LC-MS-Based Metabolomics Study of Marine Bacterial Secondary Metabolite and Antibiotic Production in Salinispora arenicola
Abstract
:1. Introduction
2. Results
2.1. Production of Rifamycins at Three Time Points of Bacterial Growth
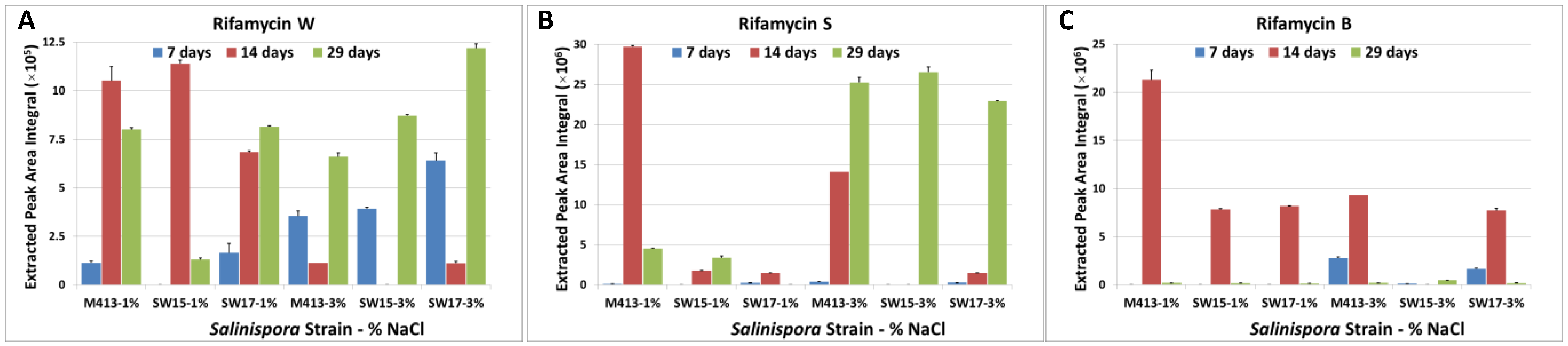
2.2. Comparison of Secondary Metabolite Production in Three Strains (M413, SW15 and SW17) at Two Salt Concentrations (1% and 3% NaCl) at Three Time Points (7, 14 and 29 Days)

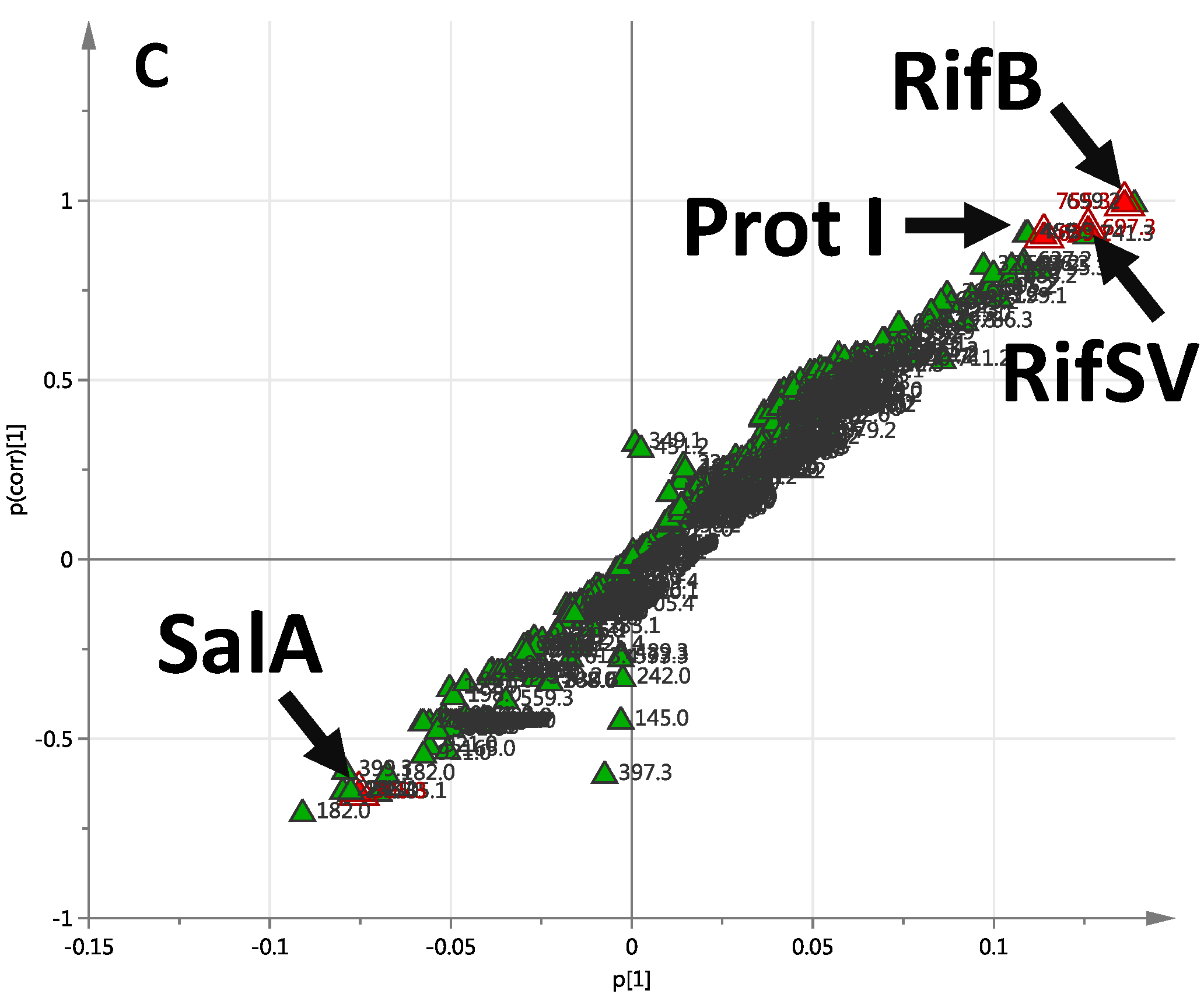
2.2.1. Day 7 Time Point

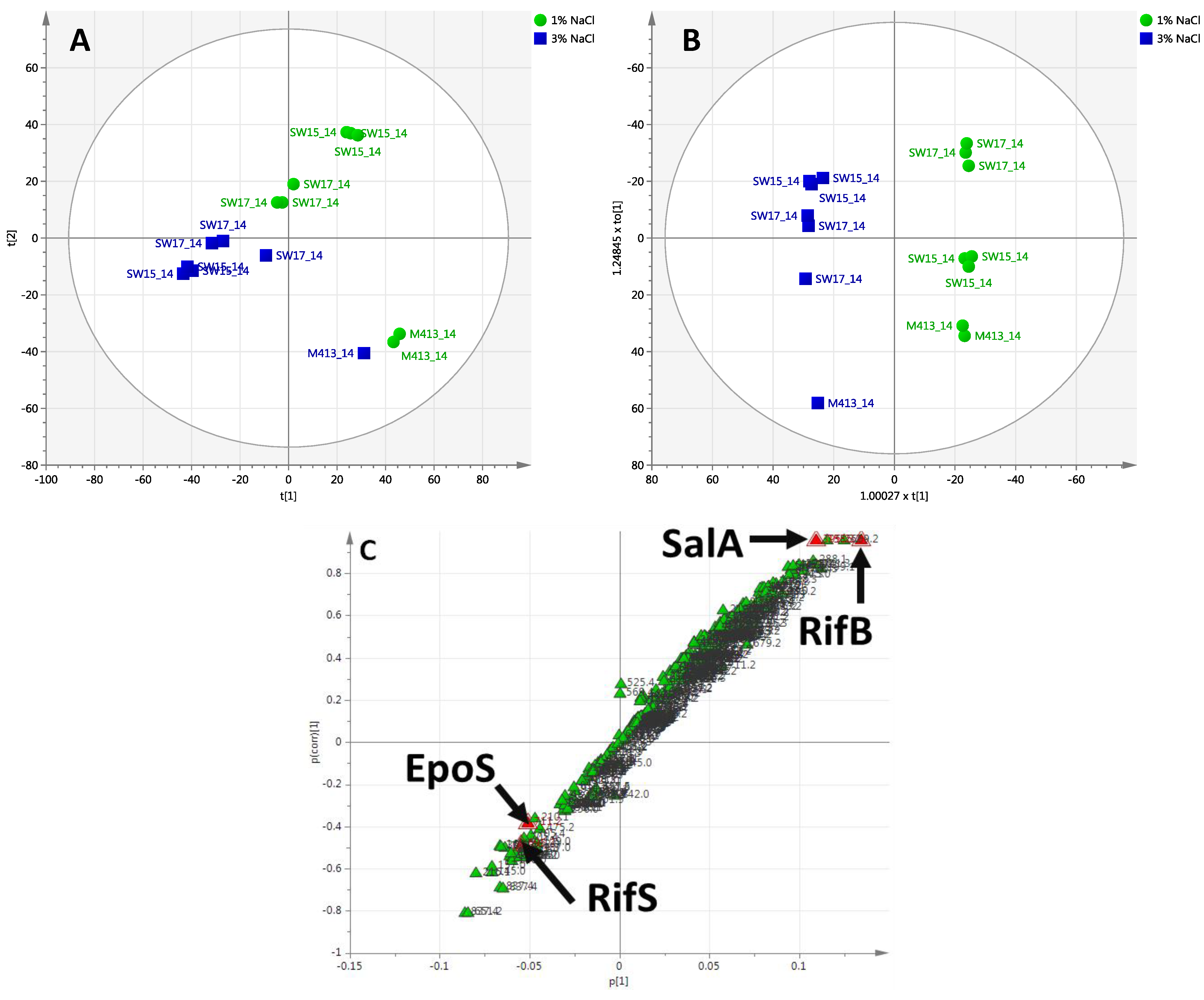
2.2.2. Day 14 Time Point
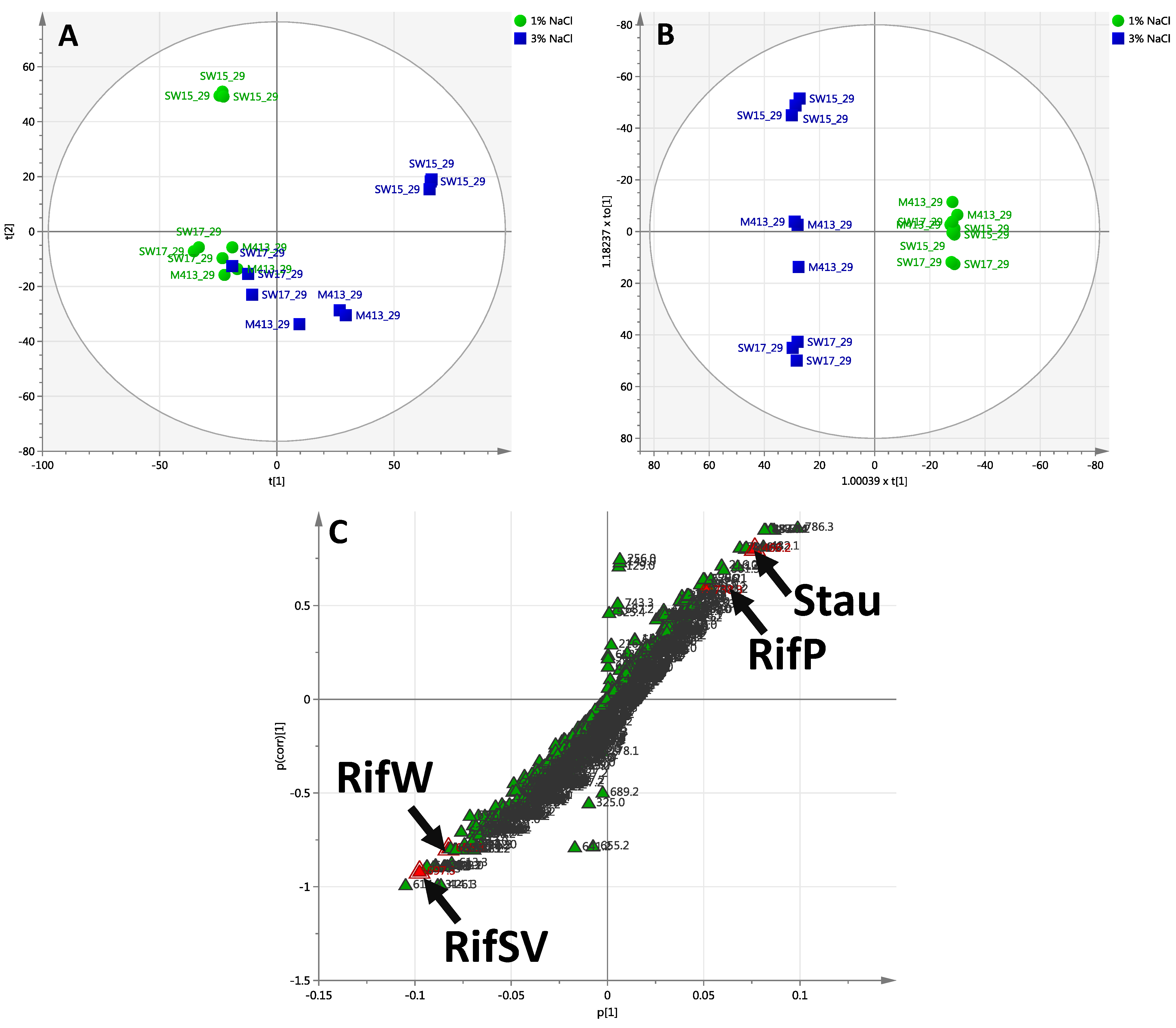
2.2.3. Day 29 Time Point
| Molecular Formula | Score * | Mass ** | Polarity | RT (min) | Mass Error (MFG, ppm) | Compound *** | Day(s) Observed |
|---|---|---|---|---|---|---|---|
| C38H49NO12S | 95.35 | 743.2975 | Negative | 15.28 | 3.97 | Awamycin | 14 |
| C39H49NO14 | 94.34 | 755.3153 | Negative | 8.80 | 0.52 | Rifamycin B | 7, 14 |
| C38H46N2O11S | 91.91 | 738.2822 | Negative | 10.16 | 3.50 | Rifamycin P | 14, 29 |
| C37H45NO13 | 90.34 | 711.2891 | Positive | 10.34 | 2.60 | 18,19-Epoxyrifamycin S | 14, 29 |
| C22H37NO6 | 89.72 | 411.2621 | Positive | 9.40 | 1.44 | Saliniketal B | 7, 14, 29 |
| C39H47NO15 | 87.34 | 769.2946 | Negative | 13.54 | 1.52 | Rifamycin Y | 14 |
| C37H47NO12 | 90.27 | 697.3098 | Negative | 22.10 | 1.81 | Rifamycin SV | 7, 14, 29 |
| C36H45NO12 | 68.75 | 683.2942 | Negative | 15.42 | 0.54 | 16,17 Dehydroxyrifamycin G | 29 |
| C37H45NO12 | 84.34 | 695.2942 | Negative | 22.61 | 3.43 | Rifamycin S | 7, 14, 29 |
| C22H37NO5 | 81.72 | 395.2672 | Positive | 12.58 | 1.44 | Saliniketal A | 7, 14, 29 |
| C12H12N2O2 | 86.05 | 216.0899 | Negative | 5.86 | 1.36 | Mansouramycin A | 7, 14 |
| C28H26N4O3 | 74.68 | 466.2005 | Positive | 18.47 | 3.19 | Staurosporine | 7, 14, 29 |
| C35H45NO11 | 95.67 | 655.2993 | Negative | 7.25 | 1.1 | Rifamycin W | 14, 29 |
| C35H45NO10 | 95.58 | 639.3043 | Negative | 9.19 | 1.0 | Protorifamycin I | 14, 29 |
