Tipping Points in Seaweed Genetic Engineering: Scaling Up Opportunities in the Next Decade
Abstract
:1. Introduction
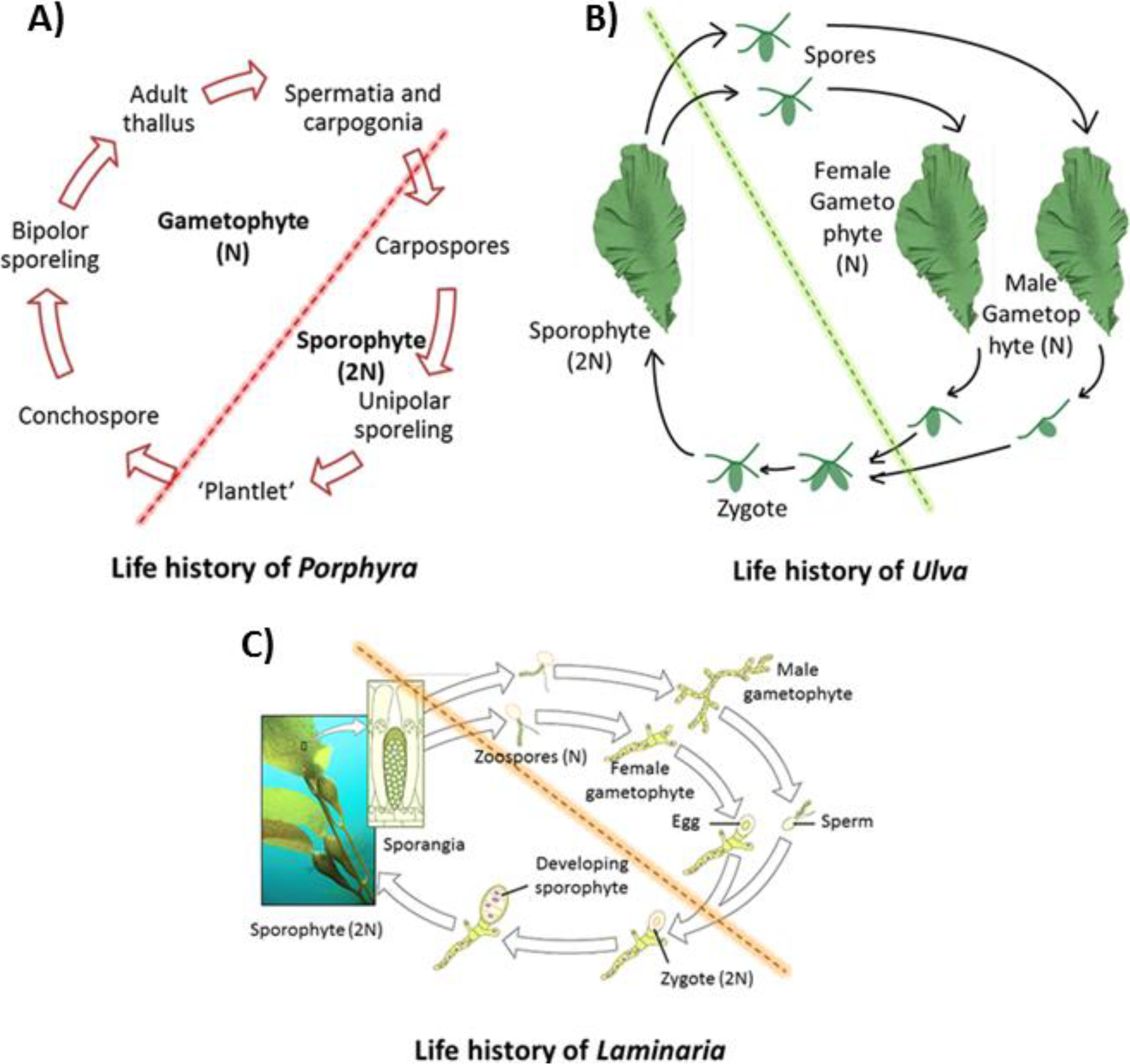
2. Seaweed Genomics and Model Organism Selection in Seaweeds
3. Genome Engineering

4. Natural Promoter Identification and Promoter Engineering
5. Transformation Methods
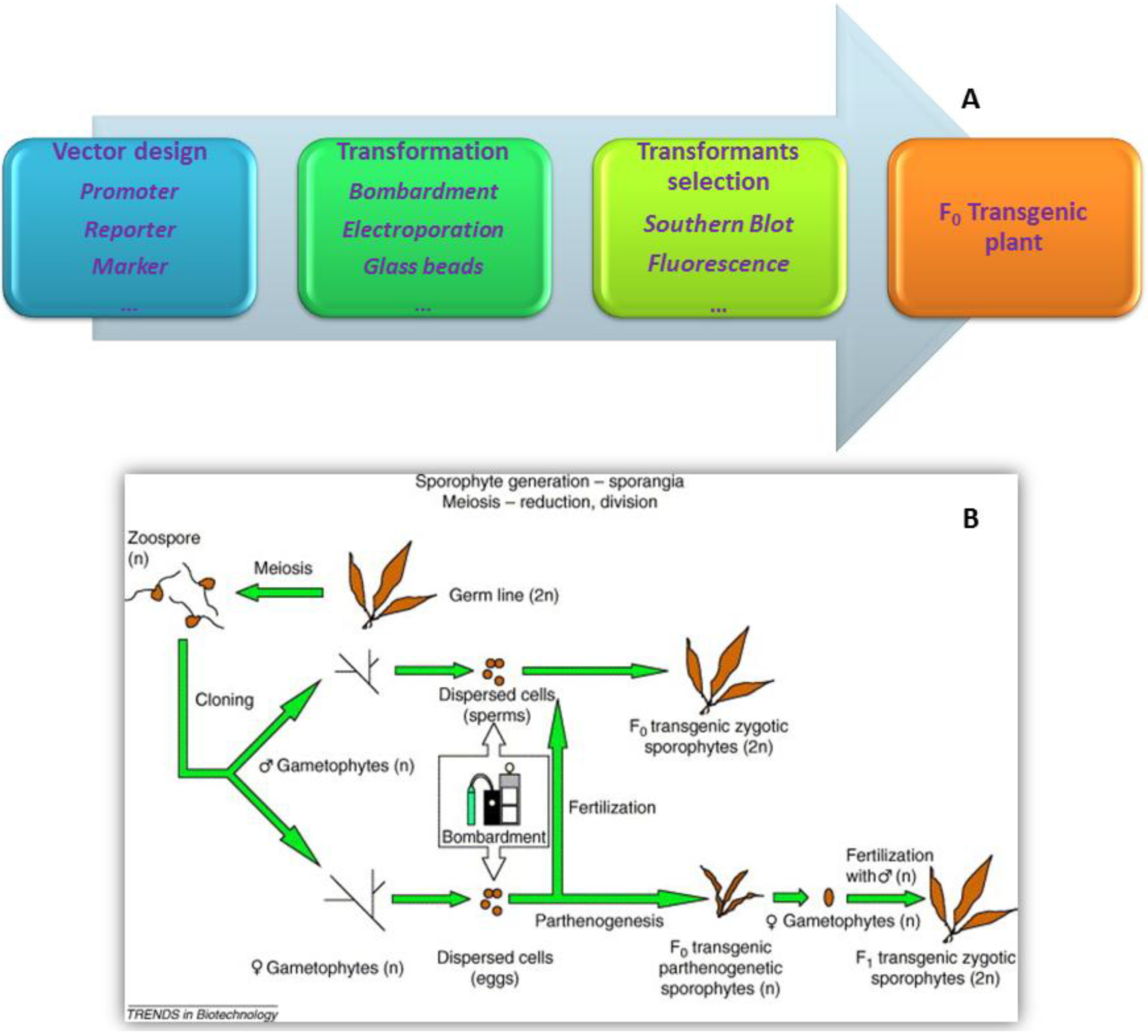
6. Biosafety Assessments of Transgenic Seaweeds
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Luning, K. Seaweeds. Their Environment, Biogeography, and Ecophysiology; John Wiley & Sons, Inc.: Toronto, Canada, 1990. [Google Scholar]
- Falkowski, P.G.; Katz, M.E.; Knoll, A.H.; Quigg, A.; Raven, J.A.; Schofield, O.; Taylor, F.J.R. The evolution of modern eukaryotic phytoplankton. Science 2004, 305, 354–360. [Google Scholar]
- Dorrell, R.G.; Smith, A.G. Do red and green make brown?: Perspectives on plastid acquisitions within chromalveolates. Eukaryot. Cell 2011, 10, 856–868. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Eukaryote kingdoms: Seven or nine? BioSystems 1981, 14, 461–481. [Google Scholar] [CrossRef]
- Simpson, A.G.B.; Roger, A.J. The real “kingdoms” of eukaryotes. Curr. Biol. 2004, 14, R693–R696. [Google Scholar] [CrossRef]
- West, J.A.; Hommersand, M.H. Rhodophyta: Life histories. In The Biology of Seaweeds; Lobban, C.S., Wynne, M., Eds.; University of California Press: Berkeley, CA, USA, 1981; pp. 133–193. [Google Scholar]
- Kohlmeyer, J. New clues to the possible origin of ascomycetes. BioScience 1975, 25, 86–93. [Google Scholar] [CrossRef]
- von Stosch, H.A. The sporophyte of Liagora farinosa Lamour. Br. Phycol. Bull. 1965, 2, 486–496. [Google Scholar] [CrossRef]
- van derMeer, J.P.; Todd, E.R. The life history of Palmaria palmata in culture. A new type for the Rhodophyta. Can. J. Bot. 1980, 58, 1250–1256. [Google Scholar] [CrossRef]
- Pedersen, P.M. Phaeophyta: Life histories. In The Biology of Seaweeds; Lobban, C.S., Ed.; University of California Press: Berkeley, CA, USA, 1981; pp. 194–217. [Google Scholar]
- Tseng, C.K. Laminaria mariculture in China. In Case Studies of Seven Commercial Seaweed Resources; Doty, M.S., Caddy, J.F., Santelices, B., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1987; p. 311. [Google Scholar]
- Lee, R.E. Phycology; Cambridge University Press: Cambridge, UK, 2008; p. 194. [Google Scholar]
- Tanner, C.E. Chlorophyta: Life histories. In The Biology of Seaweeds; Lobban, C.S., Wynne, M., Eds.; University of California Press: Berkeley, CA, USA, 1981; pp. 218–247. [Google Scholar]
- Otto, S.P.; Gerstein, A.C. The evolution of haploidy and diploidy. Curr. Biol. 2008, 18, R1121–R1124. [Google Scholar]
- Institute, M.B.A.R. Ulva Life History. Available online: http://www.mbari.org/staff/conn/botany/greens/anna/frontpages/lifecyc.htm (accessed on 25 March 2014).
- College, O. Groups of Protists. Available online: http://cnx.org/content/m44617/1.7/ (accessed on 25 March 2014).
- McHugh, D.J. A Guide to the Seaweed Industry; Food and Agriculture Organization of the United Nations: Rome, Italy, 2003; Volume 441. [Google Scholar]
- Hughes, A.; Kelly, M.; Black, K.; Stanley, M. Biogas from macroalgae: Is it time to revisit the idea? Biotechnol. Biofuels 2012, 5. [Google Scholar] [CrossRef]
