico, V.; Currenti, R.; Oriente, G.; Piatelli, M.; Tringali, C. A phloroglucinol derivative from the brown alga Zonaria tournefortii. Phytochemistry 1981, 20, 1451–1453. [Google Scholar] [CrossRef]
- Kim, S.-K.; Pangestuti, R. Biological activities and potential health benefits of fucoxanthin derived from marine brown algae. Adv. Food Nutr. Res. 2011, 64, 111–128. [Google Scholar] [PubMed]
- Tan, C.-P.; Hou, Y.-H. First evidence for the anti-inflammatory activity of fucoxanthin in high-fat-diet-induced obesity in mice and the antioxidant functions in PC12 cells. Inflammation 2014, 37, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-N.; Heo, S.-J.; Kang, S.-M.; Ahn, G.; Jeon, Y.-J. Fucoxanthin induces apoptosis in human leukemia HL-60 cells through a ROS-mediated Bcl-xL pathway. Toxicol. Vitro 2010, 24, 1648–1654. [Google Scholar] [CrossRef]
- Xia, S.; Wang, K.; Wan, L.; Li, A.; Hu, Q.; Zhang, C. Production, charaterization, and antioxidant activity of fucoxanthin from the marine diatom Odontella aurita. Mar. Drugs 2013, 11, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Wisespongpand, P.; Kuniyoshi, M. Bioactive phloroglucinols from the brown alga Zonaria diesingiana. J. Appl. Phycol. 2003, 15, 225–228. [Google Scholar] [CrossRef]
- Gerwick, W.; Fenical, W. Phenolic lipids from the related marine algae of the order Dictyotales. Phytochemistry 1982, 21, 633–637. [Google Scholar] [CrossRef]
- Ravi, B.N.; Murphy, P.T.; Lidgard, R.O.; Warren, R.G.; Wells, R.J. C18 terpenoid metabolites of the brown alga Cystophora moniliformis. Aust. J. Chem. 1982, 35, 171–182. [Google Scholar] [CrossRef]
- Brkljača, R.; Urban, S. Chemical profiling (HPLC-NMR & HPLC-MS), isolation, and identification of bioactive meroditerpenoids from the southern Australian marine brown alga Sargassum paradoxum. Mar. Drugs 2015, 13, 102–127. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).