| C36H43N5O6 | 94.23 | 641.3213 | Positive | 13.72 | 1.94 | Cycloaspeptide A | 14, 29 |
| C35H43NO11 | 67.65 | 653.2836 | Negative | 8.62 | 1.84 | (34aR)-Rifamycin W hemiacetal | 14, 29 |
3. Discussion
4. Experimental Section
4.1. Sample Preparation
4.2. UHPLC-QToF-MS Analysis
4.3. Data Processing and Molecular Formula Generation
4.4. Chemometric Analyses and Compound Identification
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2013, 30, 237–323. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.L.; Moore, B.S. A sea of biosynthesis: Marine natural products meet the molecular age. Nat. Prod. Rep. 2011, 28, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Tsueng, G.; Lam, K.S. A low-sodium-salt formulation for the fermentation of salinosporamides by Salinispora tropica strain NPS21184. Appl. Microbiol. Biotechnol. 2008, 78, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Tsueng, G.; Teisan, S.; Lam, K.S. Defined salt formulations for the growth of Salinispora tropica strain NPS21184 and the production of salinosporamide A (NPI-0052) and related analogs. Appl. Microbiol. Biotechnol. 2008, 78, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Kawasaki, S.; Unemoto, T. Roles of K+ and Na+ in pH homeostasis and growth of the marine bacterium Vibrio alginolyticus. J. Gen. Microbiol. 1992, 138, 1271–1276. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Z.; Sun, K.; Noumi, M.; Ōmura, S.; Namikoshi, M. Effect of sea water concentration on hyphal growth and antimicrobial metabolite production in marine fungi. Mycoscience 2001, 42, 455–459. [Google Scholar] [CrossRef]
- Rüger, H.-J.; Hentzschel, G. Mineral salt requirements of Bacillus globisporus subsp. marinus strains. Arch. Microbiol. 1980, 126, 83–86. [Google Scholar] [CrossRef]