- Roesijadi, G.; Jones, S.B.; Snowden-Swan, L.J.; Zhu, Y. Macroalgae as A Biomass Feedstock: A Preliminary Analysis; Pacific Northwest National Laboratory, U.S. Department of Energy: Washington, DC, USA, 2010. [Google Scholar]
- Wei, N.; Quarterman, J.; Jin, Y.S. Marine macroalgae: An untapped resource for producing fuels and chemicals. Trends Biotechnol. 2013, 31, 70–77. [Google Scholar] [CrossRef]
- FAO. Fishery and Aquaculture Statistics 2010; Food and Agriculture Organization of the United Nations: Rome, Italy, 2012. [Google Scholar]
- Murty, U.S.; Banerjee, A.K. Seaweeds: The wealth of oceans. In Handbook of Marine Macroalgae; Kim, S.-K., Ed.; John Wiley & Sons, Ltd.: West Sussex, UK, 2012; pp. 36–44. [Google Scholar]
- Qin, S.; Lin, H.Z.; Jiang, P. Advances in genetic engineering of marine algae. Biotechnol. Adv. 2012, 30, 1602–1613. [Google Scholar] [CrossRef]
- Qin, S.; Jiang, P.; Tseng, C. Transforming kelp into a marine bioreactor. Trends Biotechnol. 2005, 23, 264–268. [Google Scholar] [CrossRef]
- Lin, H.; Qin, S.; Jiang, P. Biotechnology of seaweeds: Facing the coming decade. In Handbook of Marine Macroalgae; John Wiley & Sons, Ltd.: West Sussex, UK, 2011; pp. 424–430. [Google Scholar]
- Walker, T.L.; Collet, C.; Purton, S. Algal transgenics in the genomic era. J. Phycol. 2005, 41, 1077–1093. [Google Scholar] [CrossRef]
- Purnick, P.E.M.; Weiss, R. The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 2009, 10, 410–422. [Google Scholar] [CrossRef]
- Mikami, K. Current advances in seaweed transformation. In An Integrated View of the Molecular Recognition and Toxinology—From Analytical Procedures to Biomedical Applications; Baptista, G.R., Ed.; InTech: Rijeka, Croatia, 2013; pp. 323–347. [Google Scholar]
- Hallmann, A. Algal transgenics and biotechnology. Transgenic Plant J. 2007, 1, 81–98. [Google Scholar]
- Cock, J.M.; Sterck, L.; Rouze, P.; Scornet, D.; Allen, A.E.; Amoutzias, G.; Anthouard, V.; Artiguenave, F.; Aury, J.M.; Badger, J.H.; et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 2010, 465, 617–621. [Google Scholar] [CrossRef]
- Michel, G.; Tonon, T.; Scornet, D.; Cock, J.M.; Kloareg, B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in eukaryotes. New Phytol. 2010, 188, 82–97. [Google Scholar] [CrossRef]
- Collen, J.; Porcel, B.; Carre, W.; Ball, S.G.; Chaparro, C.; Tonon, T.; Barbeyron, T.; Michel, G.; Noel, B.; Valentin, K.; et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 2013, 110, 5247–5252. [Google Scholar] [CrossRef]
- Nakamura, Y.; Sasaki, N.; Kobayashi, M.; Ojima, N.; Yasuike, M.; Shigenobu, Y.; Satomi, M.; Fukuma, Y.; Shiwaku, K.; Tsujimoto, A.; et al. The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS One 2013, 8, e57122. [Google Scholar] [CrossRef]
- Mardis, E.R. A decade’s perspective on DNA sequencing technology. Nature 2011, 470, 198–203. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013, 6, 287–303. [Google Scholar] [CrossRef]
- Legall, Y.; Brown, S.; Marie, D.; Mejjad, M.; Kloareg, B. Quantification of nuclear-DNA and G-C content in marine macroalgae by flow-cytometry of isolated-nuclei. Protoplasma 1993, 173, 123–132. [Google Scholar] [CrossRef]
