New Ikarugamycin Derivatives with Antifungal and Antibacterial Properties from Streptomyces zhaozhouensis
Abstract
:1. Introduction
2. Results and Discussion
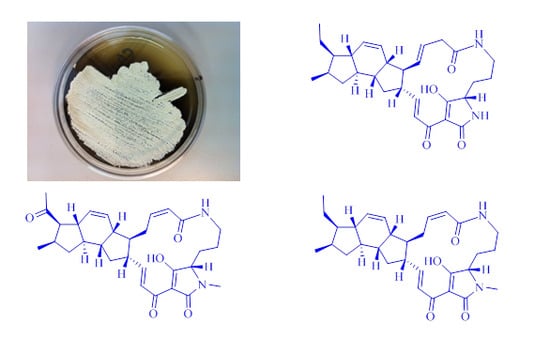
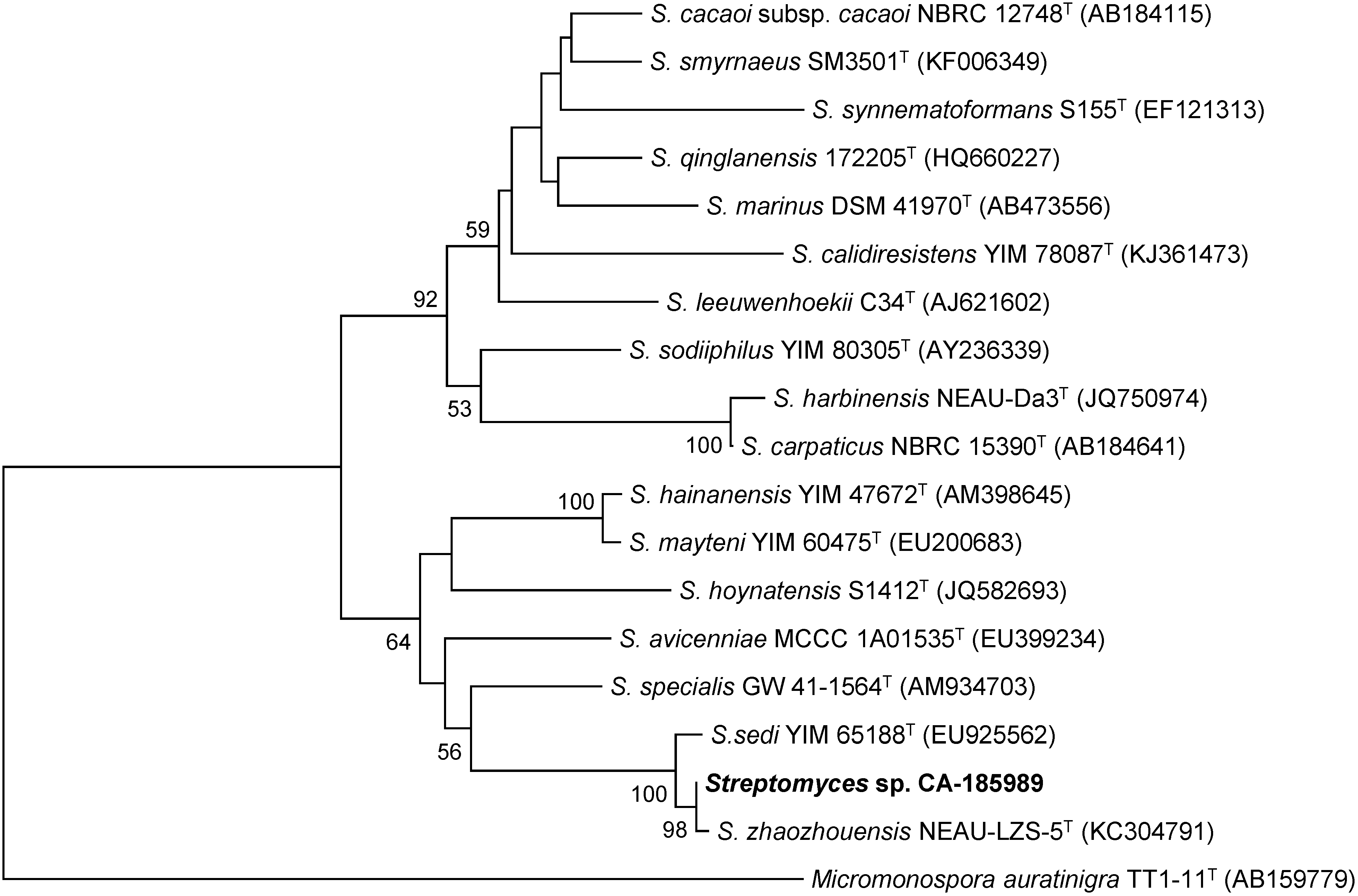
2.1. Isolation and Taxonomy of the Producing Microorganism
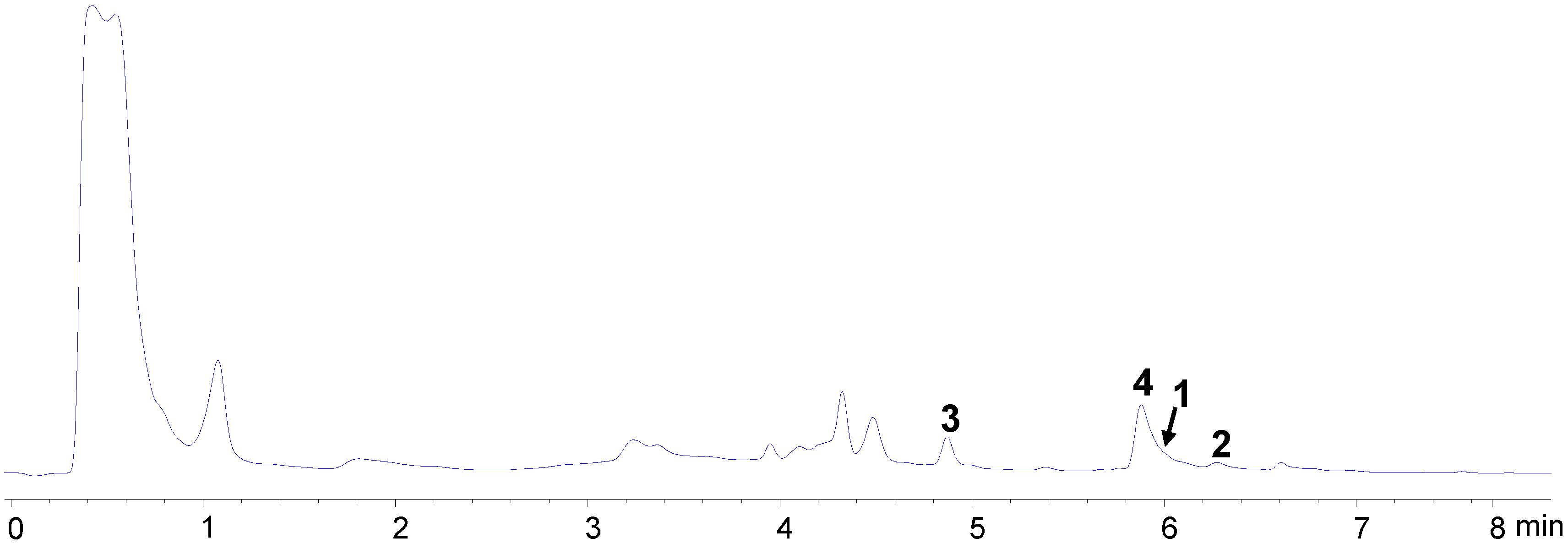
2.2. Bioassay-Guided Isolation
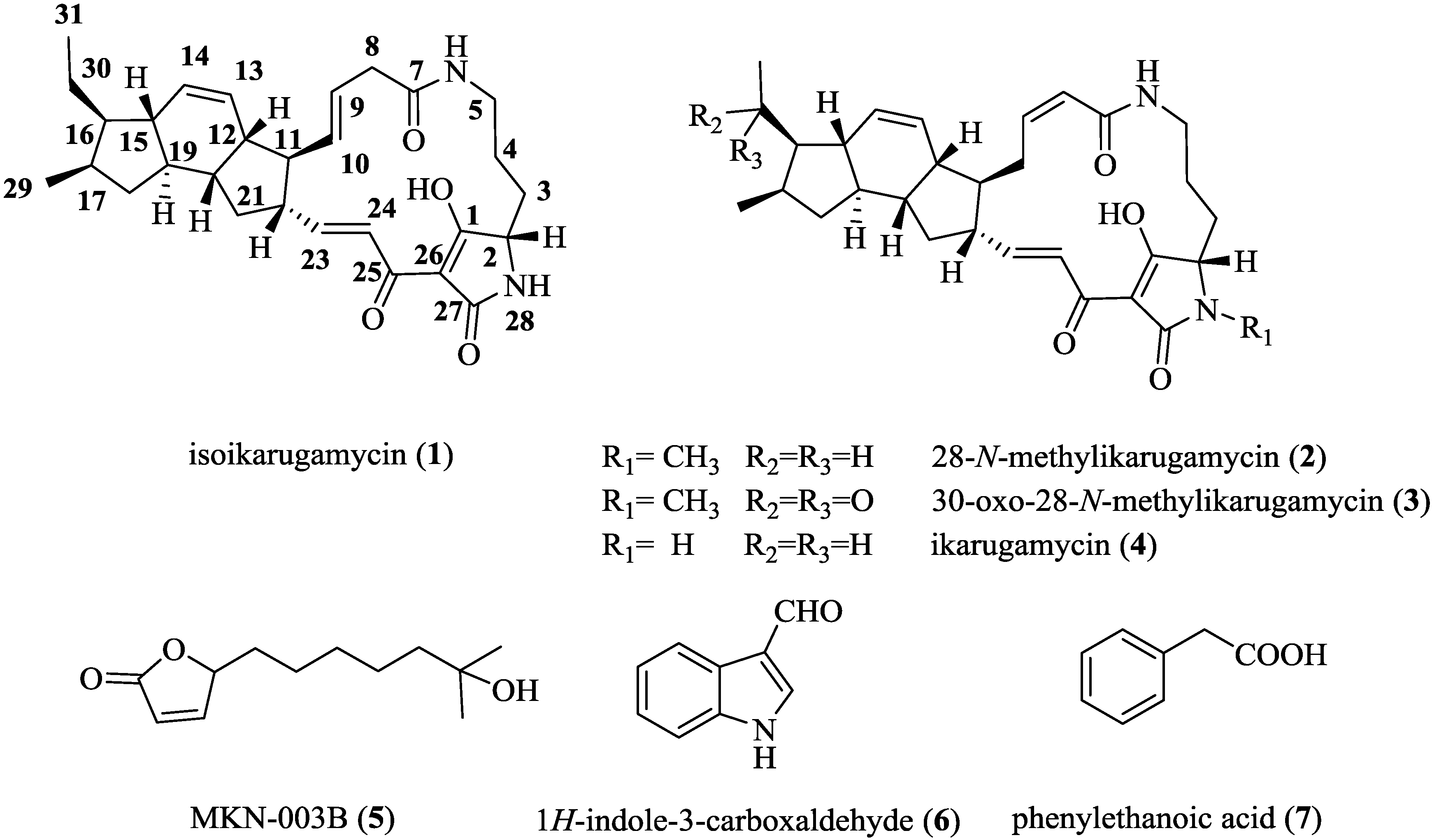
2.3. Structural Determination of the Compounds Isolated
| Position | Isoikarugamycin (1) | 28-N-Methylikarugamycin (2) | 30-Oxo-28-N-Methyl Ikarugamycin (3) | ||||
|---|---|---|---|---|---|---|---|
| δ H, Mult., (J Hz) | δ C, Mult. | δ H, Mult., (J Hz) | δ C, Mult. | δ H, Mult., (J Hz) | δ C, Mult. | ||
| 1 | 196.7, C | 195.3, C | 195.2, C | ||||
| 2 | 3.95, br s | 61.4, CH | 3.72, d (3.5) | 66.2, CH | 3.71, d (4.0) | 66.3, CH | |
| 3 | 1.92, m2.02, m | 27.0, CH2 | 1.79, d (15.3) 2.14 m | 25.0, CH2 | 1.79, d (15.8) 2.13 m | 24.9, CH2 | |
| 4 | 1.10, m1.67, m | 22.3, CH2 | 1.13, m 1.47, m | 20.6, CH2 | 1.10, m 1.47 m | 20.6, CH2 | |
