Abstract
Bioassay-guided fractionation of a culture extract of Beauveria felina EN-135, an entomopathogenic fungus isolated from a marine bryozoan, led to the isolation of a new cyclodepsipeptide, iso-isariin D (1); two new O-containing heterocyclic compounds that we have named felinones A and B (2 and 3); and four known cyclodepsipeptides (4–7). The structures were elucidated via spectroscopic analysis, and the absolute configurations of 1 and 2 were determined using single-crystal X-ray diffraction and CD, respectively. All isolated compounds were evaluated for antimicrobial activity and brine-shrimp (Artemia salina) lethality.
1. Introduction
Entomopathogenic fungi such as species from the genera Beauveria and Metarhizium have been frequently used as an alternative to chemical insecticides for agricultural pest control, and they are attracting increasing attention because of their ability to produce structurally unique and biologically active secondary metabolites []. The marine-derived fungal species Beauveria felina, which is poorly described, has proven to be a rich source of various cyclodepsipeptides, such as the destruxin, isaridin, and isariin classes, as well as polyketides and terpenoids [,,,]. As part of our efforts toward the investigation of bioactive secondary metabolites of marine-derived fungi [,,,,], Beauveria felina EN-135, an entomopathogenic fungus isolated from a marine bryozoan, attracted our attention because of the strong brine-shrimp lethality of the culture extract. Bioassay-guided fractionation of the EtOAc extract led to the isolation and identification of a new cyclodepsipeptide, iso-isariin D (1); two new O-containing heterocyclic compounds that we have termed felinones A and B (2 and 3); and four known cyclodepsipeptides: destruxin A (4), roseotoxin B (5), destruxin E chlorohydrin (6), and [β-Me-Pro] destruxin E chlorohydrin (7) (Figure 1). The brine-shrimp lethality and antimicrobial activity of these compounds were evaluated. Details of the isolation, structure elucidation, and biological activity of compounds 1–7 are reported herein.
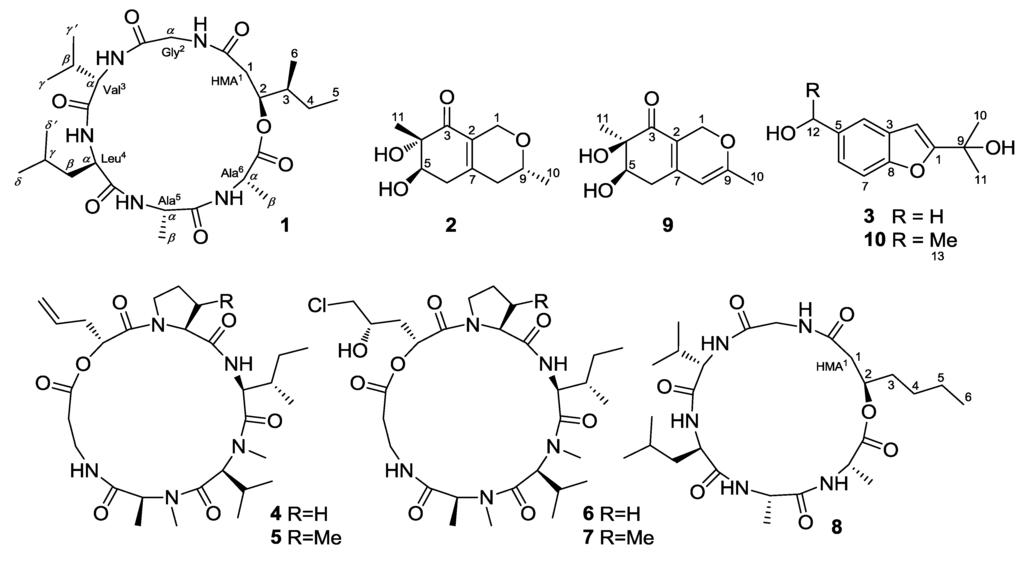
Figure 1.
Structures of the isolated compounds 1–7 and reference compounds 8–10.
2. Results and Discussion
2.1. Structure Elucidation of the New Compounds
Compound 1 was initially isolated as a colorless amorphous powder. Its molecular formula was established to be C26H45N5O7 based on HRESIMS, with seven degrees of unsaturation. Analysis of the 1H NMR spectrum of 1 revealed the presence of five amide NH protons (δH 9.30, 8.60, 8.54, and two at 8.48), four N-CHαs (δH 4.84, 4.79, 4.76, and 4.68), one O-CHβ (δH 5.63), one N-CH2α (δH 4.73 and 4.64), and eight methyl groups (δH 1.95, 1.85, 1.55, 1.53, 1.51, 1.49, 1.48, and 1.46). The 13C and DEPT NMR spectra of compound 1 exhibited signals of eight methyl groups, four methylenes (one nitrogenated), eight methines (one oxygenated and four nitrogenated), and six ester/amide carbonyl carbons (Table 1). These 1D NMR data indicated that compound 1 is a cyclic hexadepsipeptide with a β-hydroxy aliphatic acid moiety [].