- Tsueng, G.; Lam, K. A preliminary investigation on the growth requirement for monovalent cations, divalent cations and medium ionic strength of marine actinomycete Salinispora. Appl. Microbiol. Biotechnol. 2010, 86, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Unemoto, T.; Hayashi, M. Na+-translocating NADH-quinone reductase of marine and halophilic bacteria. J. Bioenerg. Biomembr. 1993, 25, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Penn, K.; Jensen, P.R. Comparative genomics reveals evidence of marine adaptation in Salinispora species. BMC Genomics 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Vidgen, M.; Hooper, J.N.A.; Fuerst, J. Diversity and distribution of the bioactive actinobacterial genus Salinispora from sponges along the Great Barrier Reef. Antonie Van Leeuwenhoek 2011, 101, 603–618. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Williams, P.G.; Oh, D.-C.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Garson, M.J.; Fuerst, J.A. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 2005, 7, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, L.; Jensen, P.R.; Freel, K.C.; Brown, R.; Jones, A.L.; Kim, B.Y.; Goodfellow, M. Salinispora pacifica sp. nov., an actinomycete from marine sediments. Antonie Van Leeuwenhoek 2013, 103, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.A.; Fenical, W.; Jensen, P.R.; Kauffman, C.A.; Mincer, T.J.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 2005, 55, 1759–1766. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Hewavitharana, A.K.; Shaw, P.N.; Fuerst, J.A. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl. Environ. Microbiol. 2006, 72, 2118–2125. [Google Scholar] [CrossRef] [PubMed]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Mincer, T.J.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 2002, 68, 5005–5011. [Google Scholar] [CrossRef] [PubMed]
- Tsueng, G.; Lam, K.S. Growth of Salinispora tropica strains CNB440, CNB476, and NPS21184 in nonsaline, low-sodium media. Appl. Microbiol. Biotechnol. 2008, 80, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Schupp, T.; Traxler, P.; Auden, J. New rifamycins produced by a recombinant strain of Nocardia mediterranei. J. Antibiot. 1981, 34, 965. [Google Scholar] [CrossRef] [PubMed]