| 5 | 2.85, br s; 3.46 br s | 38.4, CH2 | 2.64, dd (10.0, 10.0) 3.69, br s | 38.9, CH2 | 2.64, m 3.64, m | 38.7, CH2 | |
| 6-NH | 5.83, br s | 5.93, br s | 5.78, br s | ||||
| 7 | 171.5, C | 166.6, C | 166.5, C | ||||
| 8 | 2.92, dd (14.8, 9.4) 3.05, d (14.6) | 41.3, CH2 | 5.83, dd (11.3, 1.3) | 123.7, CH | 5.82, d (11.3) | 123.8, CH | |
| 9 | 5.49, dd (14.8, 9.4) | 127.7, CH | 6.08, ddd (11.3, 11.3, 2.5) | 141.5, CH | 6.06, dd (10.9, 10.9) | 141.2, CH | |
| 10 | 5.34, dd (14.8, 9.4) | 133.9, CH | 2.39, dd (11.3, 2.5) 3.43, m | 25.4, CH2 | 2.35, m 3.47, m | 25.2, CH2 | |
| 11 | 1.88, dd (9.4, 9.4) | 56.1, CH | 1.57, m | 48.2, CH | 1.55, dd (11.5, 11.5) | 48.2, CH | |
| 12 | 2.30, m | 47.9, CH | 2.50, m | 42.8, CH | 2.51 m | 42.5, CH | |
| 13 | 5.55, d (10.0) | 128.4, CH | 5.68, dd (10.0, 2.0) | 128.0, CH | 5.68, dd (10.0, 2.0) | 128.6, CH | |
| 14 | 5.88, d (10.0) | 130.9, CH | 5.94, d (10.0) | 131.6, CH | 5.74, d (10.0) | 130.0, CH | |
| 15 | 1.54, m | 47.0, CH | 1.57, m | 46.9, CH | 2.37, m | 43.0, CH | |