Table 1.
1H- (500 MHz) and 13C-NMR (125 MHz) data for compound 1 in DMSO-d6 (δ in ppm).
| Position | δH (J in Hz) | δC | Position | δH (J in Hz) | δC |
|---|---|---|---|---|---|
| HMA1 | - | - | Leu4 | - | - |
| CO | - | 169.7 C | CO | - | 171.1 C |
| 1 | 3.19, m; 2.92, d (13.6) | 37.6 CH2 | α | 4.68, m | 51.9 CH |
| 2 | 5.63, m | 74.3 CH | β | 2.13, m | 38.6 CH2 |
| 3 | 2.20, m | 38.0 CH | γ | 2.28, m | 24.1 CH |
| 4 | 2.03, m; 1.70, m | 24.8 CH2 | δ | 1.48, d (6.8) | 21.1 CH3 |
| 5 | 1.49, t (6.9) | 11.3 CH3 | δ’ | 1.53, d (6.8) | 18.8 CH3 |
| 6 | 1.46, d (6.8) | 14.1 CH3 | NH | 9.30, d (6.0) | - |
| Gly2 | - | - | Ala5 | - | - |
| CO | - | 168.9 C | CO | - | 171.6 C |
| α | 4.73, m; 4.64, dd (16.4, 3.4) | 42.4 CH2 | α | 4.84, m | 47.5 CH |
| NH | 8.54, br. s | - | β | 1.85, d (7.0) | 17.3 CH3 |
| Val3 | - | - | NH | 8.60, d (8.3) | - |
| CO | - | 171.7 C | Ala6 | - | - |
| α | 4.76, m | 58.1 CH | CO | - | 171.6 C |
| β | 2.51, m | 29.9 CH | α | 4.79, m | 48.3 CH |
| γ | 1.55, d (7.7) | 22.8 CH3 | β | 1.95, d (7.2) | 16.6 CH3 |
| γ’ | 1.51, d (7.7) | 18.6 CH3 | NH | 8.48, d (7.6) | - |
| NH | 8.48, d (7.6) | - | - | - | - |
The assignment of the proton and carbon signals to the amino-acid residues was achieved via COSY, HSQC, and HMBC experiments. Briefly, the five amide NH protons were correlated with their corresponding CHα protons in the COSY spectrum. Starting from the CHα signals, the five amino-acid residues were identified as glycine (Gly2), valine (Val3), leucine (Leu4), and two alanines (Ala5 and Ala6) via additional COSY and HMBC analysis. The observed HMBC correlations, including CH2α (Gly2) and NH (Val3)/CO (Gly2), CHα (Val3) and NH (Leu4)/CO (Val3), CHα (Leu4) and NH (Ala5)/CO (Leu4), β-Me (Ala5) and NH (Ala6)/CO (Ala5), and NH (Ala6)/CO (Ala6), established the amino-acid sequence Gly2-Val3-Leu4-Ala5-Ala6 (Figure 2). The remainder of the 1H and 13C NMR signals were unambiguously assigned to a 2-hydroxy-3-methylpentanoic acid moiety (HMA1) based on the COSY and HMBC experiments (Figure 2). The HMBC cross-peaks from CH2-1 (HMA1) and CH2α (Gly2) to CO (HMA1) indicated the presence of the hexadepsipeptide sequence cyclo(HMA1-Gly2-Val3-Leu4-Ala5-Ala6) in compound 1, which was consistent with the seven calculated degrees of unsaturation.
Compound 1 is considered to be a cyclohexadepsipeptide of the isariin class [,,], and a literature search revealed that the structure of this family of compounds has been primarily determined via spectroscopic analysis and the absolute configuration has been determined by analyzing the amino-acid derivatives, but none of these cyclohexadepsipeptides has been unambiguously determined via X-ray crystallography. We performed crystallization of 1 alongside the spectroscopic studies. Although compound 1 was initially obtained as a colorless amorphous powder, single crystals suitable for X-ray analysis were obtained after many attempts. The structure and absolute configuration of 1 could thus be further determined based on single-crystal X-ray diffraction using Cu Kα radiation (Figure 3). The result not only confirmed the peptide sequence but also allowed the identification of the amino-acid residues as l-Val3, d-Leu4, l-Ala5, and l-Ala6, along with 2S-hydroxy-3S-methylpentanoic acid (HMA1). Most reported cyclodepsipeptides in the isariin class have a 2-hydroxy aliphatic acid moiety characterized with a straight chain [,,]; only one derivative, iso-isariin B, has a branch chain []. Compound 1 was the second cyclodepsipeptide of the isariin class to be identified that has this type of unusual HMA moiety, which differentiated it from isariin D (8). Thus, the structure of 1 was elucidated and named iso-isariin D. The HMA residue of iso-isariin D (1) in B. felina is most likely related to the polyketide biosynthesis pathway, and the amino-acid sequence might be obtained from the nonribosomal peptide biosynthesis pathway [,].