- Yao, C.; Jun, L. Chinese scientists sequence genome of kelp, seafood species. Available online: http://english.peopledaily.com.cn/202936/8442860.html (accessed on 25 March 2014).
- Konotchick, T.; Dupont, C.L.; Valas, R.E.; Badger, J.H.; Allen, A.E. Transcriptomic analysis of metabolic function in the giant kelp, Macrocystis pyrifera, across depth and season. New Phytol. 2013, 198, 398–407. [Google Scholar] [CrossRef]
- Dittami, S.M.; Scornet, D.; Petit, J.L.; Segurens, B.; Da Silva, C.; Corre, E.; Dondrup, M.; Glatting, K.H.; Konig, R.; Sterck, L.; et al. Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol. 2009, 10, R66. [Google Scholar] [CrossRef]
- Heesch, S.; Cho, G.Y.; Peters, A.F.; Le Corguille, G.; Falentin, C.; Boutet, G.; Coedel, S.; Jubin, C.; Samson, G.; Corre, E.; et al. A sequence-tagged genetic map for the brown alga Ectocarpus siliculosus provides large-scale assembly of the genome sequence. New Phytol. 2010, 188, 42–51. [Google Scholar] [CrossRef]
- Peters, A.F.; Marie, D.; Scornet, D.; Kloareg, B.; Cock, J.M. Proposal of Ectocarpus siliculosus (Ectocarpales, Phaeophyceae) as a model organism for brown algal genetics and genomics. J. Phycol. 2004, 40, 1079–1088. [Google Scholar] [CrossRef]
- Coelho, S.M.; Scornet, D.; Rousvoal, S.; Peters, N.T.; Dartevelle, L.; Peters, A.F.; Cock, J.M. Ectocarpus: A model organism for the brown algae. Cold Spring Harb. Protoc. 2012, 2012, 193–198. [Google Scholar]
- Coelho, S.M.; Scornet, D.; Rousvoal, S.; Peters, N.; Dartevelle, L.; Peters, A.F.; Cock, J.M. Genetic crosses between Ectocarpus strains. Cold Spring Harb. Protoc. 2012, 2012, 262–265. [Google Scholar]
- Coelho, S.M.; Scornet, D.; Rousvoal, S.; Peters, N.; Dartevelle, L.; Peters, A.F.; Cock, J.M. Extraction of high-quality genomic DNA from Ectocarpus. Cold Spring Harb. Protoc. 2012, 2012, 365–368. [Google Scholar]
- Coelho, S.M.; Scornet, D.; Rousvoal, S.; Peters, N.; Dartevelle, L.; Peters, A.F.; Cock, J.M. Immunostaining of Ectocarpus cells. Cold Spring Harb. Protoc. 2012, 2012, 369–372. [Google Scholar]
- Coelho, S.M.; Scornet, D.; Rousvoal, S.; Peters, N.; Dartevelle, L.; Peters, A.F.; Cock, J.M. Isolation and regeneration of protoplasts from Ectocarpus. Cold Spring Harb. Protoc. 2012, 2012, 361–364. [Google Scholar]
- Coelho, S.M.; Scornet, D.; Rousvoal, S.; Peters, N.T.; Dartevelle, L.; Peters, A.F.; Cock, J.M. How to cultivate Ectocarpus. Cold Spring Harb. Protoc. 2012, 2012, 258–261. [Google Scholar]
- Blouin, N.A.; Brodie, J.A.; Grossman, A.C.; Xu, P.; Brawley, S.H. Porphyra: A marine crop shaped by stress. Trends Plant Sci. 2011, 16, 29–37. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.D.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef]
- Voytas, D.F. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 2013, 64, 327–350. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Wang, H.H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 2013, 9, 641. [Google Scholar] [CrossRef]
- Sizova, I.; Greiner, A.; Awasthi, M.; Kateriya, S.; Hegemann, P. Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 2013, 73, 873–882. [Google Scholar] [CrossRef]