| 16 | 1.35, m | 46.9, CH | 1.37, m | 47.2, CH | 2.68, m | 58.8, CH | |
| 17 | 2.26, m | 33.0, CH | 2.26, ddd (7.4, 7.4, 7.4) | 33.0, CH | 2.62, m | 33.7, CH | |
| 18 | 0.69, ddd (12.0, 12.0, 6.8) 2.11, m | 38.4, CH2 | 0.69, ddd (12.0, 12.0, 6.8) 2.08, m | 38.4, CH2 | 0.82, m 2.15, m | 38.9, CH2 | |
| 19 | 1.13, m | 48.7, CH | 1.16, m | 48.8, CH | 1.20, m | 47.7, CH | |
| 20 | 2.10, m | 42.3, CH | 2.08, m | 41.7, CH | 2.13, m | 41.0, CH | |
| 21 | 1.37, m 2.30, m | 37.1, CH2 | 1.24, m 2.16, m | 36.7, CH2 | 1.23, m 2.15, m | 36.5, CH2 | |
| 22 | 2.55, m | 50.6, CH | 2.49, m | 49.5, CH | 2.51, m | 49.3, CH | |
| 23 | 6.84, dd (15.7, 9.0) | 152.6, CH | 6.75, dd (15.5, 10.3) | 153.2, CH | 6.72, dd (15.4, 10.2) | 151.7, CH | |
| 24 | 7.04, d (15.7) | 123.2, CH | 7.12, d (15.5) | 122.2, CH | 7.12, d (15.5) | 122.4, CH | |
| 25 | 174.7, C | 172.9, C | 172.9, C | ||||
| 26 | 100.0, C | 100.7, C | 100.8, C | ||||
| 27 | 175.5, C | 173.4, C | 173.4, C | ||||