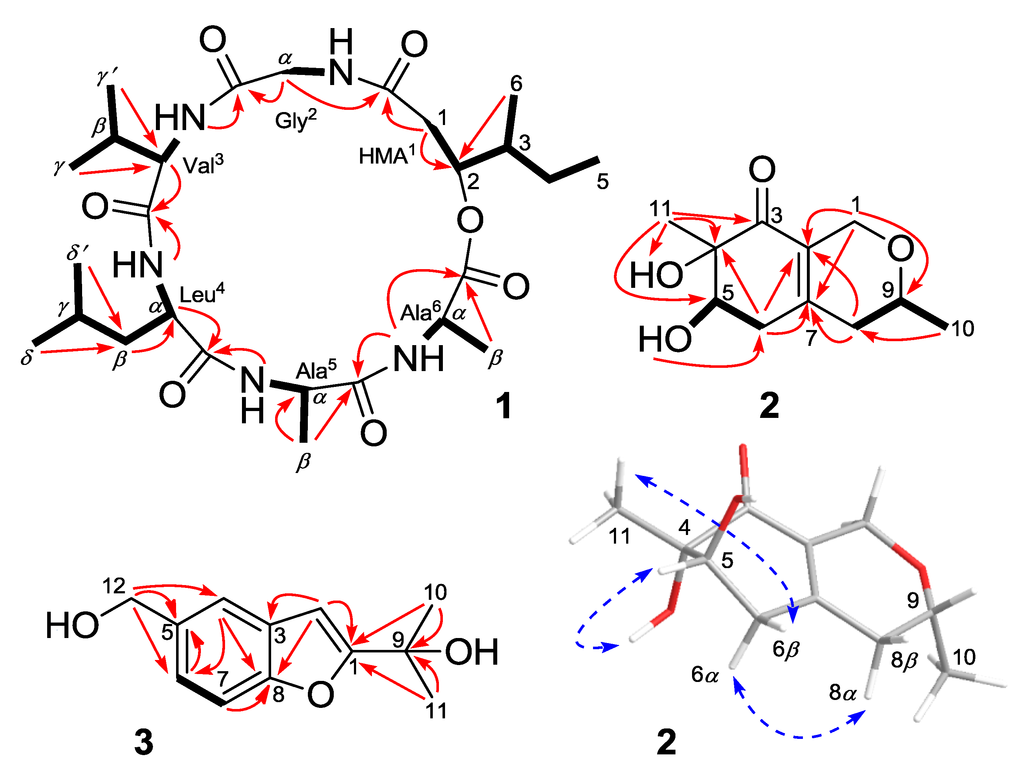
Figure 2.
Key 1H–1H COSY (bold lines), HMBC (red arrows), and NOE (dotted blue arrows) correlations of compounds 1–3.
Compound 2 was obtained as a yellowish solid. The molecular formula C11H16O4 was determined via HRESIMS, implying four degrees of unsaturation (one less than the reference compound 9) []. Detailed analyses of the 1D NMR data (Table 2) indicated the presence of one carbonyl carbon, three additional quaternary carbons (two sp2 and one oxygenated sp3), two oxygenated sp3 methines, three methylenes (one oxygenated sp3), and two methyl groups. Comparison between the NMR data of 2 and 9 indicated that their planar structures were very similar except that the signals of the double bond of C-8/C-9 at δC 102.1 (CH) and 165.7 (C) in 9 were replaced by signals of a single bond at δC 36.9 (CH2) and 68.6 (CH) in the 13C-NMR spectrum of 2. Accordingly, the proton signal at δH 5.18 (s, H-8) of 9 disappeared in the 1H NMR spectrum of 2. Instead, additional CH2 and OCH signals at δH 2.07 (m, H-8α), 2.20 (m, H-8β), and 3.54 (m, H-9) were observed in the 1H-NMR spectrum of 2. The correlations from H-8 to H-9 and from H-9 to H-10 in the COSY spectrum of 2 supported the above deduction (Figure 2).