- Jiang, W.Z.; Zhou, H.B.; Bi, H.H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of crispr/cas9/sgrna-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Mahfouz, M.M.; Li, L.X.; Shamimuzzaman, M.; Wibowo, A.; Fang, X.Y.; Zhu, J.K. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. USA 2011, 108, 2623–2628. [Google Scholar]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Liu, W.S.; Yuan, J.S.; Stewart, C.N. Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 2013, 14, 781–793. [Google Scholar] [CrossRef]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Schornack, S.; Lahaye, T. TAL effectors: Finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010, 13, 394–401. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Voytas, D.F. TAL effectors: Customizable proteins for DNA targeting. Science 2011, 333, 1843–1846. [Google Scholar] [CrossRef]
- Mussolino, C.; Morbitzer, R.; Lutge, F.; Dannemann, N.; Lahaye, T.; Cathomen, T. A novel tale nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011, 39, 9283–9293. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, F.; Li, X.H.; Baller, J.A.; Qi, Y.P.; Starker, C.G.; Bogdanove, A.J.; Voytas, D.F. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013, 161, 20–27. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, 7879. [Google Scholar] [CrossRef]
- Weeks, D.P. Homologous recombination in Nannochloropsis: A powerful tool in an industrially relevant alga. Proc. Natl. Acad. Sci. USA 2011, 108, 20859–20860. [Google Scholar] [CrossRef]
- Borchers, A.; Wright, D.; Spalding, M.H. Development of tal nucleases for genome modification in Chlamydomonas. Available online: http://www.cbirc.iastate.edu/files/2012/09/Development-of-TAL-Nucleases-for-Genome-Modification-in-Chlamydomonas.pdf (accessed on 25 March 2014).
- Brueggeman, A.J. Transcriptomic Analyses of the CO2-Concentrating Mechanisms and Development of Molecular Tools for Chlamydomonas Reinhardtii. Ph.D. Thesis, 2013. [Google Scholar]
- Rusk, N. CRISPRs and epigenome editing. Nat. Methods 2014, 11, 28–28. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Xu, T.; Li, Y.; Van Nostrand, J.D.; He, Z.; Zhou, J. Cas9-based tools for targeted genome editing and transcriptional control. Appl. Environ. Microbiol. 2014, 80, 1544–1552. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.R.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Jiang, P.; Qin, S.; Tseng, C.K. Expression of the lacZ reporter gene in sporophytes of the seaweed Laminaria japonica (Phaeophyceae) by gametophyte-targeted transformation. Plant Cell Rep. 2003, 21, 1211–1216. [Google Scholar] [CrossRef]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Shan, Q.W.; Wang, Y.P.; Li, J.; Zhang, Y.; Chen, K.L.; Liang, Z.; Zhang, K.; Liu, J.X.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.-W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Mali, P.; Braff, J.L.; Moosburner, M.; Yaung, S.J.; Church, G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 2013, 10, 1116–1121. [Google Scholar] [CrossRef]