| 28-NR1 | 2.93, s | 26.3, CH3 | 2.93,s | 26.3, CH3 | |||
| 29 | 0.88, d (7.2) | 17.7, CH3 | 0.87, d (7.2) | 17.7, CH3 | 0.85, d (7.0) | 18.8, CH3 | |
| 30 | 1.35, m 1.46, m | 21.6, CH2 | 1.35, m 1.46, m | 21.6, CH2 | 210.2, C | ||
| 31 | 0.93, t (7.2) | 13.2, CH3 | 0.93, t (7.2) | 13.4, CH3 | 2.16, s | 31.4, CH3 | |
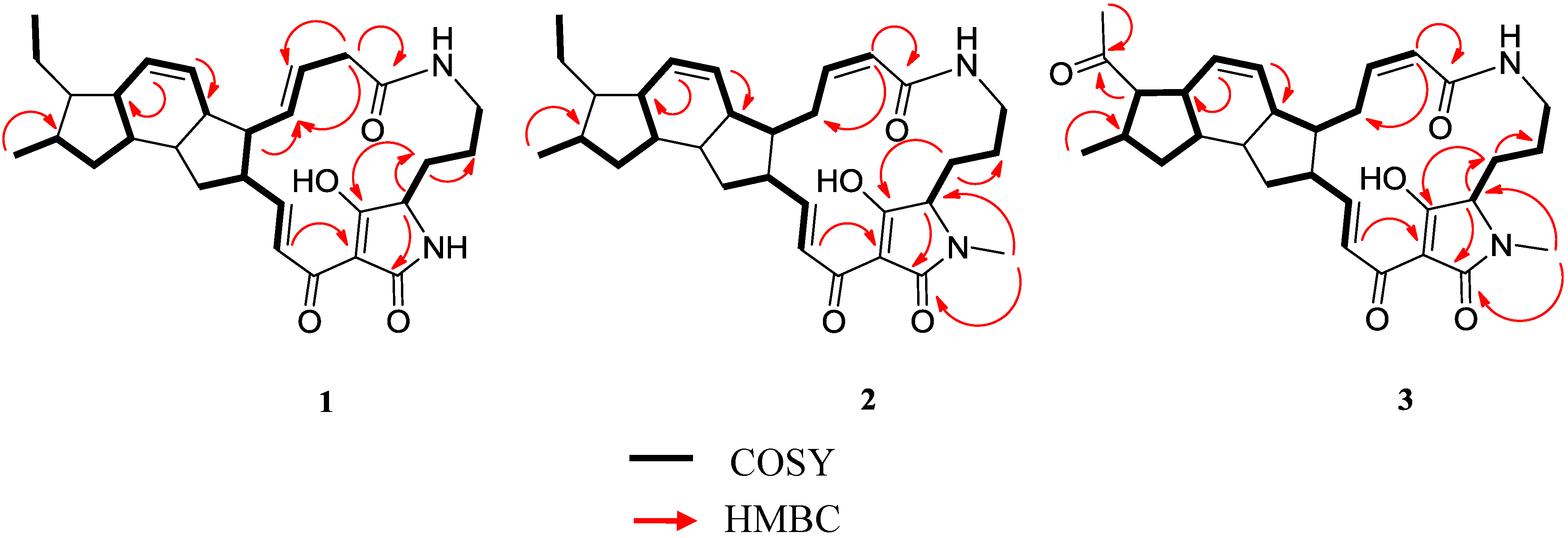
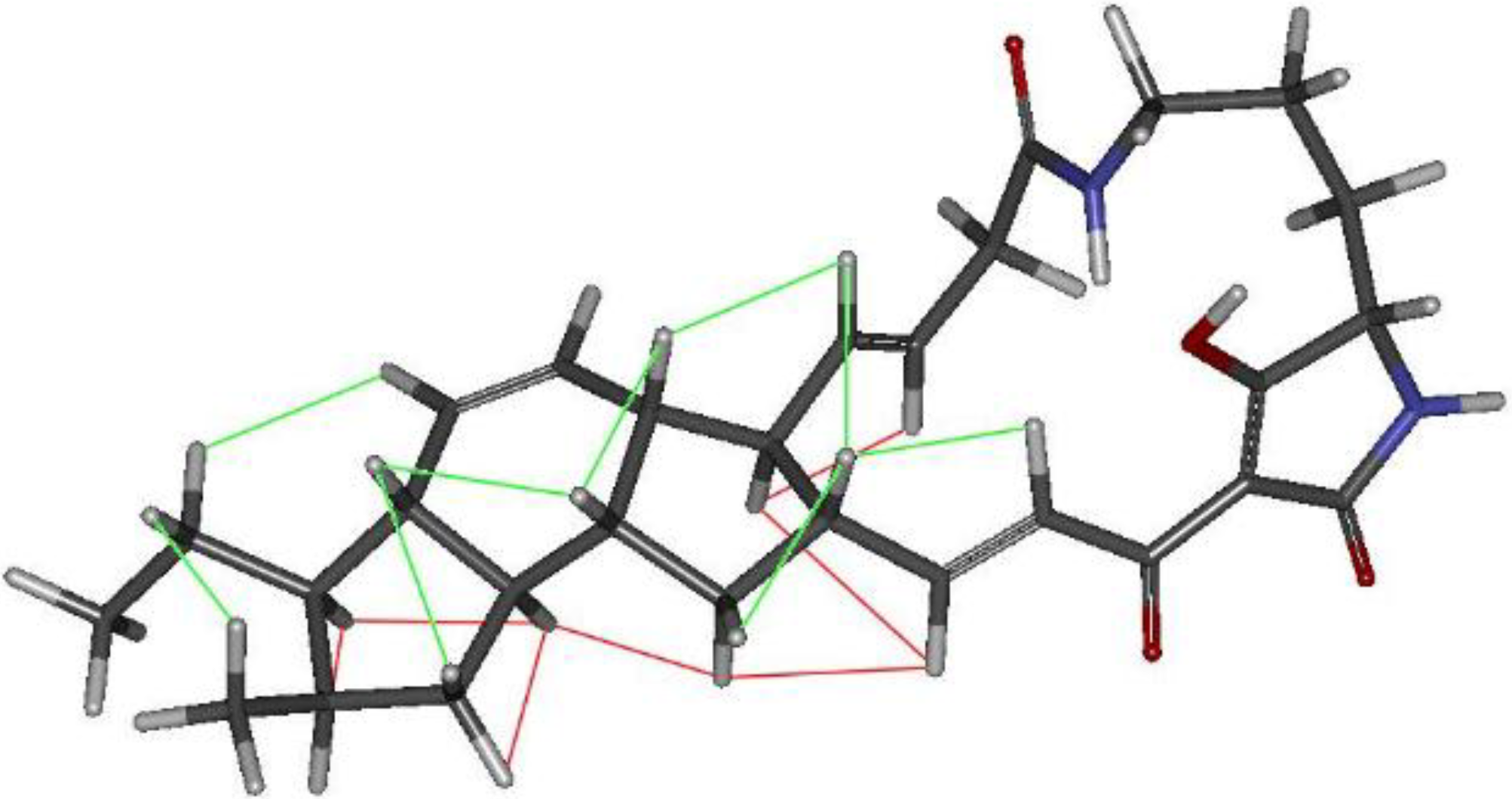
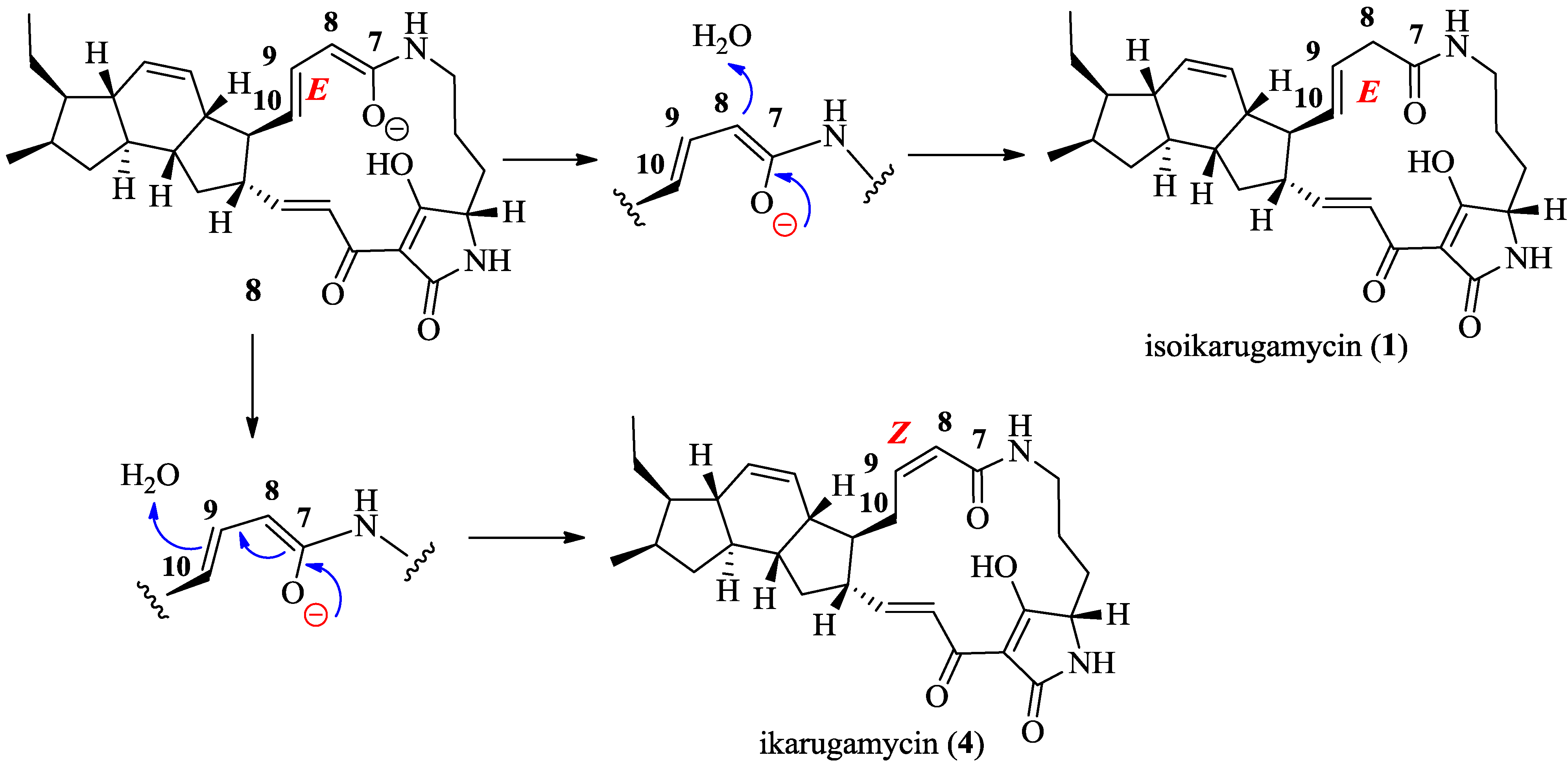
2.4. Antifungal and Antibacterial Activities
| Compound | MIC, μg/mL | |||
|---|---|---|---|---|
| MRSA MB5393 | C. albicans MY1055 | A. fumigatus ATCC46645 | E. coli MB2884 | |
| isoikarugamycin (1) | 2–4 | 2–4 | 4–8 | >64 |
| 28-N-methylikaguramycin (2) | 1–2 | 4 | 4–8 | >64 |
| 30-oxo-28-N-methylikarugamycin (3) | 32–64 | >64 | >64 | >64 |
| Ikarugamycin (4) | 2–4 | 4 | 4–8 | >64 |
| MKN-003B (5) | >64 | >64 | >64 | >64 |
| 1 H-indole-3-carboxaldehyde (6) | >64 | >64 | >64 | >64 |
| Phenylethanoic acid (7) | >64 | >64 | >64 | >64 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fermentation of the Producing Microorganism