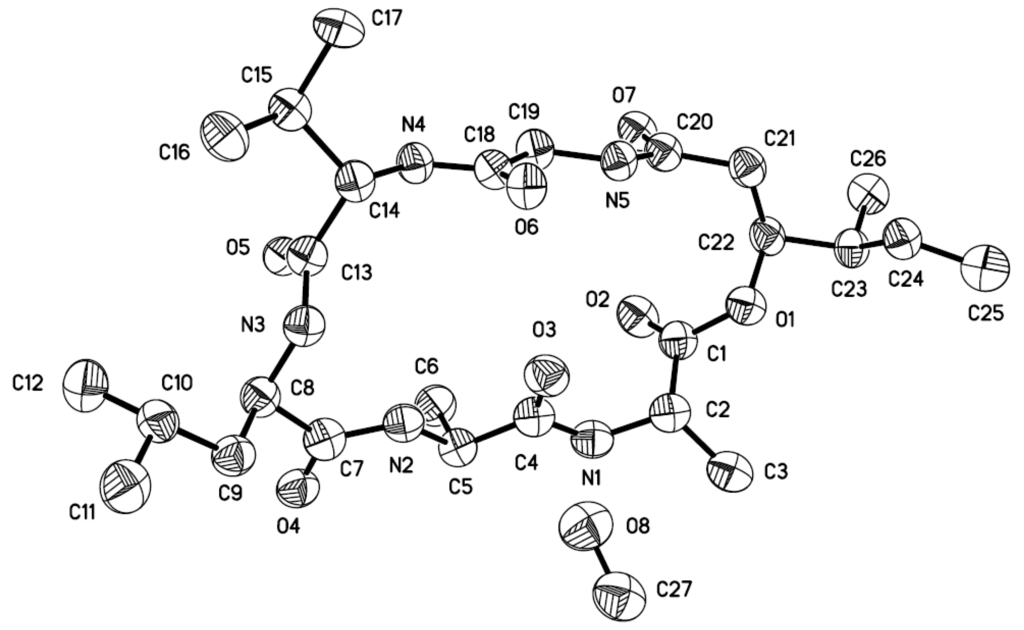
Figure 3.
X-ray structure of compound 1 (Note: A different numbering system is used for the structure described in the text).

Table 2.
1H- (500 MHz) and 13C-NMR (125 MHz) data for compounds 2 and 3 (δ in ppm).
| Compound 2 | Compound 3 | |||||
|---|---|---|---|---|---|---|
| Position | δH (J in Hz) a | δH (J in Hz) b | δC a | Position | δH (J in Hz) b | δC b |
| 1 | 4.29, d (15.5) 4.00, d (15.5) | 4.43, d (15.8) 4.04, d (15.8) | 62.9 CH2 | 1 | - | 164.1 C |
| 2 | - | - | 127.3 C | 2 | 6.61, s | 100.0 CH |
| 3 | - | - | 199.1 C | 3 | - | 128.5 C |
| 4 | - | - | 76.1 C | 4 | 7.52, s | 119.2 CH |
| 5 | 3.74, m | 3.91, dd (9.5, 5.3) | 71.4 CH | 5 | - | 135.8 C |
| 6α 6β | 2.52, m 2.14, m | 2.59, dd (18.1, 5.3) 2.35, dd (18.1, 9.5) | 36.7 CH2 | 6 | 7.24, d (8.4) | 123.1 CH |
| 7 | - | - | 151.6 C | 7 | 7.39, d (8.4) | 110.2 CH |
| 8α 8β | 2.07, m 2.20, m | 2.12, dd (18.3, 10.1) 2.24, br. s (18.3) | 36.9 CH2 | 8 | - | 154.2 C |
| 9 | 3.54, m | 3.60, ddd (10.1, 6.2, 3.5) | 68.6 CH | 9 | - | 68.4 C |
| 10 | 1.16, d (6.2) | 1.21, d (6.2) | 20.9 CH3 | 10 | 1.61, s | 27.6 CH3 |
| 11 | 1.08, s | 1.18, s | 18.0 CH3 | 11 | 1.61, s | 27.6 CH3 |
| 4-OH | 5.05, br. s | - | - | 12 | 4.65, s | 64.1 CH2 |
| 5-OH | 5.07, d (4.2) | - | - | - | - | - |
a Measured in DMSO-d6; b Measured in CD3OD.
The relative configuration of compound 2 was determined by analyzing the NOESY data and 1H-NMR J-values []. The observed NOE correlations from OH-4 to H-5 and from 11-Me to H-6β indicated that they were on the same face of the molecule, respectively (Figure 2). However, the NOEs and coupling constants for H-6 and H-8 were difficult to measure due to overlap with other protons, but a spectrum in CD3OD afforded better resolution of these signals. (Table 2). The large coupling constants observed between H-5 and H-6β (9.5 Hz) and between H-9 and H-8α (10.1 Hz) indicated trans relationships. The key correlation between H-6α and H-8α in the NOESY spectrum measured in CD3OD finally established the relative configuration of 2 as shown in Figure 2.
The absolute configuration of 2 was determined by the application of CD. The CD spectrum displayed a negative Cotton Effect (CE) at 219 nm and a positive CE at 248 nm, which was nearly identical to that of (S)-2-acetyl-3,6-dihydroxycyclohex-2-enone, suggesting the presence of an S-configuration at C-4, which was opposite to that of pestafolide A [,]. The cyclohexenone ring of pestafolide A showed the same relative configuration as that of 2, but a reverse absolute stereochemistry, which was probably due to a different stereochemistry-selective biosynthetic pathway in Pestalotiopsis foedan []. The Mosher’s method was also tried to further determine the absolute configuration of 2, but unsatisfied, probably due to the unstable MTPA esters. Therefore, the absolute configuration of 2 was tentatively assigned to be 4S, 5R, and 9R, and this compound was named felinone A.