- Fu, Y.F.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Podevin, N.; Devos, Y.; Davies, H.V.; Nielsen, K.M. Transgenic or not? No simple answer! EMBO Rep. 2012, 13, 1057–1061. [Google Scholar] [CrossRef]
- Keeling, P.J.; Burger, G.; Durnford, D.G.; Lang, B.F.; Lee, R.W.; Pearlman, R.E.; Roger, A.J.; Gray, M.W. The tree of eukaryotes. Trends Ecol. Evol. 2005, 20, 670–676. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Price, D.C.; Chan, C.X.; Qiu, H.; Rose, N.; Ball, S.; Weber, A.P.M.; Arias, M.C.; Henrissat, B.; Coutinho, P.M.; et al. Genome of the red alga Porphyridium purpureum. Nat. Commun. 2013, 4, 1941. [Google Scholar]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.G.; Allen, A.E.; Apt, K.E.; Bechner, M.; et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Cook, C.; Martin, L.; Bastow, R. Opportunities in plant synthetic biology. J. Exp. Bot. 2014, 65. [Google Scholar] [CrossRef]
- Schlabach, M.R.; Hu, J.K.; Li, M.; Elledge, S.J. Synthetic design of strong promoters. Proc. Natl. Acad. Sci. USA 2010, 107, 2538–2543. [Google Scholar] [CrossRef]
- Gagniuc, P.; Ionescu-Tirgoviste, C. Eukaryotic genomes may exhibit up to 10 generic classes of gene promoters. BMC Genomics 2012, 13, 512. [Google Scholar] [CrossRef]
- Huang, W.; Nevins, J.R.; Ohler, U. Phylogenetic simulation of promoter evolution: Estimation and modeling of binding site turnover events and assessment of their impact on alignment tools. Genome Biol. 2007, 8, R225. [Google Scholar]
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003, 72, 449–479. [Google Scholar] [CrossRef]
- Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 40–51. [Google Scholar] [CrossRef]
- Molina, C.; Grotewold, E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics 2005, 6, 25. [Google Scholar] [CrossRef]
- Kuang, M.; Wang, S.; Li, Y.; Shen, D.; Zeng, C. Transient expression of exogenous GUS gene in Porphyra yezoensis (Rhodophyta). Chin. J. Oceanol. Limnol. 1998, 16, 56–61. [Google Scholar] [CrossRef]
- Liu, H.; Yu, W.; Dai, J.; Gong, Q.; Yang, K.; Zhang, Y. Increasing the transient expression of GUS gene in Porphyra yezoensis by 18s rDNA targeted homologous recombination. J. Appl. Phycol. 2003, 15, 371–377. [Google Scholar] [CrossRef]
- Qin, S.; Jiang, P.; Tseng, C.K. Molecular biotechnology of marine algae in China. Hydrobiologia 2004, 512, 21–26. [Google Scholar] [CrossRef]
- Feng, S.Y.; Xue, L.X.; Liu, H.T.; Lu, P.J. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol. Biol. Rep. 2009, 36, 1433–1439. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef]
- Mikami, K.; Hirata, R.; Takahashi, M.; Uji, T.; Saga, N. Transient transformation of red algal cells: Breakthrough toward genetic transformation of marine crop Porphyra species. In Genetic Transformation; Alvarez, M., Ed.; InTech: Rijeka, Croatia, 2011; pp. 241–258. [Google Scholar]
- Li, F.C.; Qin, S.; Jiang, P.; Wu, Y.; Zhang, W. The integrative expression of GUS gene driven by FCP promoter in the seaweed Laminaria japonica (Phaeophyta). J. Appl. Phycol. 2009, 21, 287–293. [Google Scholar] [CrossRef]
- Hirata, R.; Takahashi, M.; Saga, N.; Mikami, K. Transient gene expression system established in Porphyra yezoensis is widely applicable in Bangiophycean algae. Mar. Biotechnol. 2011, 13, 1038–1047. [Google Scholar] [CrossRef]