3.3. Extraction and Bioassay Guided Isolation
3.4. Antifungal and Antibacterial Assays
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jensen, P.R.; Mincer, T.J.; Williams, P.G.; Fenical, W. Marine actinomycete diversity and natural product discovery. Antonie Leeuwenhoek 2005, 87, 43–48. [Google Scholar] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [PubMed]
- Manivasagana, P.; Venkatesan, J.; Sivakumar, K.; Kim, S. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2014, 169, 267–278. [Google Scholar]
- Hairong, H.; Chongxi, L.; Junwei, Z.; Wejun, L.; Tong, P.; Lingyu, Y.; Xiangjing, W.; Wensheng, X. Streptomyces zhaozhuensis sp. nov., an actinomycete isolated from candelabra aloe (Aloe arborescens Mill). Int. J. Syst. Evol. Microbiol. 2014, 64, 1096–1101. [Google Scholar]
- Jomon, K.; Kuroda, Y.; Ajisaka, M.; Sakai, H. A new antibiotic, Ikarugamycin. J. Antibiot. 1972, 25, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Hirata, Y. The structure of Ikarugamycin, an acyltetramic acid antibiotic possessing a unique as-hydrindacene skeleton. B. Chem. Soc. Jpn. 1977, 50, 1813–1820. [Google Scholar] [CrossRef]