Felinone B (3) was determined to have the molecular formula C12H14O3 (six degrees of unsaturation) via HRESIMS. The 1D NMR spectra of 3 indicated the presence of four aromatic and one oxygenated sp3 quaternary carbons, four sp2 methines, one oxygenated sp3 methylene, and two methyl groups (Table 2). The structure of 3 was very similar to that of 10, a compound previously isolated from Smallanthus fruticosus []. The primary difference between compounds 3 and 10 was that the 13-Me and 12-OCH proton signals (δH 1.54 and 4.99) of 10 [] were absent in the 1H NMR spectrum of 3 and, instead, an oxygenated methylene signal at δH 4.65 (s, H-12) was observed. Detailed analyses of the COSY and HMBC spectra further confirmed the structure of compound 3 as shown in Figure 2.
In addition to the three novel compounds 1–3, four known cyclodepsipeptides—destruxin A (4) [], roseotoxin B (5) [], destruxin E chlorohydrin (6), and [β-Me-Pro] destruxin E chlorohydrin (7) [] (Figure 1)—were also isolated and identified from the culture extract of B. felina EN-135. Their structures were determined via spectroscopic analysis and comparison with previously published reports.
2.2. Biological Activities of the Isolated Compounds
The isolated compounds 1–7 were evaluated for brine-shrimp lethality and antimicrobial activity. Among them, the hexadepsipeptides 1 and 4–7 exhibited potent lethality against brine shrimp (Artemia salina), with LD50 values of 26.58, 5.34, 0.73, 2.16, and 1.03 μΜ, respectively, which were notably stronger than that of the positive control colchicine (with an LD50 value of 88.4 μΜ). Compounds 2 and 3 exhibited weak activity with lethal rates of 61.4% and 59.6%, respectively, at a concentration of 100 μg/mL. The antimicrobial activities against six bacteria (Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Vibrio alginolyticus, Vibrio anguillarum, and Edwardsiella tarda) and four plant pathogenic fungi (Physalospora piricola, Alternaria brassicae, Colletotrichum gloeosporioides, and Cucumber fusarium) were also evaluated. Only compound 3 showed inhibitory activity higher than that of the chloramphenicol control (MIC value of 4 μg/mL) against P. aeruginosa; this compound was found to have an MIC value of 32 μg/mL.
3. Experimental Section
3.1. General
The optical rotations were determined using an Optical Activity AA-55 polarimeter. UV spectra were measured using a Lengguang Gold S54 spectrophotometer (Shanghai Lengguang Technology Co. Ltd., Shanghai, China). The 1H, 13C, and 2D NMR spectra were acquired using a Bruker Advance 500 spectrometer (Bruker BioSpin Group, Karlsruhe, Germany). Mass spectra were obtained using a VG Autospec 3000 mass spectrometer (VG instruments, London, UK). Semi-preparative HPLC was performed using a Dionex UltiMate U3000 system (Dionex Corporation, Sunnyvale, CA, USA) with an Agilent Prep RP-18 column (21.2 × 250 mm, 10 μm) with UV detection. Column chromatography (CC) was performed using silica gel (200–300 mesh, Qingdao Haiyang Chemical Factory, Qingdao, China), Lobar LiChroprep RP-18 (40–63 μm, Merck, Darmstadt, Germany), and Sephadex LH-20 (18–110 μm, Merck).
3.2. Fungal Material
The fungus Beauveria felina EN-135 was isolated from an unidentified marine bryozoan and identified via sequence analysis of the ITS region of its rDNA as previously described []. The sequence data derived from the fungus, which was similar (99%) to the sequence of Beauveria felina CBS 250.34 (compared to accession No. AY261369), was deposited in GenBank with the accession No. HQ891664. The strain was preserved at the Key Laboratory of Experimental Marine Biology at the Institute of Oceanology of the Chinese Academy of Sciences.
3.3. Fermentation
The fungal strain was statically fermented at r.t. for 40 days on rice solid medium containing rice (100 g/flask), peptone (0.6 g/flask), and sea water (100 mL/flask) in 1 L Erlenmeyer flasks (×60).
3.4. Extraction and Isolation
The fermented rice medium was exhaustively extracted using EtOAc to obtain a crude extract (25.1 g), which was subjected to silica-gel vacuum liquid chromatography (VLC) and eluted with mixed solvents of increasing polarity (petroleum ether-EtOAc, 20:1 to 1:1, followed by CHCl3-MeOH, 40:1 to 1:1) to yield nine fractions (Frs. 1–9). Frs. 7 and 8 exhibited potent lethality against brine shrimp, with LD50 values of 17.27 and 11.67 μg/mL, respectively. Fr. 7 (3.4 g) was separated into five subfractions (Frs. 7. 1–5) via CC on RP-18 (MeOH/H2O, from 1:9 to 1:0). Fr. 7.1 (179.2 mg) was subjected to CC on Sephadex LH-20 (MeOH) and was further purified via semi-preparative HPLC (30% MeOH/H2O, 16 mL/min) to yield compounds 2 (tR = 11.6 min, 5.1 mg) and 3 (tR = 16.3 min, 18.6 mg). Fr. 7.3 (429.4 mg) was subjected to CC on silica gel eluted with CHCl3-MeOH (100:1 to 20:1) and Sephadex LH-20 (acetone) and was further purified via semi-preparative HPLC (40% MeCN/H2O, 16 mL/min) to yield compounds 4 (tR = 12.8 min, 16.0 mg) and 5 (tR = 15.8 min, 15.2 mg). Fr. 8 (2.3 g) was separated via CC on RP-18 (MeOH/H2O, from 1:9 to 1:0) to yield four subfractions (Frs. 8. 1–4). Fr. 8.2 (387.6 mg) was subjected to CC over silica gel eluted with CHCl3-MeOH (50:1 to 10:1) and was purified via semi-preparative HPLC (65% MeOH/H2O, 16 mL/min) to yield compounds 6 (tR 13.5 min, 39.1 mg) and 7 (tR 17.3 min, 19.5 mg). Fr. 8.3 (108.7 mg) was separated via CC on Sephadex LH-20 (acetone) and then via semi-preparative HPLC (45% MeCN/H2O, 16 mL/min) to yield compound 1 (tR 12.8 min, 10.4 mg).