- Aristoff, P.A.; Garcia, G.A.; Kirchhoff, P.D.; Hollis, S.H. Rifamycins-obstacles and opportunities. Tuberculosis 2010, 90, 94–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Fuerst, J.A. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: Culture-dependent and culture-independent approaches. Environ. Microbiol. 2006, 8, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.K.; Hewavitharana, A.K.; Webb, R.; Shaw, P.N.; Fuerst, J.A. Developmental cycle and pharmaceutically relevant compounds of Salinispora actinobacteria isolated from Great Barrier Reef marine sponges. Appl. Microbiol. Biotechnol. 2013, 97, 3097–3108. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.K.; Hodson, M.P.; Hewavitharana, A.K.; Bose, U.; Shaw, P.N.; Fuerst, J.A. Effects of salinity on antibiotic production in sponge-derived Salinispora actinobacteria. J. Appl. Microbiol. 2014, 117, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Hewavitharana, A.K.; Vidgen, M.E.; Ng, Y.K.; Shaw, P.N.; Fuerst, J.A.; Hodson, M.P. Discovering the recondite secondary metabolome spectrum of Salinispora species: A study of inter-species diversity. PLoS One 2014, 9, e91488. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Hodson, M.P.; Dear, G.J.; Roberts, A.D.; Haylock, C.L.; Ball, R.J.; Plumb, R.S.; Stumpf, C.L.; Griffin, J.L.; Haselden, J.N. A gender-specific discriminator in Sprague-Dawley rat urine: The deployment of a metabolic profiling strategy for biomarker discovery and identification. Anal. Biochem. 2007, 362, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Goodacre, R.; Neyses, L.; Mamas, M. Integration of metabolomics in heart disease and diabetes research: Current achievements and future outlook. Bioanalysis 2011, 3, 2205–2222. [Google Scholar] [CrossRef] [PubMed]