- Takahashi, M.; Uji, T.; Saga, N.; Mikami, K. Isolation and regeneration of transiently transformed protoplasts from gametophytic blades of the marine red alga Porphyra yezoensis. Electron. J. Biotechnol. 2010, 13. [Google Scholar] [CrossRef]
- Blanvillain, R.; Gallois, P. Promoter trapping system to study embryogenesis. In Methods in Molecular Biology; Suarez, M.F., Bozhkov, P.V., Eds.; Humana Press: Totowa, NJ, USA, 2008; Volume 427, pp. 121–135. [Google Scholar]
- Vila, M.; Diaz-Santos, E.; de la Vega, M.; Rodriguez, H.; Vargas, A.; Leon, R. Promoter trapping in microalgae using the antibiotic paromomycin as selective agent. Mar. Drugs 2012, 10, 2749–2765. [Google Scholar] [CrossRef]
- Coelho, S.M.; Simon, N.; Ahmed, S.; Cock, J.M.; Partensky, F. Ecological and evolutionary genomics of marine photosynthetic organisms. Mol. Ecol. 2013, 22, 867–907. [Google Scholar] [CrossRef]
- Levine, M.; Tjian, R. Transcription regulation and animal diversity. Nature 2003, 424, 147–151. [Google Scholar] [CrossRef]
- Blazeck, J.; Alper, H.S. Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 2013, 8, 46–58. [Google Scholar] [CrossRef]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef]
- Dorokhov, Y.L.; Skulachev, M.V.; Ivanov, P.A.; Zvereva, S.D.; Tjulkina, L.G.; Merits, A.; Gleba, Y.Y.; Hohn, T.; Atabekov, J.G. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA 2002, 99, 5301–5306. [Google Scholar] [CrossRef]
- Babiskin, A.H.; Smolke, C.D. Synthetic RNA modules for fine-tuning gene expression levels in yeast by modulating RNase III activity. Nucleic Acids Res. 2011, 39, 8651–8664. [Google Scholar]
- Puigbo, P.; Guzman, E.; Romeu, A.; Garcia-Vallve, S. Optimizer: A web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007, 35, W126–W131. [Google Scholar] [CrossRef]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef]
- Lithwick, G.; Margalit, H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 2003, 13, 2665–2673. [Google Scholar] [CrossRef]
- Surzycki, R.; Greenham, K.; Kitayama, K.; Dibal, F.; Wagner, R.; Rochaix, J.D.; Ajam, T.; Surzycki, S. Factors effecting expression of vaccines in microalgae. Biologicals 2009, 37, 133–138. [Google Scholar] [CrossRef]
- Potvin, G.; Zhang, Z.S. Strategies for high-level recombinant protein expression in transgenic microalgae: A review. Biotechnol. Adv. 2010, 28, 910–918. [Google Scholar] [CrossRef]
- Mayfield, S.P.; Schultz, J. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 2004, 37, 449–458. [Google Scholar] [CrossRef]
- Lim, J.-M.; Ahn, J.-W.; Hwangbo, K.; Choi, D.-W.; Park, E.-J.; Hwang, M.S.; Liu, J.R.; Jeong, W.-J. Development of cyan fluorescent protein (CFP) reporter system in green alga Chlamydomonas reinhardtii and macroalgae Pyropia sp. Plant Biotechnol. Rep. 2013, 7, 407–414. [Google Scholar] [CrossRef]
- Norholm, M.H.H.; Toddo, S.; Virkki, M.T.I.; Light, S.; von Heijne, G.; Daley, D.O. Improved production of membrane proteins in Escherichia coli by selective codon substitutions. Febs Lett. 2013, 587, 2352–2358. [Google Scholar] [CrossRef]