Iso-isariin D (1): colorless crystal; mp 258–259 °C;  : −22.2 (c 0.36, MeOH); UV (MeOH) λmax (log ε) 206 (3.64) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 540 [M + H]+; HRESIMS m/z 540.3392 [M + H]+ (calcd for C26H46N5O7, 540.3392).
: −22.2 (c 0.36, MeOH); UV (MeOH) λmax (log ε) 206 (3.64) nm; 1H and 13C NMR data, see Table 1; ESIMS m/z 540 [M + H]+; HRESIMS m/z 540.3392 [M + H]+ (calcd for C26H46N5O7, 540.3392).
Felinone A (2): yellowish solid; 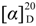 : +104.8 (c 0.21, MeOH); UV (MeOH) λmax (log ε) 200 (3.45), 240 (3.77) nm; CD ëmax (Δε) 219 (−13.89), 248 (+27.06) nm; 1H and 13C NMR data, see Table 2; ESIMS m/z 213 [M + H]+; HRESIMS m/z 213.1123 [M + H]+ (calcd for C11H17O4, 213.1121).
: +104.8 (c 0.21, MeOH); UV (MeOH) λmax (log ε) 200 (3.45), 240 (3.77) nm; CD ëmax (Δε) 219 (−13.89), 248 (+27.06) nm; 1H and 13C NMR data, see Table 2; ESIMS m/z 213 [M + H]+; HRESIMS m/z 213.1123 [M + H]+ (calcd for C11H17O4, 213.1121).
Felinone B (3): yellowish solid; 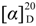 : −16.7 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 209 (4.38), 247 (4.09), 279 (3.47), 286 (3.48) nm; 1H and 13C NMR data, see Table 2; ESIMS m/z 229 [M + Na]+; HRESIMS m/z 229.0828 [M + Na]+ (calcd for C12H14O3Na, 229.0835).
: −16.7 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 209 (4.38), 247 (4.09), 279 (3.47), 286 (3.48) nm; 1H and 13C NMR data, see Table 2; ESIMS m/z 229 [M + Na]+; HRESIMS m/z 229.0828 [M + Na]+ (calcd for C12H14O3Na, 229.0835).
3.5. X-ray Crystallographic Analysis of Compound 1
The crystallographic data were collected using a Bruker D8-advance X-ray diffractometer (Bruker AXS Corporation, Karlsruhe, Germany) equipped with graphite monochromatic Cu-Kα radiation (λ = 1.54178 Å) at 293(2) K []. The absorption data were obtained using the program SADABS []. The structure was analyzed via direct methods using the SHELXTL software package []. All non-hydrogen atoms were refined anisotropically. The H atoms were located via geometrical calculations, and their positions and thermal parameters were fixed during structure refinement. The structure was refined using full-matrix least-squares techniques [].
Crystal data for compound 1: C26H45N5O7·CH3OH, F.W. = 571.71, one molecule containing a CH3OH solvent molecule in the unit, orthorhombic, space group P2(1)2(1)2(1), unit-cell dimensions a = 9.5437(7) Å, b = 15.5388(13) Å, c = 22.315(2) Å, α = β = γ = 90°, V = 3309.3(5) Å3, Z = 4, dcalcd = 1.147 mg/m3, crystal dimensions 0.24 × 0.10 × 0.06 mm3, μ = 0.696 mm−1, F(000) = 1240. The 17936 measurements yielded 5791 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement yielded R1 = 0.1104 and wR2 = 0.1978[I > 2σ(I)]. The Flack parameter was 0.0 (13) in the final refinement for all 5791 reflections with 372 Friedel pairs.
3.6. Brine-Shrimp Lethality
The brine-shrimp (Artemia salina) lethality of the isolated compounds was determined as previously described []. Colchicine was used as a positive control.