- Hewavitharana, A.K.; Shaw, P.N.; Kim, T.K.; Fuerst, J.A. Screening of rifamycin producing marine sponge bacteria by LC-MS-MS. J. Chromatogr. B 2007, 852, 362–366. [Google Scholar] [CrossRef]
- Bose, U.; Hodson, M.P.; Shaw, P.N.; Fuerst, J.A.; Hewavitharana, A.K. Two peptides, cycloaspeptide A and nazumamide A, from marine actinobacteria Salinispora. Nat. Prod. Commun. 2014, 9, 545–546. [Google Scholar] [PubMed]
- MacLeod, R.A. The question of the existence of specific marine bacteria. Bacteriol. Rev. 1965, 29, 9. [Google Scholar] [PubMed]
- Wilson, M.C.; Gulder, T.A.M.; Mahmud, T.; Moore, B.S. Shared biosynthesis of the saliniketals and rifamycins in Salinispora arenicola is controlled by the sare1259-encoded cytochrome P450. J. Am. Chem. Soc. 2010, 132, 12757–12765. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, U.; Saxena, B.; Chisti, Y. Biotransformations of rifamycins: Process possibilities. Biotechnol. Adv. 1992, 10, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Freel, K.C.; Nam, S.J.; Fenical, W.; Jensen, P.R. Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl. Environ. Microbiol. 2011, 77, 7261–7270. [Google Scholar] [CrossRef] [PubMed]
- Mitova, M.; Tommonaro, G.; de Rosa, S. A novel cyclopeptide from a bacterium associated with the marine sponge Ircinia muscarum. Zeitschrift Fuer Naturforschung. C 2003, 58, 740–745. [Google Scholar]
- Jones, A.M.; Berkelmans, R. Flood impacts in Keppel Bay, southern Great Barrier Reef in the aftermath of cyclonic rainfall. PLoS One 2014, 9, e84739. [Google Scholar] [CrossRef] [PubMed]
- Hodson, M.P.; Dear, G.J.; Griffin, J.L.; Haselden, J.N. An approach for the development and selection of chromatographic methods for high-throughput metabolomic screening of urine by ultra pressure LC-ESI-ToF-MS. Metabolomics 2009, 5, 166–182. [Google Scholar] [CrossRef]