- Welch, M.; Villalobos, A.; Gustafsson, C.; Minshull, J. You’re one in a googol: Optimizing genes for protein expression. J. R. Soc. Interface 2009, 6, S467–S476. [Google Scholar] [CrossRef]
- Mizukami, Y.; Hado, M.; Kito, H.; Kunimoto, M.; Murase, N. Reporter gene introduction and transient expression in protoplasts of Porphyra yezoensis. J. Appl. Phycol. 2004, 16, 23–29. [Google Scholar] [CrossRef]
- Huang, X.; Weber, J.C.; Hinson, T.K.; Mathieson, A.C.; Minocha, S.C. Transient expression of the GUS reporter gene in the protoplasts and partially digested cells of Ulva lactuca L (Chlorophyta). Bot. Mar. 1996, 39, 467–474. [Google Scholar]
- Wang, J.F.; Jiang, P.; Cui, Y.L.; Guan, X.Y.; Qin, S. Gene transfer into conchospores of Porphyra haitanensis (Bangiales, Rhodophyta) by glass bead agitation. Phycologia 2010, 49, 355–360. [Google Scholar] [CrossRef]
- Jackson, M.A.; Anderson, D.J.; Birch, R.G. Comparison of Agrobacterium and particle bombardment using whole plasmid or minimal cassette for production of high-expressing, low-copy transgenic plants. Transgenic Res. 2013, 22, 143–151. [Google Scholar] [CrossRef]
- Cheney, D.; Metz, B.; Stiller, J. Agrobacterium-mediated genetic transformation in the macroscopic marine red alga Porphyra yezoensis. J. Phycol. 2001, 37. [Google Scholar] [CrossRef]
- Henry, E.C.; Meints, R.H. Recombinant viruses as transformation vectors of marine macroalgae. J. Appl. Phycol. 1994, 6, 247–253. [Google Scholar] [CrossRef]
- Muller, D.G.; Kapp, M.; Knippers, R. Viruses in marine brown algae. Adv. Virus Res. 1998, 50, 49–67. [Google Scholar] [CrossRef]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef]
- Murray, A.G.; Jackson, G.A. Viral dynamics—A model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 1992, 89, 103–116. [Google Scholar] [CrossRef]
- Fuhrman, K.M.; Pinter, G.A.; Berges, J.A. Dynamics of a virus-host model with an intrinsic quota. Math. Comput. Model. 2011, 53, 716–730. [Google Scholar] [CrossRef]
- Middelboe, M. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 2000, 40, 114–124. [Google Scholar]
- Short, S.M. The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 2012, 14, 2253–2271. [Google Scholar] [CrossRef]
- Charrier, B.; Coelho, S.M.; Le Bail, A.; Tonon, T.; Michel, G.; Potin, P.; Kloareg, B.; Boyen, C.; Peters, A.F.; Cock, J.M. Development and physiology of the brown alga Ectocarpus siliculosus: Two centuries of research. New Phytol. 2008, 177, 319–332. [Google Scholar]
- Kawata, Y.; Yano, S.; Kojima, H.; Toyomizu, M. Transformation of spirulina platensis strain c1 (Arthrospira sp PCC9438) with Tn5 transposase-transposon DNA-cation liposome complex. Mar. Biotechnol. 2004, 6, 355–363. [Google Scholar] [CrossRef]
- Taton, A.; Lis, E.; Adin, D.M.; Dong, G.; Cookson, S.; Kay, S.A.; Golden, S.S.; Golden, J.W. Gene transfer in Leptolyngbya sp. strain BL0902, a cyanobacterium suitable for production of biomass and bioproducts. PLoS One 2012, 7, e30901. [Google Scholar]
- Hohmann-Marriott, M.F.; Blankenship, R.E. Evolution of photosynthesis. In Annual Review of Plant Biology; Merchant, S.S., Briggs, W.R., Ort, D., Eds.; Annual Reviews: Palo Alto, CA, USA, 2011; Volume 62, pp. 515–548. [Google Scholar]