3.7. Antimicrobial Assay
The antibacterial activities against E. coli, S. aureus, P. aeruginosa, V. alginolyticus, V. anguillarum, and E. tarda along with the antifungal activities against P. piricola, A. brassicae, C. gloeosporioides, and C. fusarium were investigated using the disk diffusion and double dilution methods as previously described [,]. Chloramphenicol and amphotericin B were used as positive controls for the antibacterial and antifungal bioassays, respectively.
4. Conclusions
One novel cyclodepsipeptide, iso-isariin D (1), and two novel O-containing heterocyclic compounds, felinones A and B (2 and 3), were isolated from a culture of Beauveria felina EN-135. In addition, four known destruxin cyclodepsipeptides (4–7) were also identified. The structures were elucidated via spectroscopic analysis, and the absolute configurations of 1 and 2 were determined using single-crystal X-ray diffraction and CD, respectively. Compounds 1 and 4–7 exhibited potent brine-shrimp lethality with LD50 values of 26.58, 5.34, 0.73, 2.16, and 1.03 μΜ, respectively, whereas compound 3 showed inhibitory activity against P. aeruginosa with an MIC value of 32 μg/mL.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 31270403) and by the Ministry of Science and Technology (No. 2013AA092901 and No. 2010CB833802).
Author Contributions
F.-Y. Du performed the experiments for the isolation, structure elucidation, and antimicrobial evaluation of compounds 1–7 and prepared the manuscript; X.-M. Li collected the bryozoan sample and performed the 1D and 2D NMR experiments; P. Zhang participated part of the compounds isolation and determined the optical rotations, UV data, and brine shrimp lethality; C.-S. Li contributed to the isolation and structure elucidation as well as manuscript discussing; B.-G. Wang supervised the research work and revised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Molnar, I.; Gibson, D.M.; Krasnoff, S.B. Secondary metabolites from entomopathogenic hypocrealean fungi. Nat. Prod. Rep. 2010, 27, 1241–1275. [Google Scholar] [CrossRef]
- Zenitani, S.; Tashiro, S.; Shindo, K.; Nagai, K.; Suzuki, K.; Imoto, M. Gerfelin, a novel inhibitor of geranylgeranyl diphosphate synthase from Beauveria felina QN22047, I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 2003, 56, 617–621. [Google Scholar] [CrossRef]
- Lira, S.P.; Vita-Marques, A.M.; Seleghim, M.H.R.; Bugni, T.S.; Labarbera, D.V.; Sette, L.D.; Sponchiado, S.R.P.; Ireland, C.M. New destruxins from the marine-derived fungus Beauveria felina. J. Antibiot. 2006, 59, 553–563. [Google Scholar] [CrossRef]
- Langenfeld, A.; Blond, A.; Gueye, S.; Herson, P.; Nay, B.; Dupont, J.; Prado, S. Insecticidal cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2011, 74, 825–830. [Google Scholar] [CrossRef]
- Chung, Y.M.; EI-Shazly, M.; Chuang, D.W.; Hwang, T.L.; Asai, T.; Oshima, Y.; Ashour, M.L.; Wu, Y.C.; Chang, F.R. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, induces the production of anti-inflammatory cyclodepsipeptides from Beauveria felina. J. Nat. Prod. 2013, 76, 1260–1266. [Google Scholar] [CrossRef]
- Gao, S.S.; Li, X.M.; Du, F.Y.; Li, C.S.; Proksch, P.; Wang, B.G. Secondary metabolites from a marine-derived endophytic fungus Penicillium chrysogenum QEN-24S. Mar. Drugs 2011, 9, 59–70. [Google Scholar]
- Wang, M.H.; Li, X.M.; Li, C.S.; Ji, N.Y.; Wang, B.G. Secondary metabolites from Penicillium pinophilum SD-272, a marine sediment-derived fungus. Mar. Drugs 2013, 11, 2230–2238. [Google Scholar] [CrossRef]
- An, C.Y.; Li, X.M.; Li, C.S.; Wang, M.H.; Xu, G.M.; Wang, B.G. Aniquinazolines A—D, four new quinazolinone alkaloids from marine-derived endophytic fungus Aspergillus nidulans. Mar. Drugs 2013, 11, 2682–2694. [Google Scholar] [CrossRef]
- Li, C.S.; Li, X.M.; Gao, S.S.; Lu, Y.H.; Wang, B.G. Cytotoxic anthranilic acid derivatives from deep sea sediment-derived fungus Penicillium paneum SD-44. Mar. Drugs 2013, 11, 3068–3076. [Google Scholar] [CrossRef]
- An, C.Y.; Li, X.M.; Luo, H.; Li, C.S.; Wang, M.H.; Xu, G.M.; Wang, B.G. 4-phenyl-3, 4-dihydroquinolone derivatives from Aspergillus nidulans MA-143, an endophytic fungus isolated from the mangrove plant Rhizophora stylosa. J. Nat. Prod. 2013, 76, 1896–1901. [Google Scholar] [CrossRef]