- Sangster, T.; Major, H.; Plumb, R.; Wilson, A.J.; Wilson, I.D. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst 2006, 131, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabolomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Wold, S. Multi- and Megavariate Data Analysis—Principles and Applications; Umetrics, A.B., Ed.; Umetrics: Umeå, Sweden, 2001. [Google Scholar]
- Faulkner, D.J. Marine natural products. Nat. Prod. Rep. 1988, 5, 613–663. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, D.J. Marine natural products. Nat. Prod. Rep. 1996, 13, 75–125. [Google Scholar] [CrossRef] [PubMed]
- Bernan, V.S.; Greenstein, M.; Maiese, W.M. Marine microorganisms as a source of new natural products. Adv. Appl. Microbiol. 1997, 43, 57–90. [Google Scholar] [PubMed]
- El Sayed, K.A.; Bartyzel, P.; Shen, X.; Perry, T.L.; Zjawiony, J.K.; Hamann, M.T. Marine natural products as antituberculosis agents. Tetrahedron 2000, 56, 949–953. [Google Scholar] [CrossRef]
- Faulkner, D.J. Marine natural products. Nat. Prod. Rep. 2001, 18, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Proksch, P.; Edrada, R.; Ebel, R. Drugs from the seas–current status and microbiological implications. Appl. Microbiol. Biotechnol. 2002, 59, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Proksch, P.; Ebel, R.; Edrada, R.A.; Wray, V.; Steube, K. Bioactive natural products from marine invertebrates and associated fungi. Prog. Mol. Subcell. Biol. 2003, 37, 117–142. [Google Scholar] [PubMed]
- Newman, D.J.; Cragg, G.M. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 2004, 67, 1216–1238. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2004, 21, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Simmons, T.L.; Andrianasolo, E.; McPhail, K.; Flatt, P.; Gerwick, W.H. Marine natural products as anticancer drugs. Mol. Cancer Ther. 2005, 4, 333–342. [Google Scholar] [PubMed]