- Blodgett, J.A.V.; Oh, D.-C.; Cao, S.; Currie, C.R.; Kolter, R.; Clardy, J. Common biosynthetic origins for polyciclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11692–11697. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zaleta-Rivera, K.; Zhu, X.; Huffman, J.; Millet, J.C.; Harris, S.D.; Yuen, G.; Li, X.-C.; Du, L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 2007, 51, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, S.; Fusetani, N.; Matsunaga, S. Cylindramide: Cytotoxic tetramic acid lactam from the marine sponge Halichondria cylindrata Tanita & Hoshino. Tetrahedron Lett. 1993, 34, 1065–1068. [Google Scholar] [CrossRef]
- Shigemori, H.; Bae, M.A.; Yazawa, K.; Sasaki, T.; Kobayashi, J. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J. Org. Chem. 1992, 57, 4317–4320. [Google Scholar] [CrossRef]
- Gunasekera, S.P.; Gunasekera, M.; McCarthy, P. Discodermide: A new bioactive macrocyclic lactam from the marine sponge Discodermia dissoluta. J. Org. Chem. 1991, 56, 4830–4833. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon: A prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Jukes, T.H.; Cantor, C. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Allison, J.B., Eds.; Academic Press: New York, NY, USA, 1969; pp. 121–132. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Zink, D.; Dufresne, C.; Liesch, J.; Martín, J. Automated LC-MS analysis of natural products: Extraction of UV, MS and retention time data for component identification and characterization. In Proceedings of the 50th ASMS Conference on Mass Spectrometry and Allied Topics, Orlando, FL, USA, 2–6 June 2002.
- Zink, D.; Dufresne, C.; Liesch, J.; Martín, J. Identification/dereplication of natural products by LC-UV-MS. In Proceedings of the Spectral Search Parameters Presented at the Small Molecule Science Conference (COSMOS), Bristol, RI, USA, 8–11 August 2005.