- Vining, L.C.; Taber, W.A. Isariin, a new depsipeptide from Isaria cretacea. Can. J. Chem. 1962, 40, 1579–1584. [Google Scholar] [CrossRef]
- Deffieux, G.; Merlet, D.; Baute, R.; Bourgeois, G.; Baute, M.A.; Neveu, A. New insecticidal cyclodepsispeptides from the fungus Isaria felina. J. Antibiot. 1981, 34, 1266–1270. [Google Scholar] [CrossRef]
- Sabareesh, V.; Ranganayaki, R.S.; Raghothama, S.; Bopanna, M.P.; Balaram, H.; Srinivasan, M.C.; Balaram, P. Identification and characterization of a library of microheterogeneous cyclohexadepsipeptides from the fungus Isaria. J. Nat. Prod. 2007, 70, 715–729. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Rozco, R.; Wijeratne, E.M.K.; Espinosa-Artiles, P.; Gunatilaka, A.A.L.; Stock, S.P.; Molnar, I. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet. Biol. 2009, 46, 353–364. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Orozco, R.; Wijeratne, E.M.K.; Gunatilaka, A.A.L.; Stock, S.P.; Molnar, I. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem. Biol. 2008, 15, 898–907. [Google Scholar] [CrossRef]
- Stark, L.M.; Pekari, K.; Sorensen, E.J. A nucleophile-catalyzed cycloisomerization permits a concise synthesis of (+)-harziphilone. Proc. Natl. Acad. Sci. USA 2004, 101, 12064–12066. [Google Scholar] [CrossRef]
- Ding, G.; Liu, S.C.; Guo, L.D.; Zhou, Y.G.; Che, Y.S. Antifungal metabolites from the plant endophytic fungus Pestalotiopsis foedan. J. Nat. Prod. 2008, 71, 615–618. [Google Scholar] [CrossRef]
- Zaitsev, V.G.; Mikhal’chuk, A.L. Enantioconvergent synthesis of (−)-(S)- and (+)-(R)-2-acetyl-3,6-dihydroxycyclohex-2-enone starting from rac-6-hydroxy-3-methoxycyclohex-2-enone. Chirality 2001, 13, 488–492. [Google Scholar] [CrossRef]
- Bohlmann, F.; Ziesche, J.; King, R.M.; Robinson, H. Neue melampolide aus Smallanthus Fruticosus. Phytochemistry 1980, 19, 973–974. [Google Scholar] [CrossRef]
- Gupta, S.; Roberts, D.W.; Renwick, J.A.A. Molecular conformation of destruxin A. Tetrahedron Lett. 1989, 30, 4189–4192. [Google Scholar] [CrossRef]
- Springer, J.P.; Cole, R.J.; Dorner, J.W.; Cox, R.H.; Richard, J.L.; Barnes, C.L.; Helm, D.V.D. Structure and conformation of roseotoxin B. J. Am. Chem. Soc. 1984, 106, 2388–2392. [Google Scholar] [CrossRef]
- Wang, S.; Li, X.M.; Teuscher, F.; Li, D.L.; Diesel, A.; Ebel, R.; Proksch, P.; Wang, B.G. Chaetopyranin, a benzaldehyde derivative, and other related metabolites from Chaetomium globosum, an endophytic fungus derived from the marine red alga Polysiphonia urceola. J. Nat. Prod. 2006, 69, 1622–1625. [Google Scholar] [CrossRef]
- Cambridge Crystallographic Data Centre as CCDC 983056. Crystallographic Data of Compound 1; Cambridge Crystallographic Data Centre: Cambridge, UK, 2014. Available online: http://www.ccdc.cam.ac.uk/data_request/cif (accessed on 23 January 2014).
- Sheldrick, G.M. SADABS, Software for Empirical Absorption Correction; University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXTL, Structure Determination Software Programs; Bruker Analytical X-ray System Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Sheldrick, G.M. SHELXL-97 and SHELXS-97,Program for X-ray Crystal Structure Solution and Refinement; University of Göttingen: Göttingen, Germany, 1997. [Google Scholar]
- Gerwick, W.H.; Proteau, P.J.; Nagle, D.G.; Hamel, E.; Blokhin, A.; Slate, D.L. Structure of curacin A, a novel antimitotic, antiproliferative, and brine shrimp toxic natural product from the marine Cyanobacterium Lyngbya majuscula. J. Org. Chem. 1994, 59, 1243–1245. [Google Scholar] [CrossRef]
- Bergeron, R.J.; Cavanaugh, P.F., Jr.; Kline, S.J.; Hughes, R.G., Jr.; Elliott, G.T.; Porter, C.W. Antineoplastic and antiherpetic activity of spermidine catecholamide iron chelators. Biochem. Biophys. Res. Commun. 1984, 121, 848–854. [Google Scholar] [CrossRef]
- Al-Burtamani, S.K.S.; Fatope, M.O.; Marwah, R.G.; Onifade, A.K.; Al-Saidi, S.H. Chemical composition, antibacterial and antifungal activities of the essential oil of Haplophyllum tuberculatum from Oman. J. Ethnopharmacol. 2005, 96, 107–112. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).