- König, G.M.; Kehraus, S.; Seibert, S.F.; Abdel-Lateff, A.; Müller, D. Natural products from marine organisms and their associated microbes. ChemBioChem 2006, 7, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2006, 23, 26–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 2006, 33, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2007, 24, 31–86. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Ali, M.S.; Hussain, S.; Jabbar, A.; Ashraf, M.; Lee, Y.S. Marine natural products of fungal origin. Nat. Prod. Rep. 2007, 24, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2008, 25, 35–94. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Disc. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Gulder, T.A.; Moore, B.S. Chasing the treasures of the sea—Bacterial marine natural products. Curr. Opin. Microbiol. 2009, 12, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2009, 26, 170–244. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2010, 27, 165–237. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bose, U.; Hewavitharana, A.K.; Ng, Y.K.; Shaw, P.N.; Fuerst, J.A.; Hodson, M.P. LC-MS-Based Metabolomics Study of Marine Bacterial Secondary Metabolite and Antibiotic Production in Salinispora arenicola. Mar. Drugs 2015, 13, 249-266. https://doi.org/10.3390/md13010249
Bose U, Hewavitharana AK, Ng YK, Shaw PN, Fuerst JA, Hodson MP. LC-MS-Based Metabolomics Study of Marine Bacterial Secondary Metabolite and Antibiotic Production in Salinispora arenicola. Marine Drugs. 2015; 13(1):249-266. https://doi.org/10.3390/md13010249
Chicago/Turabian StyleBose, Utpal, Amitha K. Hewavitharana, Yi Kai Ng, Paul Nicholas Shaw, John A. Fuerst, and Mark P. Hodson. 2015. "LC-MS-Based Metabolomics Study of Marine Bacterial Secondary Metabolite and Antibiotic Production in Salinispora arenicola" Marine Drugs 13, no. 1: 249-266. https://doi.org/10.3390/md13010249
APA StyleBose, U., Hewavitharana, A. K., Ng, Y. K., Shaw, P. N., Fuerst, J. A., & Hodson, M. P. (2015). LC-MS-Based Metabolomics Study of Marine Bacterial Secondary Metabolite and Antibiotic Production in Salinispora arenicola. Marine Drugs, 13(1), 249-266. https://doi.org/10.3390/md13010249