- Andow, D.A.; Zwahlen, C. Assessing environmental risks of transgenic plants. Ecol. Lett. 2006, 9, 196–214. [Google Scholar] [CrossRef]
- Pauwels, K.; Podevin, N.; Breyer, D.; Carroll, D.; Herman, P. Engineering nucleases for gene targeting: Safety and regulatory considerations. New Biotechnol. 2014, 31, 18–27. [Google Scholar] [CrossRef]
- Henley, W.J.; Litaker, R.W.; Noyoyeska, L.; Duke, C.S.; Quemada, H.D.; Sayre, R.T. Initial risk assessment of genetically modified (GM) microalgae for commodity-scale biofuel cultivation. Algal Res. Biomass Biofuels Bioprod. 2013, 2, 66–77. [Google Scholar]
- Gressel, J.; van der Vlugt, C.J.B.; Bergmans, H.E.N. Environmental risks of large scale cultivation of microalgae: Mitigation of spills. Algal Res. Biomass Biofuels Bioprod. 2013, 2, 286–298. [Google Scholar]
- Bennett, A.B.; Chi-Ham, C.; Barrows, G.; Sexton, S.; Zilberman, D. Agricultural biotechnology: Economics, environment, ethics, and the future. Annu. Rev. Env. Res. 2013, 38, 249–279. [Google Scholar] [CrossRef]
- Chandler, S.; Dunwell, J.M. Gene flow, risk assessment and the environmental release of transgenic plants. Crit. Rev. Plant Sci. 2008, 27, 25–49. [Google Scholar] [CrossRef]
- Rieben, S.; Kalinina, O.; Schmid, B.; Zeller, S.L. Gene flow in genetically modified wheat. PLoS One 2011, 6, e29730. [Google Scholar]
- Lin, A.; Shen, S.; Wang, J.; Yan, B. Reproduction diversity of Enteromorpha prolifera. J. Integr. Plant Biol. 2008, 50, 622–629. [Google Scholar] [CrossRef]
- Colbach, N.; Clermont-Dauphin, C.; Meynard, J.M. GENESYS: A model of the influence of cropping system on gene escape from herbicide tolerant rapeseed crops to rape volunteers—II. Genetic exchanges among volunteer and cropped populations in a small region. Agric. Ecosyst. Environ. 2001, 83, 255–270. [Google Scholar] [CrossRef]
- Colbach, N.; Clermont-Dauphin, C.; Meynard, J.M. GENESYS: A model of the influence of cropping system on gene escape from herbicide tolerant rapeseed crops to rape volunteers—I. Temporal evolution of a population of rapeseed volunteers in a field. Agric. Ecosyst. Environ. 2001, 83, 235–253. [Google Scholar] [CrossRef]
- Hutchinson, G.E. The paradox of the plankton. Am. Nat. 1961, 95, 137–145. [Google Scholar]
- Gressel, J.; van der Vlugt, C.J.B.; Bergmans, H.E.N. Cultivated microalgae spills: Hard to predict/easier to mitigate risks. Trends Biotechnol. 2014, 32, 65–69. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lin, H.; Qin, S. Tipping Points in Seaweed Genetic Engineering: Scaling Up Opportunities in the Next Decade. Mar. Drugs 2014, 12, 3025-3045. https://doi.org/10.3390/md12053025
Lin H, Qin S. Tipping Points in Seaweed Genetic Engineering: Scaling Up Opportunities in the Next Decade. Marine Drugs. 2014; 12(5):3025-3045. https://doi.org/10.3390/md12053025
Chicago/Turabian StyleLin, Hanzhi, and Song Qin. 2014. "Tipping Points in Seaweed Genetic Engineering: Scaling Up Opportunities in the Next Decade" Marine Drugs 12, no. 5: 3025-3045. https://doi.org/10.3390/md12053025
APA StyleLin, H., & Qin, S. (2014). Tipping Points in Seaweed Genetic Engineering: Scaling Up Opportunities in the Next Decade. Marine Drugs, 12(5), 3025-3045. https://doi.org/10.3390/md12053025