- Martín, J.; Pérez-Victoria, I.; González, V.; de Pedro, N.; Vicente, F.; Bills, G.; Reyes, F. Applying LC-MS de-replication strategies for the discovery of new natural products. Planta Med. 2012, 78, PI77. [Google Scholar]
- Zhang, G.; Zhang, W.; Zhang, Q.; Shi, T.; Ma, L.; Zhu, Y.; Li, S; Zhang, H; Zao, Y-L.; Shi, R.; et al. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew. Chem. Int. Ed. 2014, 53, 4840–4844. [Google Scholar] [CrossRef]
- Cho, K.; Lee, H.; Rho, J.; Kim, T.; Mo, S.; Shin, J. New lactone containing metabolites from marine-derivade bacterium of the genus Streptomyces. J. Nat. Prod. 2001, 64, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Maskey, R.P.; Pusecker, K.; Speitling, M.; Monecke, P.; Helmke, E.; Laatsch, H. 2″-Chartreusin-monoacetate, a new natural product with unusual anisotropy effects from the marine isolate Streptomyces sp. B5525, and its 4″-isomer. Z. Naturfosch 2002, 57b, 823–829. [Google Scholar]
- Wratten, S.J.; Wolfe, M.S.; Andersen, R.J.; Faulkner, D.J. Antibiotic metabolites from a marine pseudomonad. Antimicrob. Agents Chemother. 1977, 11, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Antosch, J.; Schaefers, F.; Gulder, T.A.M. Heterologous reconstitution of ikarugamycin biosynthesis in E. coli. Angew. Chem. Int. Ed. 2014, 53, 3011–3014. [Google Scholar] [CrossRef]
- Kyeremeh, K.; Acquah, S.; Sazak, A.; Houssen, W.; Tabudravu, J.; Deng, H.; Jaspars, M. Butremycin, the 3-hydroxyl derivative of ikarugamycin and a protonated aromatic tautomer of 5′-methylthioinosine from a ghanaian Micromonospora sp. K310. Mar. Drugs 2014, 12, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Sato, J.; Asaoka, T.; Nakajima, T. Ikarugamycin Derivative. Jpn. Patent JPS 6345280, 26 February 1988. [Google Scholar]
- Cao, S.; Blodgett, J.A.V.; Clardy, J. Targeted discovery of polycyclic tetramate macrolactams from an environmental Streptomyces strain. Org. Lett. 2010, 12, 4652–4654. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; da S. Sousa, T.; Crespo, G.; Palomo, S.; González, I.; Tormo, J.R.; de la Cruz, M.; Anderson, M.; Hill, R.T.; Vicente, F.; et al. Kocurin, the true structure of PM181104, an anti-methicillin-resistant Staphylococcus aureus (MRSA) thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar. Drugs 2013, 11, 387–398. [Google Scholar] [CrossRef]
- Monteiro, M.C.; de la Cruz, M.; Cantizani, J.; Moreno, C.; Tormo, J.R.; Mellado, E.; de Lucas, J.R.; Asensio, F.; Valiante, V.; Brakhage, A.A.; et al. A new approach to drug discovery: High-throughput screening of microbial natural extracts against Aspergillus fumigatus using resazurin. J. Biomol. Screen. 2012, 17, 542–549. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lacret, R.; Oves-Costales, D.; Gómez, C.; Díaz, C.; De la Cruz, M.; Pérez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New Ikarugamycin Derivatives with Antifungal and Antibacterial Properties from Streptomyces zhaozhouensis. Mar. Drugs 2015, 13, 128-140. https://doi.org/10.3390/md13010128
Lacret R, Oves-Costales D, Gómez C, Díaz C, De la Cruz M, Pérez-Victoria I, Vicente F, Genilloud O, Reyes F. New Ikarugamycin Derivatives with Antifungal and Antibacterial Properties from Streptomyces zhaozhouensis. Marine Drugs. 2015; 13(1):128-140. https://doi.org/10.3390/md13010128
Chicago/Turabian StyleLacret, Rodney, Daniel Oves-Costales, Cristina Gómez, Caridad Díaz, Mercedes De la Cruz, Ignacio Pérez-Victoria, Francisca Vicente, Olga Genilloud, and Fernando Reyes. 2015. "New Ikarugamycin Derivatives with Antifungal and Antibacterial Properties from Streptomyces zhaozhouensis" Marine Drugs 13, no. 1: 128-140. https://doi.org/10.3390/md13010128
APA StyleLacret, R., Oves-Costales, D., Gómez, C., Díaz, C., De la Cruz, M., Pérez-Victoria, I., Vicente, F., Genilloud, O., & Reyes, F. (2015). New Ikarugamycin Derivatives with Antifungal and Antibacterial Properties from Streptomyces zhaozhouensis. Marine Drugs, 13(1), 128-140. https://doi.org/10.3390/md13010128